Introduction
Melanoma, a life-threatening malignancy that arises
from melanocytes, is the fifth most common neoplasia in human and
accounts for 80% of skin cancer-associated mortality (1). It is characterized by aggressive
invasion, early metastasis and resistance to chemotherapy or
radiotherapy (2). In the past few
decades, the morbidity of melanoma has been increasing by 3–8% per
year in Western countries (3).
Although most patients with melanoma present with localized disease
that is curable with surgical resection (4,5), for
those with regional or distant metastasis the prognosis is poor,
with 10-year survival rates of 64 and 16%, respectively (6). Previous progress in understanding the
molecular mechanisms underlying melanoma has brought about the
development of new targeted therapies, including vemurafenib, which
targets a B-Raf proto-oncogene, serine/threonine kinase
(RAF1) V600 activating mutation and has produced significant
anti-tumor responses in clinical studies (7). However, immunotherapy, chemotherapy,
small molecule inhibitors or targeted therapies are not effective
in most melanoma cases (8,9). Therefore, it is necessary to
investigate the biology of melanoma initiation and progression and
develop effective therapeutic strategies.
icroRNAs (miRNAs) are a novel group of endogenous,
single-stranded and short non-coding RNA molecules of 18–25
nucleotides (10). They negatively
regulate gene expression through base-pair interactions between 5′
ends of miRNA and target regions within the 3′-untranslated region
(3′UTR) of mRNA, resulting in translation inhibition or induction
of mRNA degradation (11). At
present, >1,000 miRNAs have been identified in humans, and they
modulate more than a third of the human genome (12). Previous studies have reported that
miRNAs are involved in the pathogenesis of multiple human diseases,
including cancer, and are involved in multiple cellular processes
including cell proliferation, apoptosis, differentiation,
carcinogenesis, metastasis and drug resistance (13–15).
Abnormal expression of miRNAs is associated with human cancers, and
has been demonstrated to contribute to carcinogenesis and
progression of cancers, including melanoma (16–18).
In cancers, miRNAs act as tumor suppressors or oncogenes, and
inactivation of oncogenic miRNAs or restoration of tumor suppressor
miRNAs has great potential for cancer treatment (19–21).
In the present study, the functional involvement of
miR-455 in melanoma was investigated, and miR-455 was demonstrated
to act as a tumor suppressor in melanoma. The expression of miR-455
was downregulated in melanoma tissues compared with normal tissues,
and this was also observed in melanoma cell lines and human
epidermal melanocytes. Restoration of miR-455 expression inhibited
cell proliferation and invasion of melanoma cells. Furthermore,
insulin-like growth factor 1 receptor (IGF-1R) was identified as a
direct target gene of miR-455.
Materials and methods
Tissue samples, cell lines and
oligonucleotide transfection
The present study was approved by the Ethics
Committee of Weifang People's Hospital (Weifang, China). Written
informed consent for research purposes was provided by the
participants. A total of 20 paired melanoma tissues and adjacent
non-tumor tissues were collected from patients with melanoma who
underwent operations at Weifang People's Hospital between July 2013
and April 2015. All patients were diagnosed with melanoma and were
not treated with radiotherapy or chemotherapy prior to surgery.
Tissue specimens were snap-frozen in liquid nitrogen and stored at
−80°C until use.
Human melanoma cell lines (SKMEL1, A375, HT144,
A2058) and a human embryonic kidney cell line (HEK293T) used for
luciferase reporter assays were obtained from the American Type
Culture Collection (Manassas, VA, USA). The cells were cultured in
RPMI-1640 medium or Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
supplemented with 10% fetal bovine serum (FBS), 100 mg/ml
penicillin and 100 mg/ml streptomycin. Human epidermal melanocytes
(HEM) were purchased from ScienCell Research Laboratories, Inc.
(San Diego, CA, USA) and maintained in melanocyte medium (ScienCell
Research Laboratories, Inc.) according to the manufacturer's
protocol. All cells were cultured in a humidified 5% CO2
cell incubator at 37°C.
The miR-455 mimic, negative control (NC) mimic,
IGF-1R siRNA and scramble siRNA control were synthesized by
Guangzhou RiboBio Co., Ltd (Guangzhou, China). The sequence of the
IGF-1R siRNA was 5′-GCCGATGTGTGAGAAGACC-3′ and the scramble siRNA
control sequence was 5′-AACAGGCACACGTCCCAGCGT-3′. Cells were
transfected with oligonucleotides using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues and cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The quantity of total RNA
was determined using the ND-1000 NanoDrop spectrophotometer
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.). For
miR-455 expression, total RNA was reverse transcribed into cDNA
using the TaqMan miRNA reverse transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and RT-qPCR was
performed using TaqMan Universal Master Mix II (Applied Biosystems;
Thermo Fisher Scientific, Inc.). U6 was measured as an internal
control for miR-455 expression. The thermocycling conditions were
as follows: 95°C for 10 min; 40 cycles of denaturation at 95°C for
15 sec and annealing at 60°C for 1 min; and a final elongation step
at 72°C for 10 min. For IGF-1R mRNA expression, total RNA was
reverse transcribed into cDNA using the PrimeScript RT Reagent kit
(Takara Biotechnology Co., Ltd., Dalian, China), followed by
RT-qPCR using SYBR-Green PCR master mix (Takara Biotechnology Co.,
Ltd.). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as
an endogenous control for IGF-1R expression. The thermocycling
conditions were as follows: 95°C for 10 min; 40 cycles at 95°C for
15 sec and 60°C for 1 min. The primer sequences were: MiR-455,
forward 5′-CGAGCTTCCTTCTGCAGGT-3′ and reverse
5′-CACCACTGCCATCCCACA-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′ and
reverse: 5′-AACGCTTCACGAATTTGCGT-3′; IGF-1R, forward
5′-TGCGTGAGAGGATTGAGTTTC-3′ and reverse
5′-CTTATTGGCGTTGAGGTATGA-3′; and GAPDH, forward
5′-TGCACCACCAACTGCTTAGC-3′ and reverse 5′-GGCATGCACTGTGGTCATGAG-3′.
miR-455 and IGF-1R mRNA expression levels were quantified by
comparing Cq values (22). Each
sample was analyzed in triplicate and repeated at least 3
independent times.
Cell proliferation assay
Cell proliferation was evaluated using Cell Counting
kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). Cells transfected with miRNA or siRNA were collected,
seeded into 96-well plates at a density of 4,000 cells/well, and
further incubated for 24, 48, 72 and 96 h, respectively. CCK-8
reagent (10 µl) was added into each well, and incubated at 37°C for
an additional 2 h. Absorbance was measured at a wavelength of 450
nm using a spectrophotometer.
Transwell cell invasion assay
Transwell chambers with 8 µm-pore-size filter were
used to assess cell invasion ability. The chambers were pre-coated
with Matrigel (BD Biosciences, San Jose, CA, USA). Transfected
cells were collected and seeded into the upper compartment of the
chamber at a density of 1×105 cells suspended into 100
µl serum free DMEM.
A total of 200 µl DMEM containing 20% FBS was placed
in the bottom compartment of the chamber. Following incubation for
24 h, the non-invading cells were removed with cotton swabs. The
invading cells were fixed with 100% methanol, stained with 0.5%
crystal violet, washed with PBS and counted using an inverted light
microscope (CKX41; Olympus Corporation, Tokyo, Japan). The assays
were performed three independent times.
Target prediction of miRNAs
To predict miRNAs/mRNAs interactions, TargetScan
v.6.0 (http://www.targetscan.org/) and microRNA
(August 2010 release; www.microrna.org/) were used.
Western blot
Total protein was isolated using
radioimmunoprecipitation buffer (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) supplemented with 0.1 mg/ml
phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate and 1
mg/ml aprotinin. Total protein concentration was measured using the
bicinchoninic acid protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Equal amounts of total protein (30 µg) were
separated in 10% SDS polyacrylamide gels, transferred to
polyvinylidene difluoride membranes (Merck KGaA) and blocked with
5% milk in TBS containing 0.05% Tween-20 (TBST) at room temperature
for 2 h. Then, the membranes were probed with the following primary
antibodies: Mouse anti-human monoclonal IGF-1R (1:1,000 dilution;
cat. no. sc-81464; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
and GADPH (1:1,000 dilution; cat. no. sc-51907; Santa Cruz
Biotechnology, Inc.), at 4°C overnight. Following washing three
times with TBST, the membranes were incubated with their respective
horseradish peroxidase-conjugated secondary antibody (1:5,000
dilution; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.) at room
temperature for 2 h. Following three washes in TBST, the bands were
visualized with an enhanced chemiluminescence system (GE Healthcare
Life Sciences, Chalfont, UK). Band intensities were quantified
using the FluorChem imaging system (Alpha Innotec GmbH, Kasendorf,
Germany) from at least 3 independent experiments.
Luciferase report assay
The luciferase reporter vectors pGL3-IGF-1R-3′UTR Wt
and pGL3-IGF-1R-3′UTR Mut were synthesized by Shanghai GenePharma
Co., Ltd. (Shanghai, China). HEK293T cells were transfected with
luciferase reporter vector and miR-455 mimics or NC using
Lipofectamine 2000. Firefly and Renilla luciferase
activities were evaluated 48 h after transfection using the
Dual-Luciferase assay system (Promega Corporation, Madison, WI,
USA), and Renilla luciferase activity was normalized to
firefly luciferase activity.
Statistical analysis
The results were presented as the mean ± standard
deviation. SPSS 16.0 (SPSS, Inc., Chicago, IL, USA) was used for
statistical analysis with two-tailed Student's t-test or
one-way analysis of variance followed by Student-Newman-Keuls post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-455 is downregulated in melanoma
tissues and cell lines
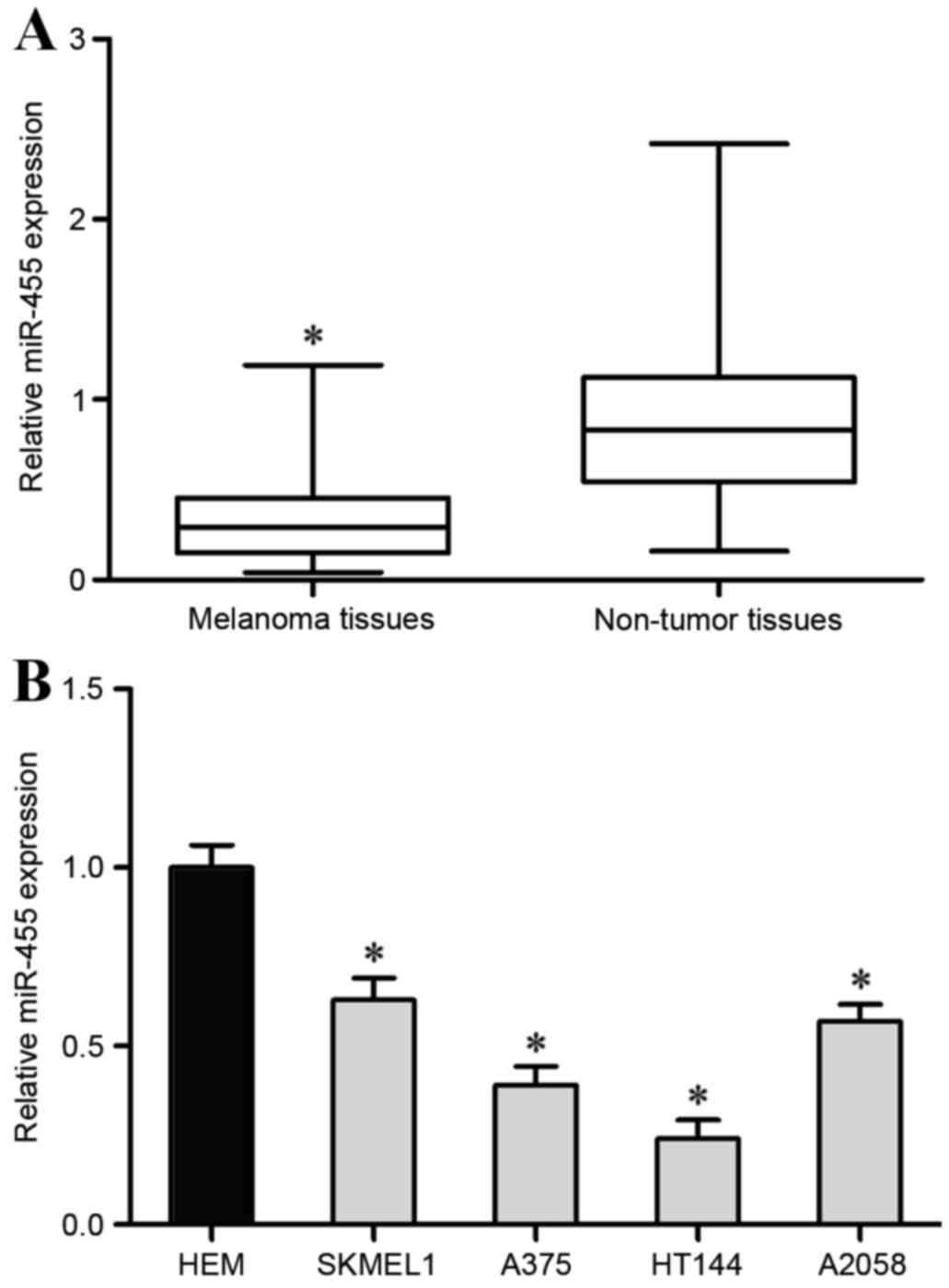
miR-455 expression levels were measured in 22 pair
of melanoma tissues and adjacent non-tumor tissues by RT-qPCR. The
results revealed that miR-455 was significantly downregulated in
melanoma tissues compared with adjacent non-tumor tissues
(P<0.05; Fig. 1A).
Subsequently, miR-455 expression was also detected in melanoma cell
lines (SKMEL1, A375, HT114, A2058) and human epidermal melanocytes
(HEM) using RT-qPCR. The results revealed that miR-455 expression
was significantly reduced in all melanoma cell lines compared with
HEM (P<0.05; Fig. 1B). The
expression levels of miR-455 were lowest in the A375 and HT114 cell
lines, therefore A375 and HT114 cell lines were selected for
subsequent experiments.
miR-455 inhibits melanoma cell
viability and invasion
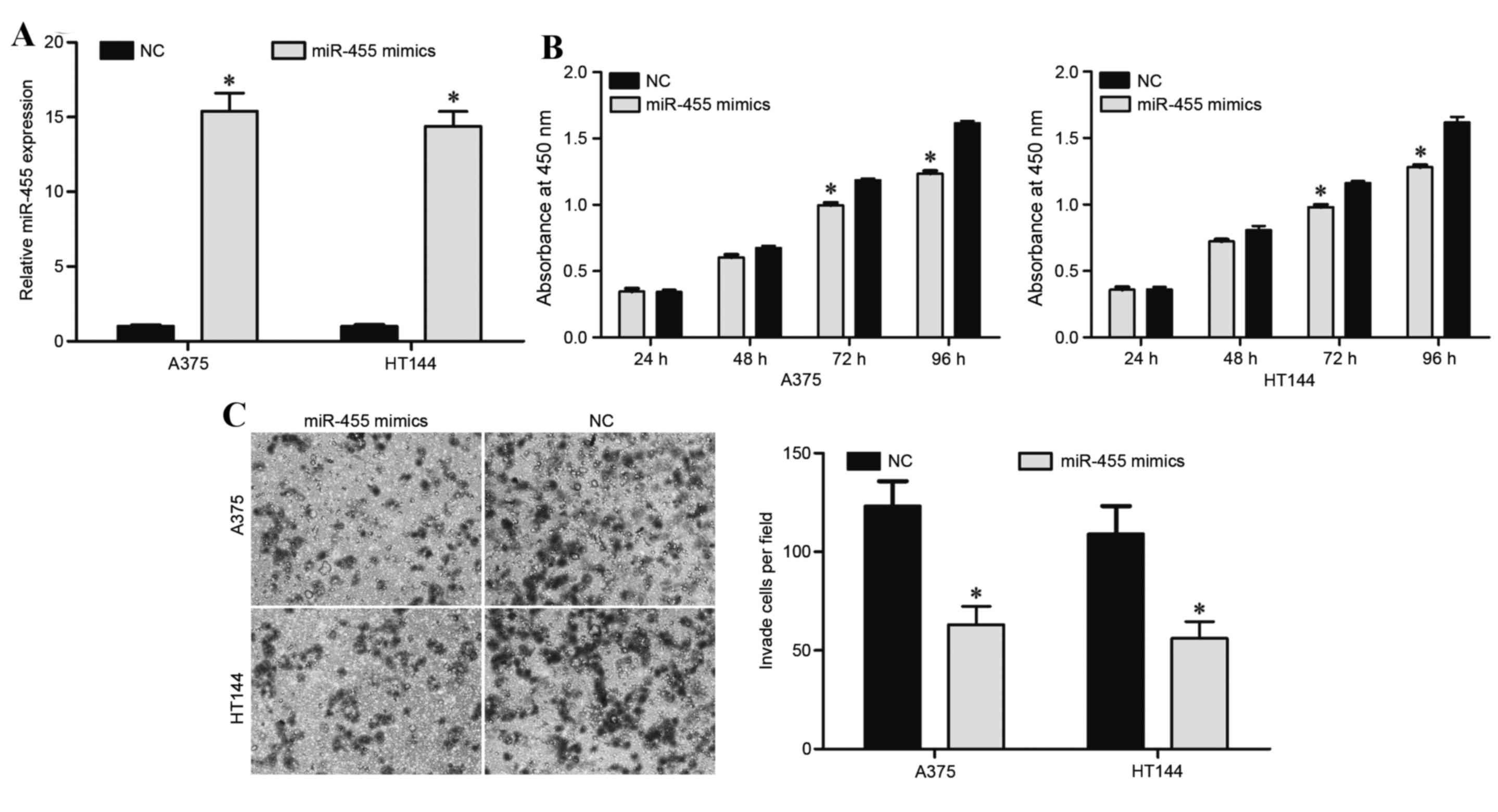
To assess the functional involvement of miR-455 in
melanoma, A375 and HT114 cells were transfected with an miR-455
mimic or NC. RT-qPCR was performed 48 h following transfection to
evaluate the transfection efficiency, and revealed that miR-455 was
significantly upregulated in A375 and HT114 cells transfected with
miR-455 mimics (P<0.05; Fig.
2A).
Cell proliferation assays were performed to evaluate
the effect of miR-455 on cell growth. As a result of miR-455
overexpression, cell proliferation in the miR-455 mimic group was
significantly decreased in A375 and HT114 cells compared with the
NC groups at 72 and 96 h (P<0.05; Fig. 2B).
Transwell cell assays were performed to assess the
effect of miR-455 on cell invasion. The results demonstrated that
miR-455 overexpression significantly inhibited A375 and HT114 cell
invasion compared with the NC groups (P<0.05; Fig. 2C). These findings suggested that
miR-455 may function as a tumor suppressor in melanoma.
IGF-1R is a direct target of
miR-455
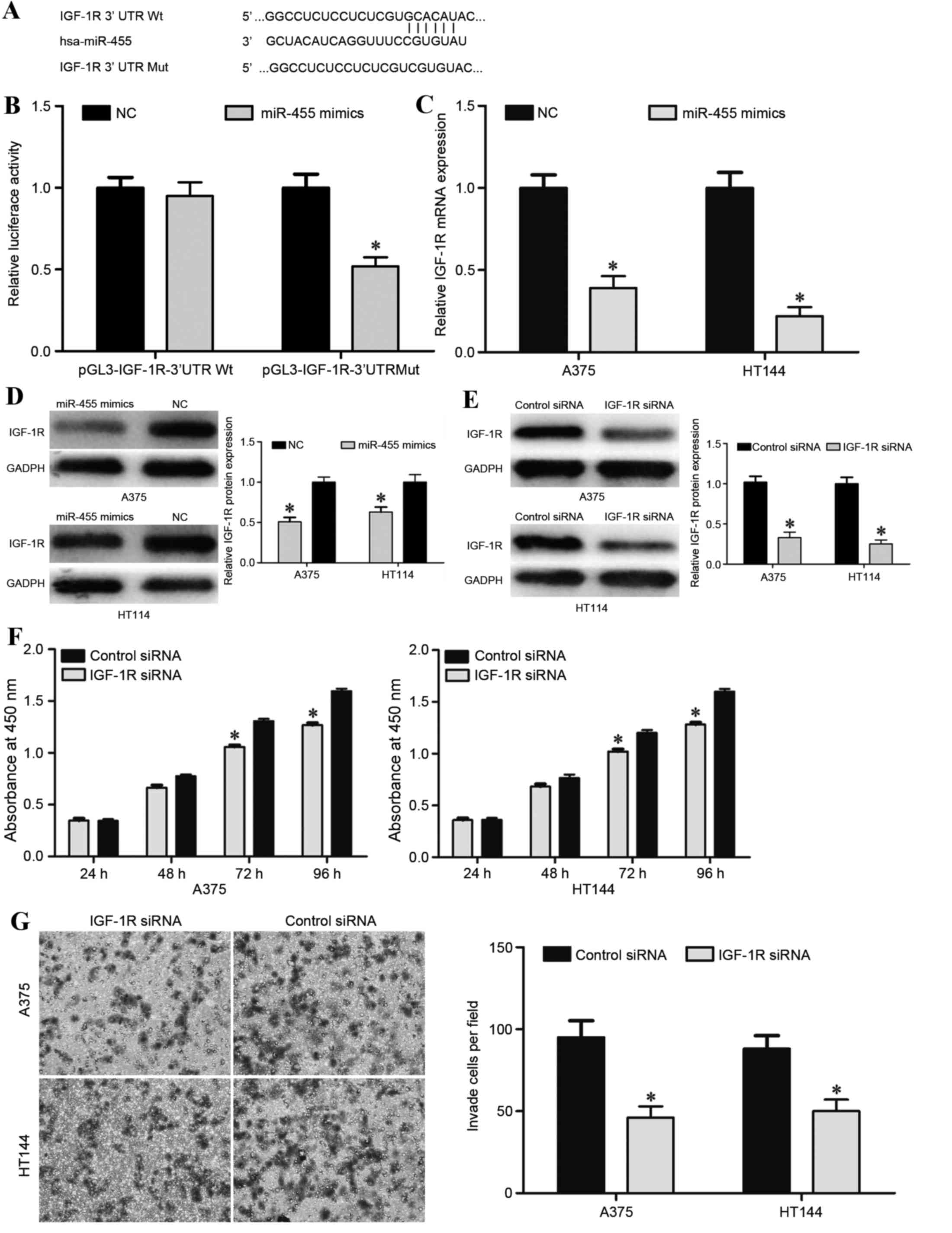
To explore the potential target genes of miR-455,
TargetScan and microRNA were used. This analysis predicted that
IGF-1R was a putative target of miR-455 (Fig. 3A). To identify the regulatory
relationship between miR-455 and IGF-1R, a luciferase reporter
assay was performed in HEK293T cells. miR-455 overexpression
significantly decreased luciferase activity compared with the NC
group following transfection with pGL3-IGF-1R-3′UTR Mut but not
pGL3-IGF-1R-3′UTR Wt, indicating that miR-455 directly targeted the
3′UTR of IGF-1R (Fig. 3B).
To directly evaluate the effect of miR-455 on IGF-1R
expression, western blot and RT-qPCR were performed. The results
demonstrated that miR-455 overexpression significantly suppressed
IGF-1R expression in A375 and HT114 cells at the mRNA (P<0.05;
Fig. 3C) and protein (P<0.05;
Fig. 3D) levels. Taken together,
these results indicated that IGF-1R was a direct downstream target
of miR-455 in melanoma.
This prompted further exploration of whether the
suppressive functions of miR-455 in melanoma were induced by
IGF-1R. For this purpose, IGF-1R siRNA or control siRNA was
transfected into A375 and HT114 cells. Western blotting was
performed 72 h following transfection to assess the transfection
efficiency, which revealed that IGF-1R was significantly
downregulated in A375 and HT114 cells transfected with IGF-1R siRNA
compared with the control siRNA group (P<0.05; Fig. 3E). Once IGF-1R expression was
effectively depressed from 72 h onwards, A375 and HT114 cells
transfected with IGF-1R siRNA exhibited significantly decreased
cell proliferation (P<0.05; Fig.
3F) and invasion (P<0.05; Fig.
3G) compared with the control siRNA group, which was similar
with the functions induced by miR-455 overexpression in melanoma
cells. Collectively, these findings demonstrated that miR-455
functions as a tumor suppressor in melanoma through directly
targeting IGF-1R.
Discussion
Melanoma has the highest fatality and malignancy of
all the skin cancers (23) and the
morbidity of melanoma was ranked 5th in males and 7th in females of
all malignancies from 1992 to 2010 (24). According to statistical analysis,
in 2015 ~73,870 novel melanoma cases and 9,940 deaths due to
melanoma occurred in the USA (25). Improvement in the prognosis of
patients with melanoma largely depends on increasing the
understanding of the molecular mechanisms underlying the
carcinogenesis and progression of this malignancy. A number of
previous studies have demonstrated that miRNAs function as a novel
group of oncogenes and tumor suppressor genes, because their
abnormal expression is associated with multiple human cancers,
including melanoma, by negatively regulating the expression of
cancer-associated genes (26–28).
Therefore, miRNAs may be novel targets for therapy in cancers.
In the present study, the expression levels of
miR-455 in melanoma tissues and cell lines were investigated.
miR-455 was significantly downregulated in melanoma tissues and
cell lines compared with adjacent non-tumor tissues and human
epidermal melanocytes. This indicated that miR-455 may be involved
in melanoma carcinogenesis and progression. Functional studies
revealed that ectopic miR-455 expression inhibited the
proliferation and invasion of melanoma cells. Furthermore, the
present study suggested that miR-455 exerts its tumor suppressive
functions by targeting IGF-1R. Taken together, these findings
indicated that miR-455 may act as a tumor suppressor in
tumorigenesis and development of melanoma through regulation of
IGF-1R.
Previous studies have demonstrated that miR-455 is
deregulated in multiple human diseases, and contributes to multiple
physiological and pathological processes including glioblastoma,
steroidogenesis, colorectal cancer, chondrogenesis and
osteoarthritis (29–33). Chai et al (33) reported that expression levels of
miR-455 were decreased in colorectal tissues, and overexpression of
miR-455 inhibited growth and invasion of colorectal cells. Using
TargetScan, luciferase report assay, RT-qPCR and western blot,
RAF1 was validated as a direct target gene of miR-455 in
colorectal cancer (33). These
findings suggested that miR-455/RAF1 may represent a new
potential therapeutic target for colorectal cancer treatment.
Since the potential molecular mechanism of miR-455
as a tumor suppressor in melanoma was unknown, the present study
aimed to identify the mechanism. Identification of target genes of
miR-455 is important for elucidation of the functions of miR-455 in
carcinogenesis and progression of melanoma, and may provide
promising therapeutic targets for patients with melanoma.
Initially, the bioinformatic analysis (using TargetScan and
microRNA) revealed that IGF-1R may be a potential target for
miR-455. Subsequently, this predication was further confirmed by
the luciferase report assay. The results demonstrated that miR-455
overexpression decreased luciferase activity in the
pGL3-IGF-1R-3′UTR Mut but not the pGL3-IGF-1R-3′UTR Wt, indicating
that miR-455 directly targeted the 3′UTR of IGF-1R. Notably, the
regulatory effect of miR-455 on IGF-1R expression in melanoma cells
was measured using RT-qPCR and western blot analysis. The data
revealed that miR-455 overexpression dramatically suppressed IGF-1R
expression at the mRNA and protein levels in melanoma cells.
Finally, IGF-1R knockdown suppressed proliferation and invasion of
melanoma cells, which was similar to the effect observed as a
result of miR-455 overexpression in melanoma cells. These findings
verified that miR-455 targeted IGF-1R to decrease melanoma cell
growth and invasion.
IGF-1R, a transmembrane tyrosine kinase receptor of
the insulin receptor family, is mainly activated by IGF1 and IGF2
in autocrine and paracrine manners (34). It activates multiple downstream
signaling cascades, including phosphoinositide 2-kinase/Akt and
mitogen activated protein kinase/extracellular signal-related
kinase signaling pathways, and is involved in cell proliferation,
differentiation, migration, invasion, metastasis and survival
(35–38). IGF-1R has been demonstrated to be
upregulated in multiple human cancers, including hepatocellular
carcinoma, non-small lung cancer, prostate cancer and melanoma
(39–42). IGF-1R has been demonstrated to be
regulated by microRNAs in cancers, including miR-497 in cervical
cancer and pancreatic cancer (43,44),
miR-332-5p and miR-181b in glioma (45,46)
and miR-133a in hepatocellular carcinoma (47). These studies indicated that
miRNAs/IGF-1R based targeted therapy may be a novel treatment for
cancer.
In conclusion, the present study confirmed the tumor
suppressor function of miR-455 in melanoma, and demonstrated that
miR-455 suppressed proliferation and invasion through directly
targeting IGF-1R. These findings suggested that the restoration of
miR-455 merits investigation as a therapeutic treatment for
patients with melanoma.
References
|
1
|
Miller AJ and Mihm MC Jr: Melanoma. N Engl
J Med. 355:51–65. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Terando A, Sabel MS and Sondak VK:
Melanoma: Adjuvant therapy and other treatment options. Curr Treat
Options Oncol. 4:187–199. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Penna E, Orso F, Cimino D, Vercellino I,
Grassi E, Quaglino E, Turco E and Taverna D: MiR-214 coordinates
melanoma progression by upregulating ALCAM through TFAP2 and
miR-148b downmodulation. Cancer Res. 73:4098–4111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Baade P and Coory M: Trends in melanoma
mortality in Australia: 1950–2002 and their implications for
melanoma control. Aust N Z J Public Health. 29:383–386. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coory M, Baade P, Aitken J, Smithers M,
McLeod GR and Ring I: Trends for in situ and invasive melanoma in
Queensland, Australia, 1982–2002. Cancer Causes Control. 17:21–27.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sosman JA, Kim KB, Schuchter L, Gonzalez
R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ,
Flaherty KT, et al: Survival in BRAF V600-mutant advanced melanoma
treated with vemurafenib. N Engl J Med. 366:707–714. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wagle N, Emery C, Berger MF, Davis MJ,
Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE,
Hahn WC, et al: Dissecting therapeutic resistance to RAF inhibition
in melanoma by tumor genomic profiling. J Clin Oncol. 29:3085–3096.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lai EC: Micro RNAs are complementary to 3′
UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh TR, Gupta A and Suravajhala P:
Challenges in the miRNA research. Int J Bioinform Res Appl.
9:576–583. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Phuah NH and Nagoor NH: Regulation of
microRNAs by natural agents: New strategies in cancer therapies.
Biomed Res Int. 2014:8045102014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ling H, Fabbri M and Calin GA: MicroRNAs
and other non-coding RNAs as targets for anticancer drug
development. Nat Rev Drug Discov. 12:847–865. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Blower PE, Verducci JS, Lin S, Zhou J,
Chung JH, Dai Z, Liu CG, Reinhold W, Lorenzi PL, Kaldjian EP, et
al: MicroRNA expression profiles for the NCI-60 cancer cell panel.
Mol Cancer Ther. 6:1483–1491. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sakurai E, Maesawa C, Shibazaki M,
Yasuhira S, Oikawa H, Sato M, Tsunoda K, Ishikawa Y, Watanabe A,
Takahashi K, et al: Downregulation of microRNA-211 is involved in
expression of preferentially expressed antigen of melanoma in
melanoma cells. Int J Oncol. 39:665–672. 2011.PubMed/NCBI
|
|
19
|
Medina PP, Nolde M and Slack FJ: OncomiR
addiction in an in vivo model of microRNA-21-induced pre-B-cell
lymphoma. Nature. 467:86–90. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Obad S, dos Santos CO, Petri A, Heidenblad
M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al:
Silencing of microRNA families by seed-targeting tiny LNAs. Nat
Genet. 43:371–378. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Saito Y, Liang G, Egger G, Friedman JM,
Chuang JC, Coetzee GA and Jones PA: Specific activation of
microRNA-127 with downregulation of the proto-oncogene BCL6 by
chromatin-modifying drugs in human cancer cells. Cancer Cell.
9:435–443. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu R, Xie H, Luo C, Chen Z, Zhou X, Xia
K, Chen X, Zhou M, Cao P, Cao K and Zhou J: Identification of FLOT2
as a novel target for microRNA-34a in melanoma. J Cancer Res Clin
Oncol. 141:993–1006. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Du B, Wang Z, Zhang X, Feng S, Wang G, He
J and Zhang B: MicroRNA-545 suppresses cell proliferation by
targeting cyclin D1 and CDK4 in lung cancer cells. PLoS One.
9:e880222014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang F, Tang J, Zhuang X, Zhuang Y, Cheng
W, Chen W, Yao H and Zhang S: MiR-196a promotes pancreatic cancer
progression by targeting nuclear factor kappa-B-inhibitor alpha.
PLoS One. 9:e878972014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin
C and Zhang W: MicroRNA-133 inhibits cell proliferation, migration
and invasion in prostate cancer cells by targeting the epidermal
growth factor receptor. Oncol Rep. 27:1967–1975. 2012.PubMed/NCBI
|
|
29
|
Ujifuku K, Mitsutake N, Takakura S,
Matsuse M, Saenko V, Suzuki K, Hayashi K, Matsuo T, Kamada K,
Nagata I and Yamashita S: MiR-195, miR-455-3p and miR-10a (*) are
implicated in acquired temozolomide resistance in glioblastoma
multiforme cells. Cancer Lett. 296:241–248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu Z, Shen WJ, Kraemer FB and Azhar S:
MicroRNAs 125a and 455 repress lipoprotein-supported
steroidogenesis by targeting scavenger receptor class B type I in
steroidogenic cells. Mol Cell Biol. 32:5035–5045. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li X, Zhang G, Luo F, Ruan J, Huang D,
Feng D, Xiao D, Zeng Z, Chen X and Wu W: Identification of
aberrantly expressed miRNAs in rectal cancer. Oncol Rep. 28:77–84.
2012.PubMed/NCBI
|
|
32
|
Swingler TE, Wheeler G, Carmont V, Elliott
HR, Barter MJ, Abu-Elmagd M, Donell ST, Boot-Handford RP,
Hajihosseini MK, Münsterberg A, et al: The expression and function
of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum.
64:1909–1919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chai J, Wang S, Han D, Dong W, Xie C and
Guo H: MicroRNA-455 inhibits proliferation and invasion of
colorectal cancer by targeting RAF proto-oncogene
serine/threonine-protein kinase. Tumour Biol. 36:1313–1321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao X, Dou W, He L, Liang S, Tie J, Liu
C, Li T, Lu Y, Mo P, Shi Y, et al: MicroRNA-7 functions as an
anti-metastatic microRNA in gastric cancer by targeting
insulin-like growth factor-1 receptor. Oncogene. 32:1363–1372.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Baserga R, Peruzzi F and Reiss K: The
IGF-1 receptor in cancer biology. Int J Cancer. 107:873–877. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cao Z, Liu LZ, Dixon DA, Zheng JZ,
Chandran B and Jiang BH: Insulin-like growth factor-I induces
cyclooxygenase-2 expression via PI3K, MAPK and PKC signaling
pathways in human ovarian cancer cells. Cell Signal. 19:1542–1553.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Christopoulos PF, Msaouel P and
Koutsilieris M: The role of the insulin-like growth factor-1 system
in breast cancer. Mol Cancer. 14:432015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gryko M, Kiśluk J, Cepowicz D, Zińczuk J,
Kamocki Z, Guzińska-Ustymowicz K, Pryczynicz A, Czyżewska J, Kemona
A and Kędra B: Expression of insulin-like growth factor receptor
type 1 correlate with lymphatic metastases in human gastric cancer.
Pol J Pathol. 65:135–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang YH, Wang ZX, Qiu Y, Xiong J, Chen YX,
Miao DS and De W: Lentivirus-mediated RNAi knockdown of
insulin-like growth factor-1 receptor inhibits growth, reduces
invasion and enhances radiosensitivity in human osteosarcoma cells.
Mol Cell Biochem. 327:257–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang YH, Han XD, Qiu Y, Xiong J, Yu Y,
Wang B, Zhu ZZ, Qian BP, Chen YX, Wang SF, et al: Increased
expression of insulin-like growth factor-1 receptor is correlated
with tumor metastasis and prognosis in patients with osteosarcoma.
J Surg Oncol. 105:235–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Scharf JG and Braulke T: The role of the
IGF axis in hepatocarcinogenesis. Horm Metab Res. 35:685–693. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kanter-Lewensohn L, Dricu A, Wang M, Wejde
J, Kiessling R and Larsson O: Expression of the insulin-like growth
factor-1 receptor and its anti-apoptotic effect in malignant
melanoma: A potential therapeutic target. Melanoma Res. 8:389–397.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Luo M, Shen D, Zhou X, Chen X and Wang W:
MicroRNA-497 is a potential prognostic marker in human cervical
cancer and functions as a tumor suppressor by targeting the
insulin-like growth factor 1 receptor. Surgery. 153:836–847. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xu JW, Wang TX, You L, Zheng LF, Shu H,
Zhang TP and Zhao YP: Insulin-like growth factor 1 receptor
(IGF-1R) as a target of MiR-497 and plasma IGF-1R levels associated
with TNM stage of pancreatic cancer. PLoS One. 9:e928472014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lian HW, Zhou Y, Jian ZH and Liu RZ:
MiR-323-5p acts as a tumor suppressor by targeting the insulin-like
growth factor 1 receptor in human glioma cells. Asian Pac J Cancer
Prev. 15:10181–10185. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi ZM, Wang XF, Qian X, Tao T, Wang L,
Chen QD, Wang XR, Cao L, Wang YY, Zhang JX, et al: MiRNA-181b
suppresses IGF-1R and functions as a tumor suppressor gene in
gliomas. RNA. 19:552–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang W, Liu K, Liu S, Ji B, Wang Y and
Liu Y: MicroRNA-133a functions as a tumor suppressor by targeting
IGF-1R in hepatocellular carcinoma. Tumour Biol. 36:9779–9788.
2015. View Article : Google Scholar : PubMed/NCBI
|