Introduction
Inflammatory responses elicit a host defense for
outer stimuli, including bacterial components and invasion of
pathogens trigger sequential innate immune responses. The main
immune cells that act in innate immune responses include
neutrophils, dendritic cells and macrophages, whose inflammatory
properties involve phagocytic action, antigen presentation and
inflammatory mediator production (1). In particular, macrophages initiate
and maintain inflammation through the production of inflammatory
mediators, including nitric oxide (NO), prostaglandin E2
(PGE2) and proinflammatory cytokines (2). The tight regulation of inflammatory
responses is important because excess inflammatory responses cause
severe inflammatory diseases, such as inflammatory bowel disease,
atherosclerosis and rheumatoid arthritis. However, tightly
regulated inflammation can rescue the human body from infectious
diseases (3,4). Therefore, the discovery of candidates
having anti-inflammatory properties is a valuable strategy for the
treatment of severe inflammatory states.
Spilanthes acmella Murr. (S. acmella),
of the family Compositae, is a small plant found in India, Sri
Lanka and other tropical countries (5). This plant has been traditionally used
to treat many inflammatory diseases, including toothache, urinary
calculi, wound, itching and psoriasis, and as a diuretic in India
and Sri Lanka (5,6). Recent scientific approaches proved
that the pharmacological effects of S. acmella noted its use
of an anesthetic, antipyretic, anti-inflammatory, analgesic,
antifungal, antimalarial, diuretic and antinociceptive, through the
use of various animal models (7–10).
Phytochemical analyses have revealed that S. acmella
contains spilanthol, an isobutylamide, as a major component and
secondary metabolites including β-sitosterol, stigmasterol,
α-amyrin, limonene, β-caryophyllene, myrecene and vanillic acid as
minor components (9,11–13).
Of these, spilanthol has been reported to exhibit antimalarial and
anti-inflammatory properties, however, the in-depth mechanism study
was not pursued (2,14).
The ethnopharmacological anti-inflammatory effects
of S. acmella remain unclear at the molecular level, as most
studies researched its pharmacological effects. A previous study
with S. acmella extract was limited to the preliminary
evaluation of the anti-inflammatory and analgesic effects in
experimental animal models (15).
In the present study, the authors investigated the
anti-inflammatory effects of the methanol extract of S.
acmella (MSA) and its precise regulatory molecular action on
the inflammatory signaling pathways, mitogen-activated protein
kinase (MAPK) and nuclear factor-κB (NF-κB), in murine
macrophages.
Materials and methods
MSA preparation
A methanol extract (cat. no. KRIB0043997) of the
S. acmella from Lam Dong (Vietnam) was obtained from the
International Biological Material Research Center [Korea Research
Institute of Bioscience & Biotechnology (KRIBB), Daejeon,
Korea]. The concentrated methanol extract was manufactured by
standard protocol of KRIBB. Briefly, the leaves and stem of plants
(>1 kg dry weight) were dried at room temperature (RT), treated
with methanol (HPLC grade), and sonicated several times at 50°C for
3 days. The extracts were filtrated to remove solid substances and
concentrated with reduced pressure at 50°C. To ensure dissolution
of both polar and non-polar compounds in the extract and to avoid
evaporation, dimethoxysulfoxide (DMSO) was used to make stock
solution (200 mg/ml) of the extract. The stock extract was stored
at −20°C before use.
Cell culture and reagents
RAW 264.7 macrophages (ATCC, Manassas, VA, USA), a
mouse monocytic cell line, were maintained in Dulbecco's modified
Eagle's medium containing 10% fetal bovine serum (both GE
Healthcare Bio-Sciences, Pittsburgh, PA, USA), 50 U/ml penicillin
and 50 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) at 37°C in humidified air containing 5%
CO2. Rabbit anti-inhibitor of κBα (IκBα; sc-371) and
anti-GAPDH (sc-25778) antibodies were purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). Rabbit anti-inducible NO
synthase (iNOS; 2982), anti-cyclooxygenase (COX)-2 (4842),
anti-p-IκBα (Ser32/36; 9246), anti-p38 (9212), anti-p-p38
(Thr180/Tyr182; 9211), anti-extracellular signal-regulated kinase
(ERK; 9102), anti-c-Jun N-terminal kinase (JNK; 9252), anti-p-JNK
(Thr183/Tyr185; 9251), anti-transforming growth factor
beta-activated kinase 1 (TAK1; 4505), anti-p-TAK1 (Thr184/187;
4508) antibodies, and mouse anti-p-ERK (Thr202/Tyr204; 9106) were
purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Goat anti-rabbit IgG (LF-SA8002) and goat anti-mouse IgG
(LF-SA8001) were purchased from AbFrontier Co., Ltd. (Seoul,
Korea). DMSO was purchased from Sigma-Aldrich; Merck KGaA
(Darmstadt, Germany). Ez-cytox solution was purchased from Daeil
Lab Service Co., Ltd. (Seoul, Korea). Ready-SET-Go! ELISA kits for
the detection of IL-6 (88–7064) and tumor necrosis factor (TNF)-α
(88–7324) were from eBioscience (San Diego, CA, USA). The
PGE2 ELISA kit (510410) was from Cayman Chemical Company
(Ann Arbor, MI, USA). Accuzol reagent was from Bioneer Corporation
(Daejeon, Korea) and TOPscript cDNA synthesis kit was from
Enzynomics Co., Ltd. (Daejeon, Korea). The iTaq Universal
SYBR-Green Supermix was obtained from Bio-Rad Laboratories, Inc.
(Hercules, CA, USA).
Measurement of cell viability
RAW 264.7 macrophages were pretreated with MSA (50,
100, 200, 300 and 400 µg/ml) for 2 h and further incubated for 24 h
at 37°C in the absence or presence of LPS (1 µg/ml). Following
incubation, Ez-cytox solution (1/10 dilution of culture medium) was
added to each well and incubated for 1 h. Supernatants were
transferred to new 96-well plates and the absorbance was measured
at 450 nm using Synergy H1 Microplate reader (BioTek Instruments,
Inc., Winnoski, VT, USA).
Measurement of NO production
Cells were seeded at 96-well plates
(4.0×104 cells/well) and incubated at 37°C overnight.
Cells were pretreated with various concentrations of MSA (50, 100,
200 and 300 µg/ml) for 2 h prior to LPS treatment. Following
stimulation with LPS (1 µg/ml) for 24 h, the supernatants (100 µl)
were transferred to new 96-well plate and 100 µl Griess reagent (1%
sulfanilamide, 0.1% N-1-naphthylenediamine dihydrochloride and 2.5%
phosphoric acid) was added to each well. NaNO2 solution
(2.5, 5, 10, 25, 50 and 100 M) was used to generate standard curve
for calculating the quantity of NO in supernatants. The absorbance
was measured at 540 nm using Synergy H1 Microplate reader (BioTek
Instruments, Winooski, VT, USA).
ELISA
Cells were seeded at 96-well plates
(4.0×104 cells/well) and incubated at 37°C overnight.
Cells were pretreated with various concentrations of MSA (50, 100,
200 and 300 µg/ml) for 2 h prior to LPS treatment. Following
stimulation with LPS (1 µg/ml) for 24 h, the supernatants were
collected and diluted according to predetermined dilution rate for
each proinflammatory cytokines. The production of proinflammatory
cytokines, including IL-6 and TNF-α, was measured using
Ready-SET-Go! ELISA kits for each cytokines according to
manufacturer's protocol. Briefly, the 96-well plate was coated with
coating solution for overnight at 4°C, washed with 1X
phosphate-buffered saline/0.05% Tween 20 (PBST) for 3 times, and
treated with 1X Assay Diluent (from the ELISA kit) for 1 h at room
temperature. Following the emptying of the wells, diluted
supernatants and standard solutions were added to each well. At 2 h
after treatment at RT, the plate was washed with 1X PBST for 3
times and detection Ab solution (also from the ELISA kit) diluted
in 1X Assay Diluent was added to plate. The plate was washed
following a 1 h treatment, horseradish peroxidase-streptavidin
solution was added for 30 min, and washed with 1X PBST 5 times. A
solution of 3,3′,5,5′-Tetramethylbenzidine was added to the plate
and incubated for 10 min at the dark. An additional 1 N
H3PO4 was added to the plate to stop the
reaction and absorbance of each well was measured using Synergy H1
Microplate reader at 450 nm.
The production of PGE2 was measured using
PGE2 ELISA kit according to manufacturer's protocol.
Briefly, 96-well plate pre-coated with goat anti-mouse IgG was
incubated with tracer, antibody and either standards or samples for
16 h. Then, the plate was washed with supplied washing buffer 5
times to remove unbound reagents and developed with Ellman's
reagent for 1 h. The absorbance of each well was measured at 405 nm
and the obtained values were analyzed by performing 4-parameter
logistic fit.
RNA preparation and cDNA
synthesis
RAW 264.7 macrophages were seeded in a 12-well plate
(8×105 cells/well) and incubated at 37°C overnight.
Cells were pretreated with MSA (50, 100, 200 and 300 µg/ml) for 2 h
and additionally stimulated with LPS (1 µg/ml) for 3 h. Total RNA
was prepared from the cells using Accuzol (Bioneer Corporation) and
reverse-transcribed into cDNA using a TOPscript cDNA synthesis kit,
according to the manufacturer's protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
PCR amplification of the cDNA was performed using
iTaq Universal SYBR-Green Supermix according to the manufacturer's
protocol. The PCR was run for 40 cycles of denaturation at 94°C for
5 sec and annealing/extension at 60°C for 30 sec using a CFX
Connect real-time thermal cycler (Bio-Rad Laboratories, Inc.).
Based on the 2−ΔΔCq method (16), the results were normalized to multi
reference genes, β-actin and GAPDH, and were expressed as the ratio
of gene expressions to LPS treated group (100%). PCR primers used
in these experiments were listed in a previous report of the
authors (17). The sequences of
PCR primers used in this study were: Mouse iNOS (sense,
5′-TGGCCACCAAGCTGAACT-3′; antisense,
5′-TCATGATAACGTTTCTGGCTCTT-3′), COX-2 (sense,
5′-GATGCTCTTCCGAGCTGTG-3′; antisense, 5′-GGATTGGAACAGCAAGGATTT-3′),
TNF-α (sense, 5-CTGTAGCCCACGTCGTAGC-3′; antisense,
5′-TTGAGATCCATGCCGTTG-3′), IL-6 (sense,
5′-TCTAATTCATATCTTCAACCAAGAGG-3′; antisense,
5′-TGGTCCTTAGCCACTCCTTC-3′), IL-1β (sense,
5′-TTGACGGACCCCAAAAGAT-3′; antisense, 5′-GATGTGCTGCTGCGAGATT-3′),
β-actin (sense, 5′-CGTCATACTCCTGCTTGCTG-3′; antisense,
5′-CCAGATCATTGCTCCTCCTGA-3′) and GAPDH (sense,
5′-GCTCTCTGCTCCTCCTGTTC-3′; antisense,
5′-ACGACCAAATCCGTTGACTC-3′).
Semiquantitative reverse transcription
(RT)-PCR
PCR primers used in these experiments were listed in
a previous report of the authors (17). The sequences of PCR primers used in
the study were as follows: Mouse iNOS (sense,
5′-GCATGGAACAGTATAAGGCAAACA-3′; antisense,
5′-GTTTCTGGTCGATGTCATGAGCAA-3′), COX-2 (sense,
5′-GCATGGAACAGTATAAGGCAAACA-3′; antisense,
5′-GTTTCTGGTCGATGTCATGAGCAA-3′), TNF-α (sense,
5′-GTGCCAGCCGATGGGTTGTACC-3′; antisense,
5-′AGGCCCACAGTCCAGGTCACTG-3′), IL-6 (sense,
5′-TCTTGGGACTGATGCTGGTGAC-3′; antisense,
5′-CATAACGCACTAGGTTTGCCGA-3′), IL-1β (sense,
5′-AGCTGTGGCAGCTACCTGTG-3′; antisense, 5′-GCTCTGCTTGTGAGGTGCTG-3′)
and GAPDH (sense, 5′-GTCTTCACCACCATGGAGAAGG-3′; antisense,
5′-CCTGCTTCACCACCTTCTTGCC-3′). The PCR was run for 17–25 cycles of
94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec by using a
Bioer thermal cycler (Bioer Technology Co., Hangzhou, China).
Following amplification, 10 µl of the PCR products were separated
in 1.5% (w/v) agarose gels and stained with ethidium bromide.
Preparation of total cell lysates
RAW 264.7 macrophages pretreated with MSA were
further stimulated with LPS (1 µg/ml) for the optimized time for
the detection of target proteins (IκBα and TAK1 for 3 min; MAPKs
for 15 min; iNOS and COX-2 for 24 h). Following stimulation for the
indicated times, cells were washed 3 times with ice-cold PBS. Lysis
buffer, containing 0.5% NP-40, 0.5% Triton X-100, 150 mM NaCl, 20
mM Tris-HCl (pH 8.0), 1 mM EDTA, 1% glycerol, 1 mM
phenylmethylsulfonyl fluoride, 10 mM NaF and 1 mM
Na3VO4, was added to the washed wells and
then collected in each microtube after 10 min. Following
centrifugation at 15,814 × g for 30 min at 4°C, the supernatants
were prepared in new microtubes.
Immunoblotting analysis
Protein concentration was measured using the
Bradford reagent (Bio-Rad Laboratories, Inc.). Briefly, optical
density was measured at 595 nm and the concentration of each lysate
was calculated by applying the values to the bovine serum albumin
standard plot. After boiling the mixture of lysates and sample
buffers, aliquots of the samples (20 µg) were separated by 10%
SDS-PAGE and transferred to nitrocellulose membranes with transfer
buffer [192 mM glycine, 25 mM Tris-HCl (pH 8.8), and 20% methanol
(v/v)]. Following blocking with 5% non-fat dried milk, each
membrane was incubated overnight at 4°C with primary antibodies
(1:1,000 dilution for all primary antibodies). Each membrane was
incubated for an additional 1 h with secondary
peroxidase-conjugated IgG (1:5,000) at room temperature. After
washing 5 times with 1X PBST, the target proteins were detected
using Pierce ECL Western Blotting Substrate for enhanced
chemiluminescence (Thermo Fisher Scientific, Inc.). Protein levels
were quantified by scanning the immunoblots and analyzing them with
LabWorks software version 4.6 (UVP, LLC; Analytik Jena AG, Upland,
CA, USA).
Statistical analysis and experimental
replicates
The data are represented as means ± standard error
of the mean. Comparisons between multiple experimental groups were
performed using one-way analysis of variance followed by Dunnett's
post-hoc test using GraphPad Prism (version 3.0; GraphPad Software,
Inc., La Jolla, CA, USA) and P<0.01 was considered statistically
significant. The data from nine replicates were analyzed, including
three independent experiments with three replicates in each.
Results
MSA inhibits the production of
proinflammatory mediators in RAW 264.7 macrophages
Since the inhibitory effect of an anti-inflammatory
reagent should be assessed under non-cytotoxic concentrations, the
authors determined the maximal effective and non-cytotoxic
concentration with a cell viability assay. MSA did not induce
cytotoxicity in concentrations up to 300 µg/ml. However, clear
cytotoxicity was observed at 400 µg/ml for both groups with LPS (1
µg/ml) and without LPS, when compared to their respective 0 µg/ml
MSA groups (P<0.05; Fig. 1A).
Thereafter, the authors used 300 µg/ml and lower concentrations of
MSA throughout the research. To evaluate the anti-inflammatory
properties of MSA, the effect of MSA on the productions of NO and
PGE2, well-known proinflammatory mediators, were
measured in LPS-treated RAW 264.7 cells. As demonstrated in
Fig. 1B, MSA inhibited LPS-induced
NO production in a dose-dependent manner and PGE2
production when compared with LPS-untreated control groups. The
demonstrated results imply that MSA inhibits LPS-induced NO and
PGE2 production in macrophages.
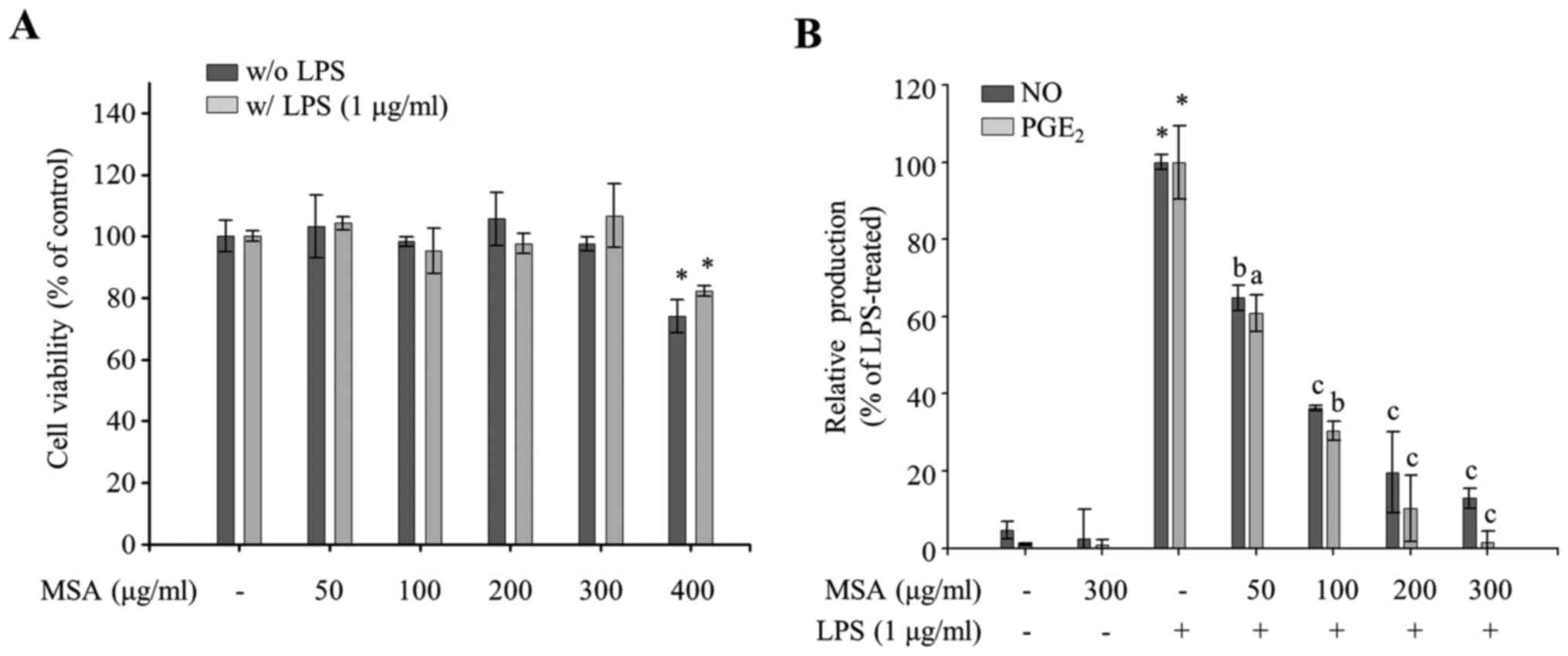 | Figure 1.Effects of MSA on cell viability and
the production of NO. (A) MSA-pretreated (50, 100, 200, 300 and 400
µg/ml) RAW 264.7 macrophages were incubated for 24 h in the
presence or absence of LPS. Cell viability of each group was
compared with that of the LPS-treated or untreated control group.
Data were represented as mean ± standard error of the mean and
analyzed using one-way analysis of variance. *P<0.01 vs.
LPS-untreated or-treated control groups. (B) RAW 264.7 macrophages
were pretreated with MSA (50, 100, 200 and 300 µg/ml) and then
incubated with LPS (1 µg/ml). Following 24 h stimulation, NO and
PGE2 levels in the supernatants were measured. Relative
production of NO and PGE2 levels to LPS-treated group
(100%) by MSA was represented as bar graphs. Data were represented
as mean ± standard error of the mean and analyzed using one-way
analysis of variance. *P<0.0001 vs. LPS-untreated control
groups. aP<0.01, bP<0.001 and
cP<0.0001 vs. LPS-treated groups. MSA, S.
Acmella; NO, nitric oxide; LPS, lipopolysaccharide;
PGE2, prostaglandin E2. |
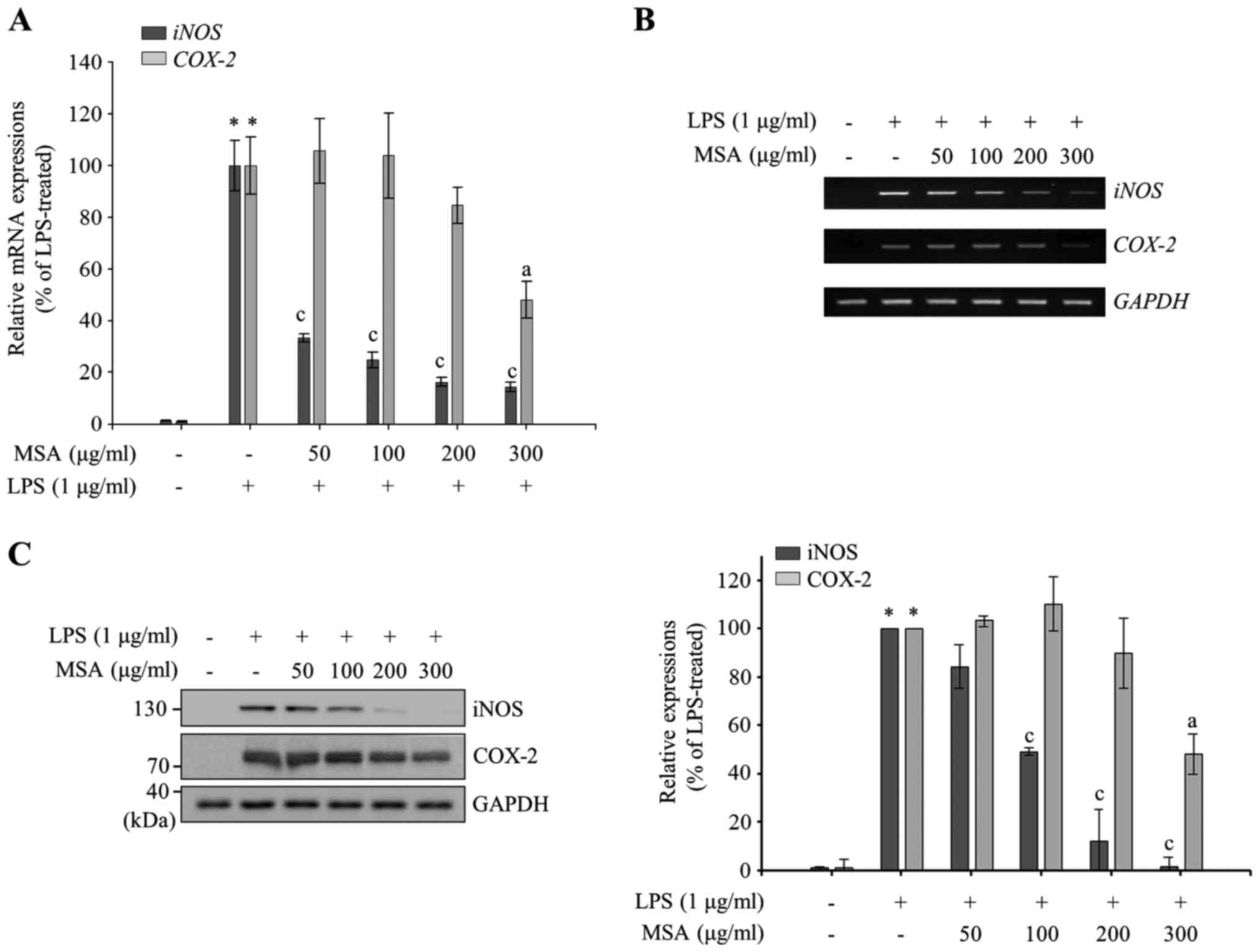
Following this, the effect of MSA was measured on
the mRNA and protein expression levels of iNOS, an NO-synthesizing
enzyme, and COX-2, a responsible enzyme for the production of
PGE2, to investigate the transcriptional regulation of
proinflammatory mediators. As presented in Fig. 2A and B, RT-qPCR and
semiquantitative RT-PCR data reveal that MSA inhibits LPS-induced
iNOS and COX-2 mRNA expression in a dose-dependent
manner. The immunoblotting analysis with iNOS and COX-2 specific
antibodies indicated an inhibitory effect of MSA on the LPS-induced
iNOS and COX-2 expression similar to PCR results (Fig. 2C), indicating that NO and
PGE2 productions are tightly regulated at the level of
transcription by MSA. It is of interest to note that iNOS
production is regulated more strongly than COX-2 by MSA.
MSA selectively inhibits production of
proinflammatory cytokines in LPS-treated RAW 264.7 macrophages
Since excessive production of inflammatory
mediators, including IL-1β, IL-6, and TNF-α as well as NO and
PGE2, in macrophages is accompanied by severe
inflammation (18–20), the effect of MSA on the production
of proinflammatory cytokines in LPS-treated RAW 264.7 macrophages
was investigated to evaluate other anti-inflammatory properties of
MSA. As demonstrated in Fig. 3B,
MSA inhibited LPS-induced production of IL-6 in a dose-dependent
manner. The mRNA expression levels of LPS-induced proinflammatory
cytokines resulting from MSA treatment were evaluated by RT-qPCR
and semiquantitative RT-PCR to investigate whether the production
of inflammatory cytokines was tightly regulated at the
transcription level. As demonstrated in Fig. 3C and D, the mRNA expressions of
IL-6 and IL-1β were decreased with MSA treatment. LPS-induced TNF-α
production (Fig. 3A) and its mRNA
expression (Fig. 3C and D) were
only slightly alleviated by MSA (P>0.05). These results
indicated that MSA inhibits the production of the proinflammatory
cytokines, including IL-1β and IL-6, whereas TNF-α production was
weakly regulated by MSA in LPS-treated macrophages.
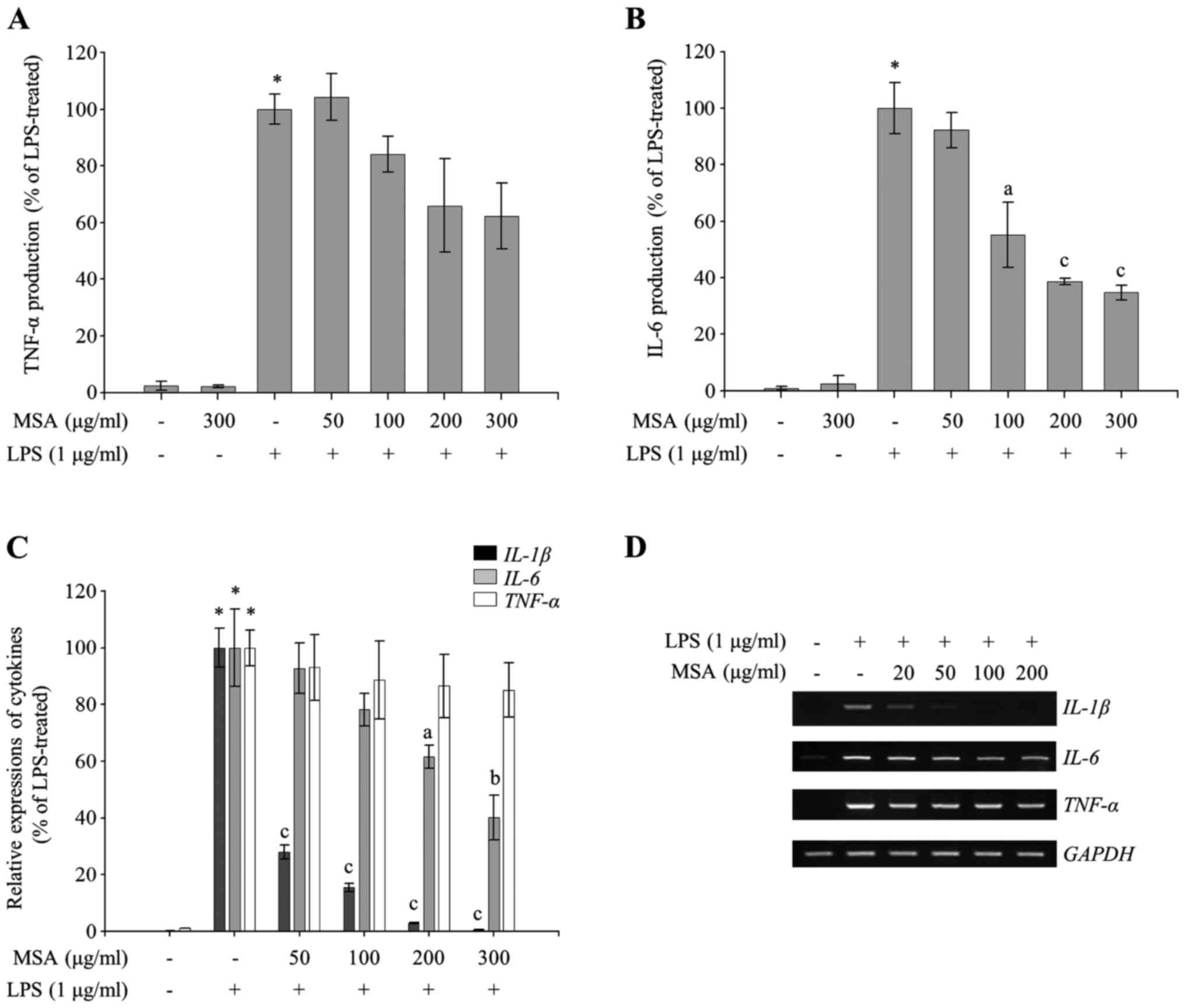 | Figure 3.Inhibitory effect of MSA on the
production of proinflammatory cytokines. RAW 264.7 macrophages were
pretreated with MSA (50, 100, 200 and 300 µg/ml) and then incubated
with LPS (1 µg/ml) for the indicated times. Following 24 h
stimulation, an ELISA was used to measure levels of (A) TNF-α and
(B) IL-6. The production of each cytokine was determined using a
standard curve. Data were represented as mean ± standard error of
the mean and analyzed using one-way analysis of variance.
*P<0.0001 vs. LPS-untreated control groups.
aP<0.01 and cP<0.0001 vs. LPS-treated
groups. (C and D) At 3 h following stimulation, total RNA was
extracted and reverse transcribed to cDNA. (C) IL-1β,
TNF-α and IL-6 were amplified and the expressions of
IL-1β, TNF-α and IL-6 in each group were
compared with those of the LPS-treated group. Data were represented
as mean ± standard error of the mean and analyzed using one-way
analysis of variance. *P<0.0001 vs. LPS-untreated control
groups. aP<0.01, bP<0.001 and
cP<0.0001 vs. LPS-treated groups. (D) IL-1β,
TNF-α and IL-6 were amplified by PCR and detected
using a gel documentation system. GAPDH was used as a loading
control. MSA, S. Acmella; LPS, lipopolysaccharide; TNF-α,
tumor necrosis factor-α; IL, interleukin. |
MSA inhibits phosphorylation of IκBα
and MAPKs in LPS-treated RAW 264.7 macrophages
The major regulatory signaling pathways for the
production of inflammatory mediators are NF-κB and MAPK. To assess
whether MSA regulates NF-κB activation, MSA-mediated IκBα
phosphorylation levels and total IκBα levels were measured in
LPS-treated RAW 264.7 cells. The p-IκBα levels were reduced by MSA
in a dose-dependent manner and IκBα levels was increased by the MSA
treatment (Fig. 4A), suggesting
that MSA inhibits IκBα phosphorylation at Ser-32/36 and thus causes
degradation of IκBα. In addition, the phosphorylation levels in the
phosphorylation loops of three MAPKs (ERK, JNK, and p38) were
measured; these are involved in another major inflammatory
signaling pathway by MSA in LPS-treated RAW 264.7 cells. This is
because the phosphorylation in the phosphorylation loops of MAPKs
leads to the activation of MAPKs. As indicated in Fig. 4B, MSA inhibited the phosphorylation
of all MAPKs without changing total MAPK levels, although its
effects on MAPKs are weaker than on IκBα.
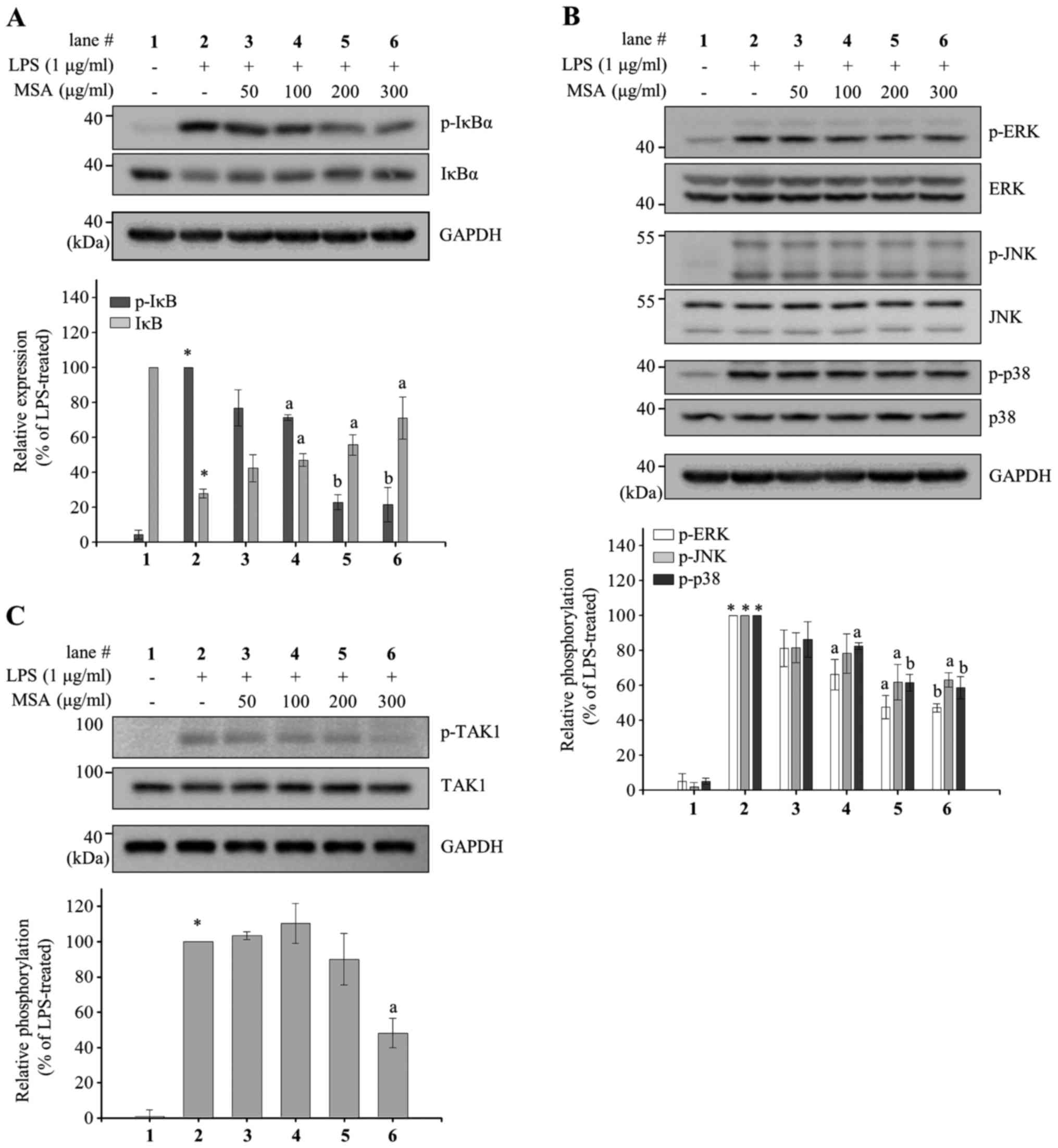 | Figure 4.Inhibitory effects of MSA on NF-κB
and MAPK. RAW 264.7 macrophages were pretreated with various
concentrations of MSA (50, 100, 200 and 300 µg/ml) for 2 h and then
incubated with LPS for 3 min (for detection of IκBα and TAK1) or 15
min (MAPKs). Total cell lysates were prepared and subjected to
immunoblot analyses. The expression levels of (A) p-IκBα, IκBα, (B)
p-JNK, JNK, p-ERK, ERK, p-p38, p38, (C) p-TAK1 and TAK1 were
detected using specific antibodies. Relative expression levels of
IκBα and p-IκBα were normalized to GAPDH levels. Levels of
phosphorylated MAPKs and TAK1 were normalized to the corresponding
MAPK and TAK1 levels. Quantitative analyses of phosphorylation and
protein levels are shown as bar graphs following normalization.
Data are represented as mean ± standard error of the mean and
analyzed using one-way analysis of variance. *P<0.0001 vs.
LPS-untreated control groups. aP<0.01 and
bP<0.001 vs. LPS-treated groups. MSA, S.
Acmella; NF-κB; nuclear factor-κB; MAPK, mitogen-activated
protein kinase; LPS, lipopolysaccharide; IκBα, inhibitor of κBα;
TAK1, transforming growth factor beta-activated kinase 1; JNK,
c-Jun N-terminal kinase; ERK, extracellular signal-regulated
kinase. |
Further investigations were conducted to elucidate
the action point of MSA in LPS-treated RAW 264.7 cells. Since MSA
inhibits both NF-κB and MAPKs, the authors investigated the
regulation of TAK1 phosphorylation level by MSA treatment in
LPS-stimulated RAW 264.7 cells. As demonstrated in Fig. 4C, LPS-induced phosphorylation of
TAK1 was inhibited by MSA treatment without changing total TAK1
protein levels. As presented in Fig.
4C, LPS-induced phosphorylation of TAK1 was inhibited by MSA
treatment without changing total TAK1 protein levels.
Discussion
The production of all of inflammatory mediators
measured in the current study was inhibited by MSA treatment
whereas TNF-α was not significant. Therefore, the authors assumed
that the major inflammatory signaling pathways, including NF-κB and
MAPKs, are involved in the MSA-mediated regulation of the
production of inflammatory mediators, although each inflammatory
mediator has different regulatory factors for those activation.
LPS-induced NF-κB activation in macrophages is primarily mediated
by IκBα phosphorylation at Ser-32/36, followed by IκBα degradation
and the translocation of released cytoplasmic p50 and p65 complex
to the nucleus (21–23). The phosphorylation levels in the
phosphorylation loops of three MAPKs (ERK, JNK and p38) that are
involved in another major inflammatory signaling pathway (24) in LPS-treated macrophages leads to
the activation of MAPKs, which results in the transcriptional
activation of activator protein-1, a transcription factor that
binds to the promoter of inflammatory mediators (25). As indicated in Fig. 4A and B, both NF-κB and MAPK
activation were tightly regulated by MSA treatment, indicating that
MSA exhibits its anti-inflammatory properties in macrophages
through inhibiting the activation of major inflammatory signaling
pathways, NF-κB and MAPKs.
LPS binding to toll-like receptor 4 leads to the
recruitment of accessory molecules including myeloid
differentiation primary response 88, interleukin-1
receptor-associated kinase 1 and TNF receptor associated factor 6,
and this complex formation induces phosphorylation and activation
of transforming growth factor β-activated kinase 1 (TAK1) (26,27).
Activated TAK1 through phosphorylation at its activation loop
induces the cascades for the activation of both NF-κB and MAPKs
(28). MSA treatment inhibited
LPS-induced phosphorylation of TAK1 (Fig. 4C). This result implies that the
inhibitory properties of MSA on NF-κB and MAPKs are due to the
suppression of TAK1 or its upstream signaling molecules.
Regulatory mechanisms of many natural extracts for
their anti-inflammatory effects are primarily focused on the
regulation of NF-κB and MAPK signaling pathways. However, the
targeted inflammatory mediators and action points of each extract
are relatively selective when compared to the inhibitory properties
of single compounds. Based on many studies for the
anti-inflammatory properties by natural extracts, the selective
inhibitory effects are due to multiple components that show
anti-inflammatory effects. A recent report has revealed that
mulberry fruit extract inhibits acute colitis by selective
inhibition of the NF-κB and ERK pathways (29). The authors reported that the
anti-inflammatory effects of mulberry fruit extract are due to its
specific compounds, including linoleic acid and ethyl linolenate.
Another study revealed that the analgesic and anti-inflammatory
effects of Litsea japonica fruit are regulated by the
inhibition of NF-κB, p38 and JNK (30). Hamabiwalactone A and
Hamabiwalactone B were demonstrated to be the major components of
the analgesic and anti-inflammatory effects. In the current study,
MSA inhibits the production of inflammatory mediators by
suppressing both NF-κB and MAPKs through the inhibition of the
upstream kinase, TAK1. Taken together, the presented results
suggested that the anti-inflammatory effects of MSA are primarily
mediated by a single compound, since the regulation was conducted
at a specific action point, TAK1.
In conclusion, MSA exhibits anti-inflammatory
properties by inhibiting the production of various inflammatory
mediators through the regulation of both NF-κB and MAPK signaling
pathways. Although more studies are required to elucidate concise
action mechanism for the anti-inflammatory effects of MSA, these
results suggested that MSA may be a valuable candidate as an
alternative medicine for the treatment of severe inflammation
states.
Acknowledgements
The present research was supported by the National
Research Foundation of Korea grant funded by the Ministry of
Science, ICT & Future Planning (grant no.
NRF-2015R1A2A2A11001446) and by a Chung-Ang University Research
Scholarship grant.
References
|
1
|
Nowarski R, Gagliani N, Huber S and
Flavell RA: Innate immune cells in inflammation and cancer. Cancer
Immunol Res. 1:77–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu LC, Fan NC, Lin MH, Chu IR, Huang SJ,
Hu CY and Han SY: Anti-inflammatory effect of spilanthol from
Spilanthes acmella on murine macrophage by down-regulating
LPS-induced inflammatory mediators. J Agric Food Chem.
56:2341–2349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jou IM, Lin CF, Tsai KJ and Wei SJ:
Macrophage-mediated inflammatory disorders. Mediators Inflamm.
2013:3164822013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cutolo M: Macrophages as effectors of the
immunoendocrinologic interactions in autoimmune rheumatic diseases.
Ann N Y Acad Sci. 876:32–42. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jayaweera DMA: Medicinal plants
(indigenous and exotic) used in Ceylon. National Science Council of
Sri Lanka Colombo: 1980
|
|
6
|
Revathi P and Parimelazhagan T:
Traditional Knowledge on Medicinal Plants Used by the Irula Tribe
of Hasanur Hills, Erode District, Tamil Nadu, India. Ethnob
Leaflets. 14:136–160. 2010.
|
|
7
|
Sathyaprasad S, Jose BK and Chandra HS:
Antimicrobial and antifungal efficacy of Spilanthes acmella
as an intracanal medicament in comparison to calcium hydroxide: An
in vitro study. Indian J Dent Res. 26:528–532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ratnasooriya WD, Pieris KP, Samaratunga U
and Jayakody JR: Diuretic activity of Spilanthes acmella
flowers in rats. J Ethnopharmacol. 91:317–320. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dubey S, Maity S, Singh M, Saraf SA and
Saha S: Phytochemistry, pharmacology and toxicology of
Spilanthes acmella: A Review. Adv Pharmacol Sci.
2013:4237502013.PubMed/NCBI
|
|
10
|
Chakraborty A, Devi BR, Sanjebam R,
Khumbong S and Thokchom IS: Preliminary studies on local anesthetic
and antipyretic activities of Spilanthes acmella Murr. In
experimental animal models. Indian J Pharmacol. 42:277–279. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Boonen J, Baert B, Burvenich C, Blondeel
P, De Saeger S and De Spiegeleer B: LC-MS profiling of
N-alkylamides in Spilanthes acmella extract and the
transmucosal behaviour of its main bio-active spilanthol. J Pharm
Biomed Anal. 53:243–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boonen J, Baert B, Roche N, Burvenich C
and De Spiegeleer B: Transdermal behaviour of the N-alkylamide
spilanthol (affinin) from Spilanthes acmella (Compositae)
extracts. J Ethnopharmacol. 127:77–84. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sharma V, Boonen J, Chauhan NS, Thakur M,
De Spiegeleer B and Dixit VK: Spilanthes acmella ethanolic
flower extract: LC-MS alkylamide profiling and its effects on
sexual behavior in male rats. Phytomedicine. 18:1161–1169. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Spelman K, Depoix D, McCray M, Mouray E
and Grellier P: The traditional medicine Spilanthes acmella,
and the alkylamides spilanthol and undeca-2E-ene-8,10-diynoic acid
isobutylamide, demonstrate in vitro and in vivo antimalarial
activity. Phytother Res. 25:1098–1101. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chakraborty A, Devi RKB, Rita S,
Sharatchandra K and Singh TI: Preliminary studies on
antiinflammatory and analgesic activities of Spilanthes
acmella in experimental animal models. Indian J Pharmacol.
36:148–150. 2004.
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cho YC, Ju A, Kim BR and Cho S:
Anti-inflammatory effects of Crataeva nurvala Buch. Ham. Are
mediated via inactivation of ERK but not NF-κB. J Ethnopharmacol.
162:140–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feghali CA and Wright TM: Cytokines in
acute and chronic inflammation. Front Biosci. 2:d12–d26. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Misko TP, Trotter JL and Cross AH:
Mediation of inflammation by encephalitogenic cells: Interferon
gamma induction of nitric oxide synthase and cyclooxygenase 2. J
Neuroimmunol. 61:195–204. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Suh N, Honda T, Finlay HJ, Barchowsky A,
Williams C, Benoit NE, Xie QW, Nathan C, Gribble GW and Sporn MB:
Novel triterpenoids suppress inducible nitric oxide synthase (iNOS)
and inducible cyclooxygenase (COX-2) in mouse macrophages. Cancer
Res. 58:717–723. 1998.PubMed/NCBI
|
|
21
|
Karin M and Delhase M: The I kappa B
kinase (IKK) and NF-kappa B: Key elements of proinflammatory
signalling. Semin Immunol. 12:85–98. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tak PP and Firestein GS: NF-kappaB: A key
role in inflammatory diseases. J Clin Invest. 107:7–11. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Verma IM, Stevenson JK, Schwarz EM, Van
Antwerp D and Miyamoto S: Rel/NF-kappa B/I kappa B family: Intimate
tales of association and dissociation. Genes Dev. 9:2723–2735.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Weinstein SL, Sanghera JS, Lemke K,
DeFranco AL and Pelech SL: Bacterial lipopolysaccharide induces
tyrosine phosphorylation and activation of mitogen-activated
protein kinases in macrophages. J Biol Chem. 267:14955–14962.
1992.PubMed/NCBI
|
|
25
|
Adcock IM: Transcription factors as
activators of gene transcription: AP-1 and NF-kappa B. Monaldi Arch
Chest Dis. 52:178–186. 1997.PubMed/NCBI
|
|
26
|
Kawai T and Akira S: The role of
pattern-recognition receptors in innate immunity: Update on
Toll-like receptors. Nat Immunol. 11:373–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kumar H, Kawai T and Akira S: Toll-like
receptors and innate immunity. Biochem Biophys Res Commun.
388:621–625. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sato S, Sanjo H, Takeda K, Ninomiya-Tsuji
J, Yamamoto M, Kawai T, Matsumoto K, Takeuchi O and Akira S:
Essential function for the kinase TAK1 in innate and adaptive
immune responses. Nat Immunol. 6:1087–1095. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qian Z, Wu Z, Huang L, Qiu H, Wang L, Li
L, Yao L, Kang K, Qu J, Wu Y, et al: Mulberry fruit prevents
LPS-induced NF-κB/pERK/MAPK signals in macrophages and suppresses
acute colitis and colorectal tumorigenesis in mice. Sci Rep.
5:173482015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koo HJ, Yoon WJ, Sohn EH, Ham YM, Jang SA,
Kwon JE, Jeong YJ, Kwak JH, Sohn E, Park SY, et al: The analgesic
and anti-inflammatory effects of Litsea japonica fruit are mediated
via suppression of NF-κB and JNK/p38 MAPK activation. Int
Immunopharmacol. 22:84–97. 2014. View Article : Google Scholar : PubMed/NCBI
|