Introduction
Melanoma is a life-threatening form of skin cancer.
In the last decade, its annual worldwide incidence has increased
more rapidly compared with other solid tumors. Approximately
160,000 cases of malignant melanoma are newly diagnosed annually
and ~48,000 patients succumb to malignant melanoma each year
worldwide (1). The incidence of
melanoma is less common compared with other types of skin cancer,
including basal and squamous cell cancer. When malignant melanoma
is diagnosed at an early stage (stage 0/1), the 5-year survival
rate is >90%, following surgical excision. However, when
malignant melanoma is diagnosed at a later stage, it is more
invasive and lethal compared with other skin cancers. The median
overall survival rate for patients with metastatic melanoma is
<1 year (2).
The poor prognosis of metastatic melanoma presents a
clinical challenge. Previous clinical studies have been conducted
to improve the efficacy of melanoma treatment (3,4). The
US Food and Drug Administration has approved seven novel agents
since 2011, including B-Raf proto-oncogene, serine/threonine kinase
(BRAF)-inhibitors (vemurafenib, dabrafenib), MEK-inhibitors
(trametinib), anti-programmed cell death protein 1 antibodies
(nivolumab, pembrolizumab), anti-cytotoxic T-lymphocyte-associated
protein 4 antibody (ipilimumab) and (peginterferon-alfa-2b); all of
which are intended for use in the most advanced cases of melanoma
(5). However, drug resistance is a
frequent problem with this type of treatment. Therefore, it is
necessary to develop novel treatment methods and identify novel
drugs directed specifically at human malignant melanoma.
The cause of melanoma is difficult to determine;
however, sun exposure and genetic susceptibility are considered to
be important risk factors. Solar ultraviolet exposure, particularly
when combined with sunburn, is the most important risk factor for
the development of cutaneous malignant melanoma (6). Previous in vitro and in
vivo studies have reported that nuclear factor-κB (NF-κB)
activity may be upregulated in dysplastic nevi and lesions of human
melanoma compared with human nevi or melanocytes in normal skin
(7–9). NF-κB is a transcription factor that
is associated with the activation of several cell processes,
including cell growth and apoptosis (10). This factor is activated in various
cell types in response to numerous stimuli, including mitogens,
inflammatory cytokines and extracellular stress. These
extracellular signals activate inhibitor of κB (IκB) kinase (IKK).
IKK, in turn, phosphorylates the IκBα protein, which leads to
ubiquitination, dissociation of IκBα from NF-κB and IκBα
degradation by proteasomes. The activated NF-κB is subsequently
translocated into the nucleus. NF-κB is an important cell cycle
regulator the activity of which may result in increased cell growth
(11).
Cyclin D1 is a protein that regulates the
G1/S phase transition. During the G1 phase,
it is synthesized rapidly and accumulates in the nucleus. Cyclin D1
regulates the activity of cyclin-dependent kinases (CDKs), leading
to phosphorylation of retinoblastoma protein (Rb) and promotion of
cell cycle progression (12). The
p16-cyclin D/CDK4-Rb pathway is altered in all melanoma cell lines
(13). Another important genetic
factor in melanoma pathogenesis is the mitogen-activated protein
kinase (MAPK) cascade (14). The
MAPK and Rb pathways interconnect at cyclin D1. The cyclin D1
promoter acts as a sensor for growth signals conveyed via the MAPK
cascade and provides a link between this pathway and the cell cycle
machinery (15). Following
treatment with BRAF inhibitors, most patients develop resistance
within 6–8 months. The overexpression of cyclin D1, which may be
observed in 15–20% of BRAF-resistant melanomas, is associated with
a higher rate of resistance to BRAF inhibitors (16,17).
Taken together, NF-κB activity upregulation and cyclin D1
overexpression have been identified as important factors
contributing to melanoma pathogenesis, and inhibition of their
expression levels may be beneficial as a treatment for
melanoma.
Fructooligosaccharides (FOS) are important
prebiotics due to their capacity to selectively stimulate the
growth and/or activity of beneficial intestinal microbiota, such as
Lactobacillus and Bifidobacterium (18). Commercially available FOS produced
from sucrose comprise 1-kestose, nystose and
1F-fructosylnystose, which are referring to inulo-type
FOS. Neo-FOS may be produced from sucrose through the catalytic
action of 6G-fructofuranosidase from
Xanthophyllomyces dendrorhous. Neo-FOS consist primarily of
neokestose (FGF) and neonystose (FGF2), in which
fructosyl units are β-(2,6)-linked to the fructofuranose residues
of sucrose (19). When compared
with inulo-type FOS, neo-FOS exert improved chemical stability and
bifidogenetic activity (20).
Our previous study suggested that the antineoplastic
effects of neokestose on the Caco-2 colorectal adenocarcinoma cell
line involved inhibition of the expression levels of NF-κB and
cytochrome c oxidase subunit II (21). The present study investigated the
mechanism of neokestose in suppressing human melanoma A2058 cell
growth. One mechanism that may contribute to the anticancer
properties of neokestose is the downregulation of NF-κB signaling.
To the best of our knowledge, the present study is the first to
suggest that neokestose may inhibit the activity of NF-κB and the
expression of cyclin D1 in the A2058 melanoma cell line in
vitro.
Materials and methods
Preparation of neokestose
Neokestose is the primary FOS produced in cultures
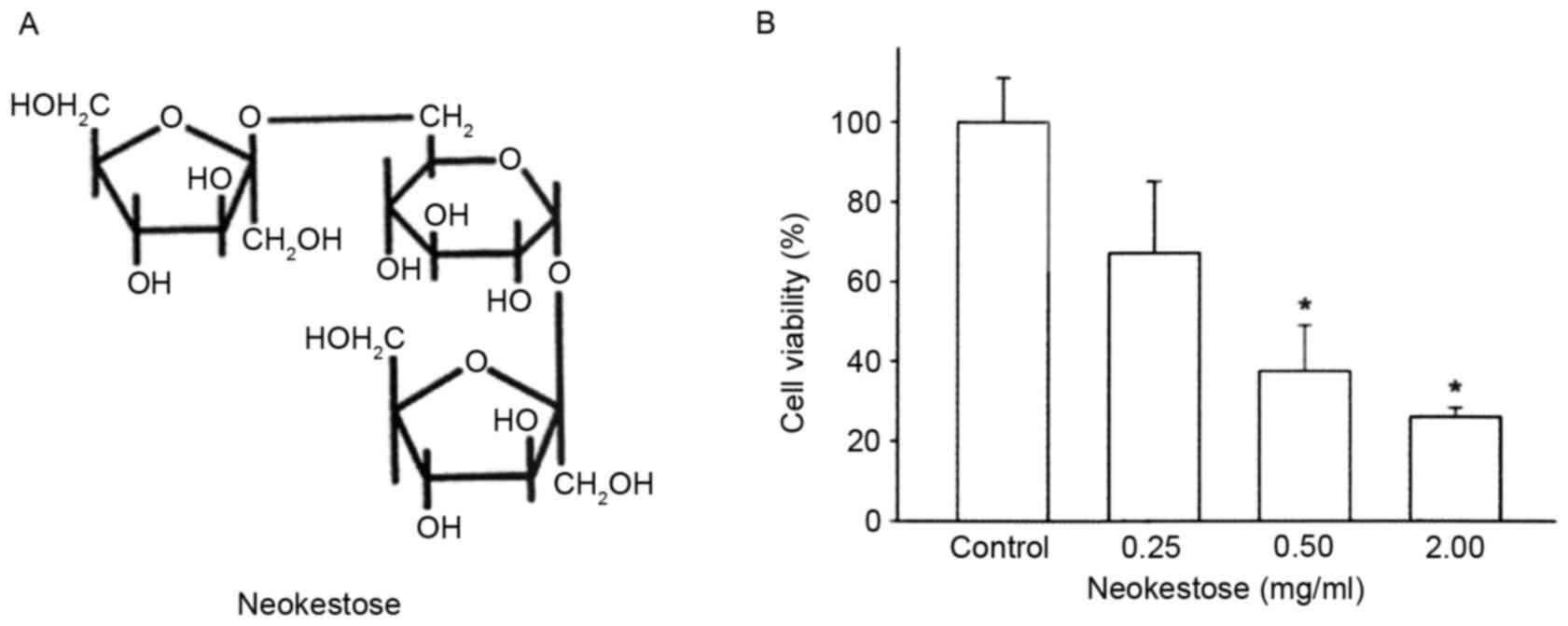
of X. dendrorhous when grown on sucrose (22). Neokestose (Fig. 1A) was purified from the high purity
neo-FOS mixture obtained as previously described (23). Neokestose was purified by
high-performance liquid chromatography on a semi-preparative ODS-AQ
column (20×250 mm; YMC, Co., Ltd., Kyoto, Japan) with a Waters 410
differential refractive index detector (Waters Corporation,
Milford, MA, USA) (21).
Cell culture
The A2058 melanoma cell line was obtained from the
Bioresource Collection and Research Center (Hsinchu, Taiwan). The
A2058 melanoma cell line was routinely maintained and subcultured
in 10 cm2 dishes at 37°C in a humidified CO2
incubator, containing 95% air and 5% CO2. The medium for
cell growth contained 10% heat-inactivated HyClone fetal bovine
serum (SH30071.03; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), 100 IU/ml penicillin and 100 IU/ml streptomycin in HyClone
Dulbecco's modified Eagle's medium (DMEM)/High Glucose solution
(SH30243.01; Thermo Fisher Scientific, Inc.). When cells reached
80% confluence, they were subcultured using 0.25% trypsin and 0.02%
EDTA in PBS. The medium was replaced every 48 h. Cells were grown
in serum-free media for 24 h prior to treatment with
neokestose.
The melanoma cells were plated onto 24-well plates
at a density of 10,000 cells/well and cultured for 24 h. The cells
were subsequently serum-starved for 24 h to synchronize cells in
the G0/G1 phase of the cell cycle. Neokestose
was dissolved in DMEM. The stock solution was diluted to final
concentrations of 0, 0.25, 0.50 and 2.00 mg/ml and the treatment
was applied for 24 h for the MTT, cell cycle and apoptosis assays.
Neokestose at 1 mg/ml was used to pretreat A2058 melanoma cells for
2 h, followed by treatment with or without 2.5 ng/ml tumor necrosis
factor-β (TNF-β; 4345-20; BioVision, Inc., Milpitas, CA, USA) for 1
h for the western blot analysis and immunocytochemistry.
MTT assay
MTT (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
is a tetrazolium salt that is cleaved by mitochondrial
dehydrogenase in living cells. The effects of neokestose (0, 0.25,
0.50 or 2.00 mg/ml) on cell viability were determined using the MTT
assay. A2058 melanoma cells were seeded at a density of
1.0×104 cells/well in 96-well plates for 2 days, the
medium was discarded and 20 µl MTT solution [0.5 mg/ml in
phosphate-buffered saline (PBS)] was added to all wells and the
cells were incubated for 3 h 37°C. Subsequently, 100 µl dimethyl
sulfoxide was added to each well to lyse the cells for 5–10 min a
37°C incubator and the plates were transferred to a microplate
reader, where absorbance was read at 595 nm. Based on a previous
cytotoxicity test (21), the
concentrations of neokestose used in the present study were 0.25,
0.50 and 2.00 mg/ml, and 0 mg/ml was used as the control. All
experiments were performed at least four times with four wells for
each concentration.
Cell cycle analysis
To analyze the cell cycle distribution, cells were
washed twice with PBS, collected by centrifugation at 725 × g for 5
min at 4°C and fixed in 70% (v/v) ethanol at 4°C for 30 min.
Following fixation, the cells were esuspended in PBS and stained
with propidium iodide (PI) solution (48 µg/ml PI and 48 µg/ml RNase
A) for 20 min at room temperature. The DNA content of the cells was
examined by flow cytometry (Cell Lab Quanta SC flow cytometer;
Beckman Coulter, Fullerton, CA, USA) and analyzed by Cell Lab
Quanta SC software version 1.0.
Apoptosis analysis
Apoptosis was quantified using the Annexin
V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit I (BD
Biosciences, San Jose, CA, USA), according to the manufacturer's
protocol. The cells were washed twice with PBS and collected by
centrifugation at 725 × g for 5 min at 25°C. Cells were resuspended
in 100 µl binding buffer and labeled with 5 µl Annexin V FITC and 5
µl PI for 15 min in the dark. Following labeling, cells were
resuspended in 400 µl binding buffer and detected using the Cell
Lab Quanta SC flow cytometer (Beckman Coulter, Fullerton, CA, USA).
The multiparametric data were analyzed using the Cell Lab Quanta SC
software version 1.0.
Western blot analysis
A2058 melanoma cells were lysed with cell extraction
buffer (M-PER Mammalian Protein Extraction reagent; Thermo Fisher
Scientific, Inc.). The Bicinchoninic Acid Protein Reagent Assay kit
(Thermo Fisher Scientific, Inc.) was used to quantify the protein
concentration. The cell protein extracts (60 µg per lane) were
separated by 12% SDS-PAGE. Following electrophoresis, the proteins
were transferred onto polyvinylidene fluoride membranes (Hybond-P;
GE Healthcare Life Sciences, Little Chalfont, UK). Membranes were
blocked with 3% bovine serum albumin (BSA; Sigma-Aldrich; Merck
KGaA) in Tris-buffered saline (20 mM Tris, pH 7.5, and 150 mM NaCl)
containing 0.1% Tween-20 (TBST) for 1 h, then the membranes were
incubated with the following primary antibodies (all diluted to
1:1,000): Anti-cyclin D1 [rabbit monoclonal antibody (mAb); 2978;
Cell Signaling Technology, Inc., Danvers, MA, USA] anti-NF-κB
(rabbit mAb; 4764; Cell Signaling Technology, Inc.), anti-p-IκBα
(rabbit mAb;2859; Cell Signaling Technology, Inc.) and
anti-β-tubulin (rabbit mAb; 2128; Cell Signaling Technology, Inc.)
for 16 h at 4°C and subsequently washed with TBST. The secondary
antibody, goat anti-rabbit immunoglobulin G horseradish
peroxidase-conjugated antibody (7074; Cell Signaling Technology,
Inc.), was incubated for 1 h at room temperature with the membranes
at a dilution of 1:2,000 in Gelatin-NET (50 mM Tris, 0.25% gelatin,
15 mM NaCl, 5 mM EDTA•2Na, 0.05% Tween 20, pH 8.0). Following
washing three times with PBS, 10 min each, the antibody complexes
were detected using the Clarity Western enhanced chemiluminescence
substrate (170–5061; Bio-Rad Laboratories, Inc., Hercules, CA, USA)
and an ImageQuant LAS 4000 imager (GE Healthcare Life
Sciences).
Immunocytochemistry
Immunocytochemistry was used to determine whether
TNF-β induced NF-κBp 65 nuclear translocation in A2058 melanoma
cells. The A2058 cells were plated onto 24-well plates at a density
of 1×104 cells/well, grown in 1 ml culture media and
then incubated for 24 h. Following pretreatment with 1 mg/ml
neokestose for 2 h, the cells were stimulated with 2.5 ng/ml TNF-β
for 1 h. Subsequently, the cells were fixed with 4%
paraformaldehyde on slides for 30 min at room temperature. In order
to improve cell adherence, the slides were coated with
poly-L-lysine (Sigma-Aldrich; Merck KGaA). Subsequently, the cells
were permeabilized with 0.5% Triton X-100 in PBS. The staining was
performed using an anti-NF-κBp 65 antibody at a dilution of 1:100
(4764; rabbit mAb; Cell Signaling Technology, Inc.) for 1 h at room
temperature. Cells were washed twice with PBS and subsequently
incubated at room temperature with a goat anti-rabbit FITC
secondary antibody (111–095-046; Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA) at a dilution of 1:100 for
1 h. Nuclei were stained using Hoechst 33342 (Invitrogen; Thermo
Fisher Scientific, Inc.). Cells were subsequently embedded in a
mounting medium following two washes with PBS. The slides were then
examined under a Leica TCS SP5 Confocal Spectral Microscope Imaging
system (Leica Microsystems, GmbH, Wetzlar, Germany).
Statistical analysis
Results of MTT assays and flow cytometric analyses
were expressed as the mean ± standard error. One-way analysis of
variance was used and the identification of significant differences
between results were performed by applying the Duncan test, with
the level of statistical significance set at P<0.05. P<0.05
was considered to indicate a statistically significant
difference.
Results
Neokestose exerts a negative effect on
A2058 cell viability
The effects of various concentrations of neokestose
(0.25, 0.50 and 2.00 mg/ml; 0 mg/ml was used as the control) were
determined on the viability of A2058 melanomacells using MTT assay.
For each experiment, at least three independent experiments were
performed and each was repeated in triplicate in 96-well plates.
Treatment of A2058 melanoma cells with increasing concentrations of
neokestose exerted a significant dose-dependent cytotoxic effect
(Fig. 1B).
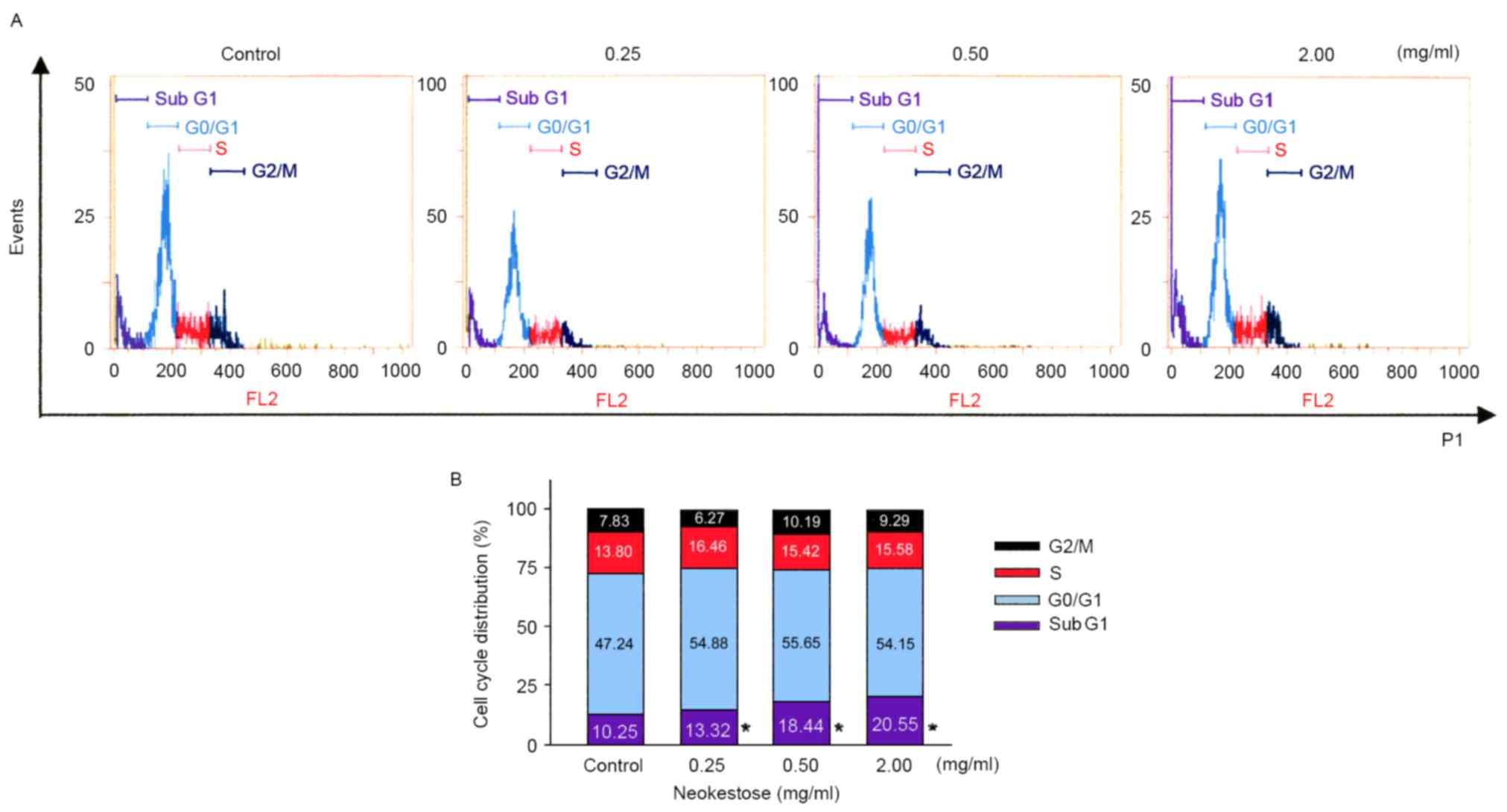
Effects of neokestose on cell cycle
distribution
A2058 melanoma cells were treated with neokestose
(0.25, 0.50 and 2.00 mg/ml; 0 mg/ml was used as the control) for 24
h and the cell cycle distribution was quantified using flow
cytometry. Neokestose treatment significantly increased the
sub-G1 phase cell population (Fig. 2).
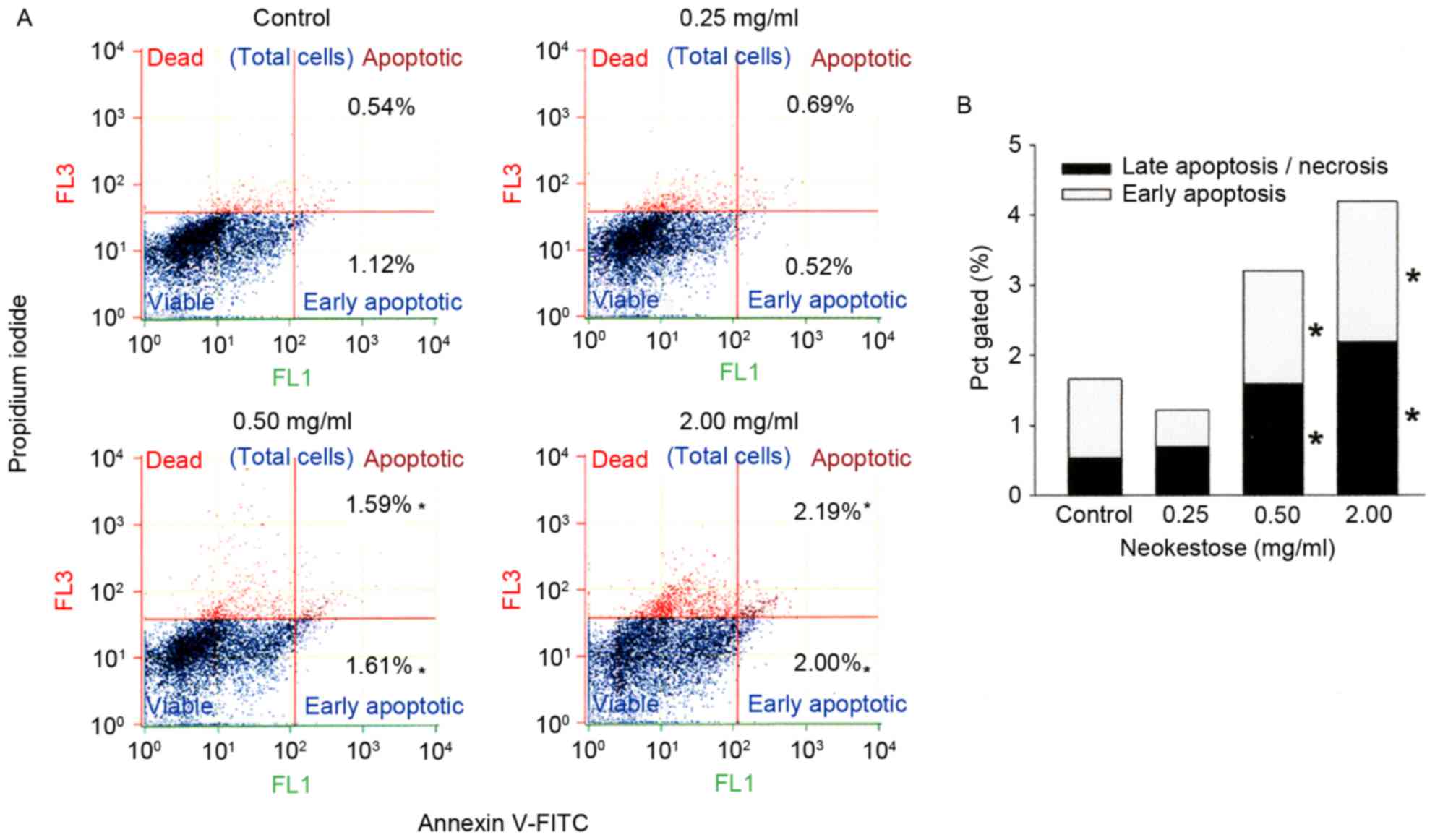
Effects of neokestose on A2058 cell
apoptosis
A2058 melanoma cells treated with neokestose were
analyzed using Annexin V-FITC/PI staining and flow cytometry in
order to investigate whether apoptosis was induced by neokestose
(Fig. 3). Neokestose treatment
significantly increased the percentage of early and late apoptotic
cells (Fig. 3B), indicating that
neokestose was able to induce both early and late stage apoptosis
of A2058 melanoma cells. In addition, neokestose exposure induced
apoptosis of A2058 melanoma cells in a dose-dependent manner
(Fig. 3).
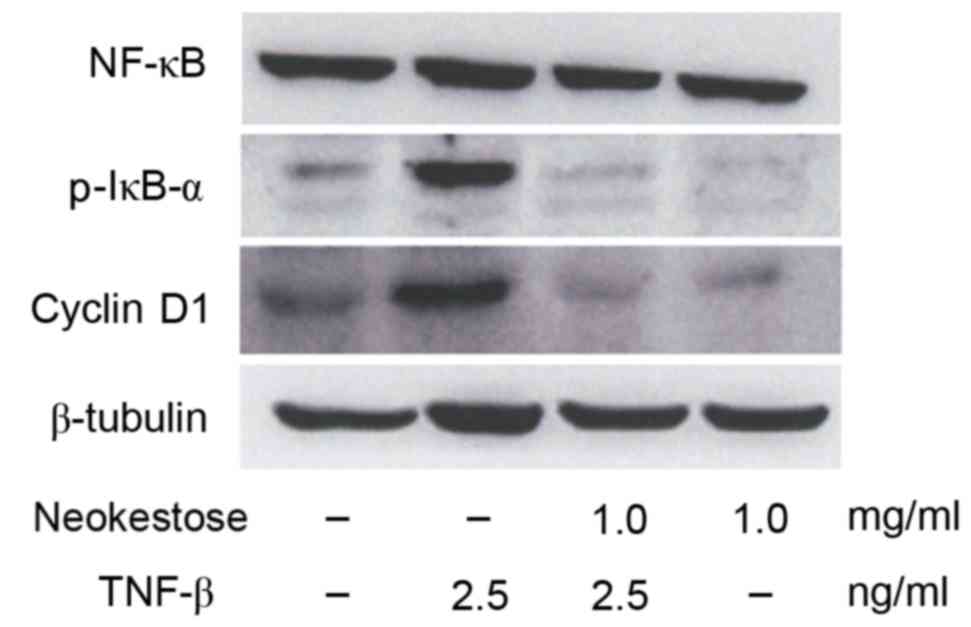
Effects of neokestose on the
expression of p-IκB and cyclin D1 in the presence of TNF-β
The present study demonstrated that exposure to 2.5
ng/ml TNF-β (an NF-κB activator) for 1 h led to increased
expression of p-IκB and cyclin D1. Fig. 4 indicates increased expression of
p-IκB and cyclin D1. Pretreatment with 1 mg/mlneokestose resulted
in reduced expression of p-IκB and cyclin D1 in TNF-β-exposed A2058
melanoma cells.
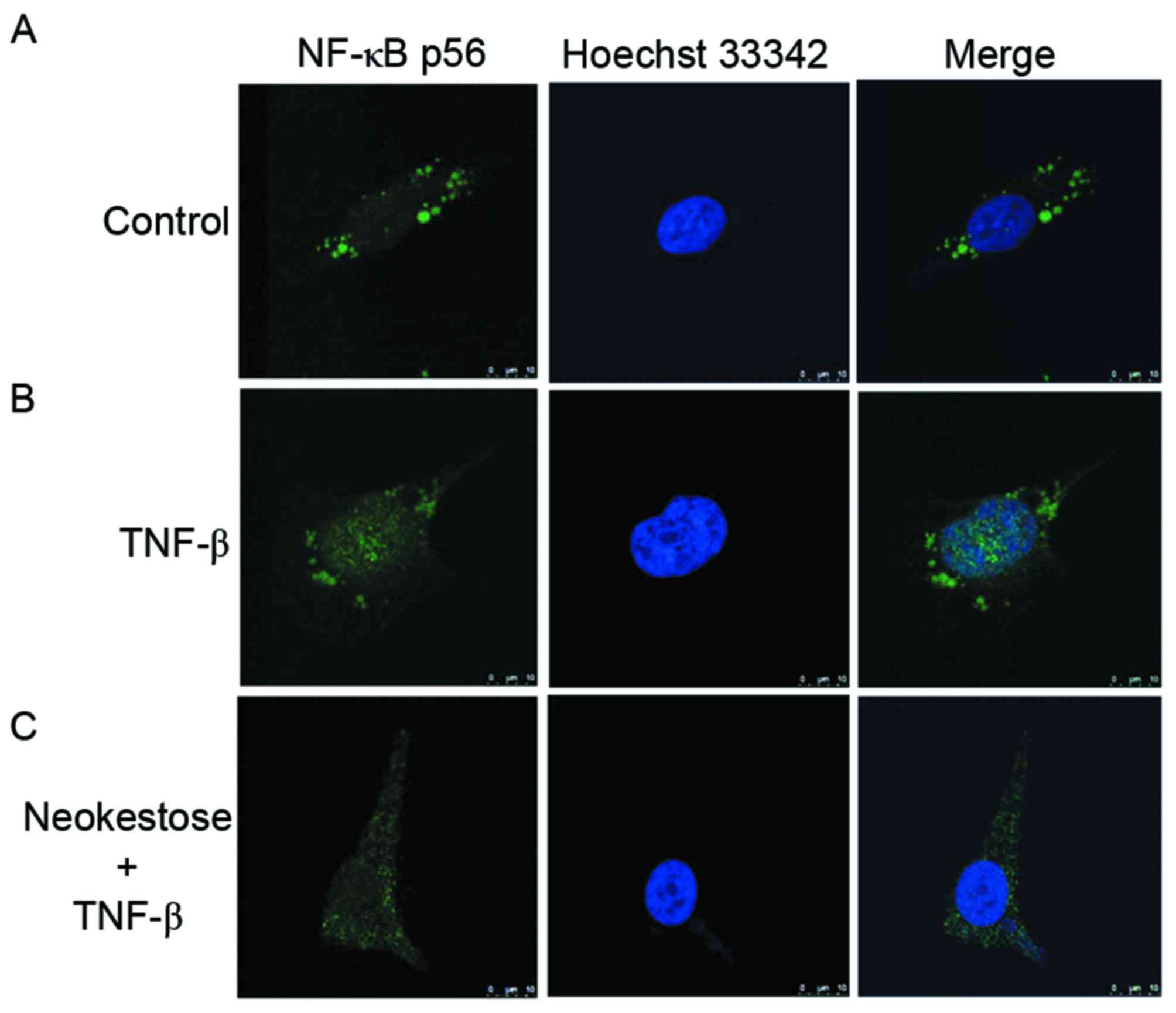
Neokestose inhibits TNF-β-induced
NF-κB p65 nuclear translocation
Following exposure to 2.5 ng/ml TNF-β for 1 h, NF-κB
nuclear translocation was increased in A2058 melanoma cells.
However, following treatment with 1 mg/ml neokestose for 2 h and
then 2.5 ng/ml TNF-β for 1 h NF-κB nuclear translocation was
reduced (Fig. 5).
Discussion
It is well known that NF-κB is conjugated to IκB
residues in the cytoplasm, where it maintains an inactive state
(24). Treatment of cells with
TNF-β may increase NF-κB activation, which leads to the
phosphorylation of IκB. Previous studies have suggested that
inhibition of this pathway may lead to the suppression of tumor
growth (21,25). NF-κB activation requires the
phosphorylation, ubiquitinationand subsequent degradation of IκB.
One primary mechanism of NF-κB inactivation is through inhibition
of IκB phosphorylation, leading to the retention of NF-κB in the
cytoplasm. Neokestose may prevent IκB phosphorylation, thus
reducing NF-κB activation.
The NF-κB signaling pathway can be potentially
targeted at various levels, including kinases, phosphatases, DNA
binding proteins, acetyl transferases and methyl transferases, in
addition to ubiquitination and nuclear translocation. It is
estimated that >700 compounds exist that inhibit NF-κB activity;
therefore, the NF-κB signaling pathways have been targeted at
various levels for effective cancer therapy (26).
It has previously been reported that specific
compounds work on the level of nuclear translocation of NF-κB; for
example, SN50 is a 41-residue synthetic peptide that may
effectively inhibit lipopolysaccharide- and TNF-α-induced NF-κB
nuclear translocation in intact cells (27). In addition, the fungal compound
dehydroxymethylepoxyquinomicin may inhibit TNF-α-induced nuclear
translocation and NF-κB activation (28). Furthermore, C086, which is a novel
analog of curcumin, may inhibit IκBα phosphorylation and suppress
the nuclear translocation of NF-κB (29).
Since the aforementioned compounds may exert adverse
side effects and are too costly to be widely implemented they may
not be used as dietary chemopreventive agents. Therefore, it is
necessary to identify cheaper and safer compounds to be used as
chemopreventive agents for cancer. In this regard neokestose is
considered to be a cheaper and safer compound, although future
in vivo studies are required before neokestose can be widely
used as a dietary chemopreventive agent.
In conclusion, the findings of the present study
revealed that neokestose may inhibit the NF-κB signaling pathway
and cyclin D1 expression in A2058 melanoma cells.
Acknowledgements
The authors would like to thank the Hsin Shen Junior
College of Medical Care and Management, Tatung University and
Taipei Medical University for their assistance in academic
resources.
References
|
1
|
Eggermont AM, Spatz A and Robert C:
Cutaneous melanoma. Lancet. 383:816–827. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mashima E, Inoue A, Sakuragi Y, Yamaguchi
T, Sasaki N, Hara Y, Omoto D, Ohmori S, Haruyama S, Sawada Y, et
al: Nivolumab in the treatment of malignant melanoma: Review of the
literature. Onco Targets Ther. 8:2045–2051. 2015.PubMed/NCBI
|
|
3
|
Robert C, Dummer R, Gutzmer R, Lorigan P,
Kim KB, Nyakas M, Arance A, Liszkay G, Schadendorf D, Cantarini M,
et al: Selumetinib plus dacarbazine versus placebo plus dacarbazine
as first-line treatment for BRAF-mutant metastatic melanoma: A
phase 2 double-blind randomised study. Lancet Oncol. 8:733–740.
2013. View Article : Google Scholar
|
|
4
|
Larkin J, Ascierto PA, Dréno B, Atkinson
V, Liszkay G, Maio M, Mandalà M, Demidov L, Stroyakovskiy D, Thomas
L, et al: Combined vemurafenib and cobimetinib in BRAF-mutated
melanoma. N Engl J Med. 20:1867–1876. 2014. View Article : Google Scholar
|
|
5
|
Niezgoda A, Niezgoda P and Czajkowski R:
Novel Approaches to treatment of advanced melanoma: A review on
targeted therapy and immunotherapy. Biomed Res Int.
2015:8513872015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gandini S, Sera F, Cattanizza MS, Pasquini
P, Picconi O, Boyle P and Melchi CF: Meta-analysis of risk factors
for cutaneous melanoma: II. Sun exposure. Eur J Cancer. 41:45–60.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dhawan P, Singh AB, Ellis DL and Richmond
A: Constitutive activation of Akt/protein kinase B in melanoma
leads to up-regulation of nuclear factor-kappaB and tumor
progression. Cancer Res. 62:7335–7342. 2002.PubMed/NCBI
|
|
8
|
McNulty SE, Tohidian NB and Meyskens FL
Jr: RelA, p50 and inhibitor of kappa B alpha are elevated in human
metastatic melanoma cells and respond aberrantly to ultraviolet
light B. Pigment Cell Res. 14:456–465. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McNulty SE, del Rosario R, Cen D, Meyskens
FL Jr and Yang S: Comparative expression of NFkappaB proteins in
melanocytes of normal skin vs. benign intradermal naevus and human
metastatic melanoma biopsies. Pigment Cell Res. 17:173–180. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmitz ML, Mattioli I, Buss H and Kracht
M: NF-kappaB: A multifaceted transcription factor regulated at
several levels. Chembiochem. 5:1348–1358. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gilmore TD: Introduction to NF-kappaB:
Players, pathways, perspectives. Oncogene. 51:6680–6684. 2006.
View Article : Google Scholar
|
|
12
|
Sherr CJ and Roberts JM: CDK inhibitors:
Positive and negative regulators of G1-phase progression. Genes
Dev. 13:1501–1512. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartkova J, Lukas J, Guldberg P, Alsner J,
Kirkin AF, Zeuthen J and Bartek J: The p16-cyclin D/Cdk4-pRb
pathway as a functional unit frequently altered in melanoma
pathogenesis. Cancer Res. 56:5475–5483. 1996.PubMed/NCBI
|
|
14
|
Chin L, Merlino G and DePinho RA:
Malignant melanoma: Modern black plague and genetic black box.
Genes Dev. 12:3467–3481. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Filmus J, Robles AI, Shi W, Wong MJ,
Colombo LL and Conti CJ: Induction of cyclin D1 overexpression by
activated ras. Oncogene. 9:3627–3633. 1994.PubMed/NCBI
|
|
16
|
Sullivan RJ and Flaherty KT: Resistance to
BRAF-targeted therapy in melanoma. Eur J Cancer. 6:1297–1304. 2013.
View Article : Google Scholar
|
|
17
|
Sauter ER, Yeo UC, von Stemm AV, Zhu W,
Litwin S, Tichansky DS, Pistritto G, Nesbit M, Pinkel D, Herlyn M
and Bastian BC: Cyclin D1 is a candidate oncogene in cutaneous
melanoma. Cancer Res. 62:3200–3206. 2002.PubMed/NCBI
|
|
18
|
Gibson GR and Roberfroid MB: Dietary
modulation of the human colonic microbiota: Introducing the
concepts of prebiotics. J Nutr. 125:1401–1412. 1995.PubMed/NCBI
|
|
19
|
Lim JS, Lee JH, Kang SW, Park SW and Kim
SW: Studies on production and physical properties of neo-FOS
produced by co-immobilized Penicillium citrinum and
neo-fructosyltransferase. Eur Food Res Technol. 225:457–462. 2007.
View Article : Google Scholar
|
|
20
|
Kilian S, Kritzinger S, Rycroft C, Gibson
G and du Preez J: The effects of the novel bifidogenic
trisaccharide, neokestose, on the human colonic microbiota. World J
Microbiol Biotechnol. 18:637–644. 2002. View Article : Google Scholar
|
|
21
|
Lee SM, Chang JY, Wu JS and Sheu DC:
Antineoplastic effect of a novel chemopreventive agent, neokestose,
on the Caco-2 cell line via inhibition of expression of nuclear
factor-κB and cyclooxygenase-2. Mol Med Rep. 12:1114–1118.
2015.PubMed/NCBI
|
|
22
|
Kritzinger SM, Kilian SG, Potgieter MA and
de Preez JC: The effect of production parameters on the synthesis
of the prebiotic trisaccharide, neokestose, by Xanthophyllomyces
dendrorhous (Phaffia rhodozyma). Enzyme Microb Technol. 32:728–737.
2003. View Article : Google Scholar
|
|
23
|
Sheu DC, Chang JY, Chen YJ and Lee CW:
Production of high-purity neofructooligosaccharides by culture of
Xanthophyllomyces dendrorhous. Bioresour Technol. 132:432–435.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ghosh S and Karin M: Missing pieces in the
NF-kappaB puzzle. Cell. 109:(Suppl). S81–S96. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luqman S and Pezzuto JM: NFkappaB: A
promising target for natural products in cancer chemoprevention.
Phytother Res. 24:949–963. 2010.PubMed/NCBI
|
|
26
|
Gupta SC, Sundaram C, Reuter S and
Aggarwal BB: Inhibiting NF-κB activation by small molecules as a
therapeutic strategy. Biochim Biophys Acta. 1799:775–787. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lin YZ, Yao SY, Veach RA, Torgerson TR and
Hawiger J: Inhibition of nuclear translocation of transcription
factor NF-kappa B by a synthetic peptide containing a cell
membrane-permeable motif and nuclear localization sequence. J Biol
Chem. 270:14255–14258. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ariga A, Namekawa J, Matsumoto N, Inoue J
and Umezawa K: Inhibition of tumor necrosis factor-alpha-induced
nuclear translocation and activation of NF-kappa B by
dehydroxymethylepoxyquinomicin. J Biol Chem. 277:24625–24630. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen C, Liu Y, Chen Y and Xu J: C086, a
novel analog of curcumin, induces growth inhibition and
down-regulation of NFκB in colon cancer cells and xenograft tumors.
Cancer Biol Ther. 12:797–807. 2011. View Article : Google Scholar : PubMed/NCBI
|