Introduction
According to the World Health Organization, 360
million people are suffering disabling hearing loss, and
approximately one-third of people over the age of 65 are affected
(http://www.who.int/pbd/deafness/estimates/en/index.html).
Noncongenital deafness has been reported to be closely associated
with the environment (1). Noise
exposure is known to induce excessive reactive oxygen species
generation in the cochlea that damages macromolecules such as DNA.
Presbycusis is also termed age-related hearing loss and is
progressive bilateral sensorineural hearing loss that is primarily
characterized by impairments in high frequency hearing (2–4). In
China, presbycusis occurs in 30% of elders over the age of 60
(5). However, the pathogenesis of
presbycusis remains unclear. Although environmental factors have
been demonstrated to be associated, they cannot explain the
differences in the onset and rate of development between different
individuals from similar environments (2).
Epigenetics is the study of inherited changes in
phenotype or gene expression that are caused by mechanisms other
than changes in the underlying DNA sequence (6). Methylations that contribute to
epigenetics can occur through DNA methylations within a gene
particularly at CpG sites. The cytosines in CpG sites are often
methylated to form 5-methylcytosine, which can influence the
transcriptional activity of the promoter. Previous studies have
demonstrated that the methylation levels of certain genes are
associated with the risks of diseases, including type 2 diabetes,
colorectal cancer, gastric carcinogenesis and breast cancer
(7–9). Pendrin is encoded by the solute
carrier family 26 member 4 (SLC26A4) gene and is an anion
exchanger that exchanges Cl− and HCO3− to
ensure that the pH of the cochlear endolymph is higher than that of
the perilymph (10–12). Abolished activation of
SLC26A4 results the acidification and enlargement of the
membranous labyrinth and causes deafness (10,13,14).
A nationwide survey in China suggested that mutations in
SLC26A4 are among the most common elements of congenital
deafness (5). The SLC26A4
mutation frequencies among Han Chinese, Hui, and Uyghur people are
14.3, 12.8 and 1.6%, respectively, which suggests that
SLC26A4 is a mutation hotspot gene across different regions
and races (5). However, the
methylation level of SLC26A4 and the importance of its
contribution to deafness remain to be thoroughly investigated.
In the present study, the contribution of the
methylation level of the CpG site in the promoter region of
SL26A4 to presbycusis was investigated.
Materials and methods
Sample collection
A total of 102 Han Chinese presbycusis patients
(male:female, 56:46; mean age, 71) and 104 Han Chinese age-matched
controls (male:female, 56:48; mean age, 70.1) were selected from
Ningbo No. 7 Hospital (Ningbo, China). The patients were diagnosed
by experienced physicians based on pure-tone threshold averages
(PTAs). The PTA of the presbycusis cases was greater than 60 dB HL,
and for the controls, this value was <26 dB HL. All of the
included individuals were from Zhenhai, Ningbo and were free of
genetic diseases, otitis media, cancer and metabolic disease
including diabetes and hyperlipidemia. The peripheral blood samples
were collected in 3.2% citrate sodium-treated tubes and then stored
at −80°C. The present study was approved by the clinical committees
of Ningbo University (Ningbo, China) and Ningbo No. 7 Hospital, and
written informed consent was obtained from all subjects.
Biochemical analyses
The website database of the University of California
Santa Cruz (UCSC) human genome browser on Human February 2009
(GRCh37/hg19) Assembly (http://www.genome.ucsc.edu) was used to confirm the
gene position and obtain the DNA sequence of SLC26A4. The
SLC26A4 gene is located on chromosome 7 and is 57,173 bp
long. The 1-kb region upstream of the transcription start site
(TSS) of SLC26A4 was selected as promoter region for
analysis. A nucleic acid extraction analyzer (Lab-Aid 820; Zsandx
Co., Ltd., Xiamen, China) was used to extract the genomic DNA from
the peripheral blood samples in order to detect epigenetic changes
in the SLC26A4 gene. The DNA concentrations were measured
with a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). The methylation of the CpG site of
SLC26A4 was measured with pyrosequencing technology combined
with sodium bisulfate DNA conversion chemistry (EpiTech Bisulfite
kits; Qiagen China Co., Ltd., Shanghai, China) and polymerase chain
reaction (PCR) amplification (PyoMark PCR Kit; Qiagen China Co.,
Ltd.). The PCR protocol and sequencing primers were designed with
PyroMark Assay Design software that automatically selected the
appropriate CpG sites with high scores within a 70-nt fragment. The
revealed sequences were as follows: PCR primers forward,
5′-TTTTTTATGTGGTATGAGAGTAT-3′ and reverse
5′-CATCCCCTTACTAATCTCAA-3′; and sequence primer,
5′-TGTGGTAGGTTTTTAGAG-3′.
RNA isolation, cDNA synthesis and
reverse transcription-quantitative PCR (RT-qPCR)
The total RNAs were extracted from the peripheral
blood using TRIzol (Life Technologies; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. To synthesize the
cDNA, 1 µg total RNA was used according to the instructions of the
manufacturer of the GoScript™ Reverse Transcription System kit
(Promega Corporation, Madison, WI, USA). To quantify the expression
of the SLC26A4 gene, RT-qPCR was performed using
SYBR® Premix Ex Taq™ II (Perfect Real Time; Takara
Biotechnology Co., Ltd., Dalian, China) in an Mx3005P QPCR System
(Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA)
according to the manufacturer's protocol. The expression of the
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene was
used as an internal control. The primers were as follows:
SLC26A4, forward 5′-TGGTGGCTTGCAGATTGGAT-3′ and reverse
5′-AGCTGTGAGACCAGCACTTG-3′; GAPDH, forward
5′-AAGGTGAAGGTCGGAGTCAA-3′ and reverse 5′-AATGAAGGGGTCATTGATGG-3′.
The data were analyzed with the ΔCq method (15). All experiments were performed in
triplicate.
Statistical analysis
All statistical data were analyzed using SPSS
software, version 18.0 (SPSS, Inc., Chicago, IL, USA) and GraphPad
Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). Two-tailed
Student's t-tests were applied for the comparisons of the
presbycusis cases and controls.
Results
Methylation of the CpG3 site of
SLC26A4 was significantly elevated in the patients with
presbycusis
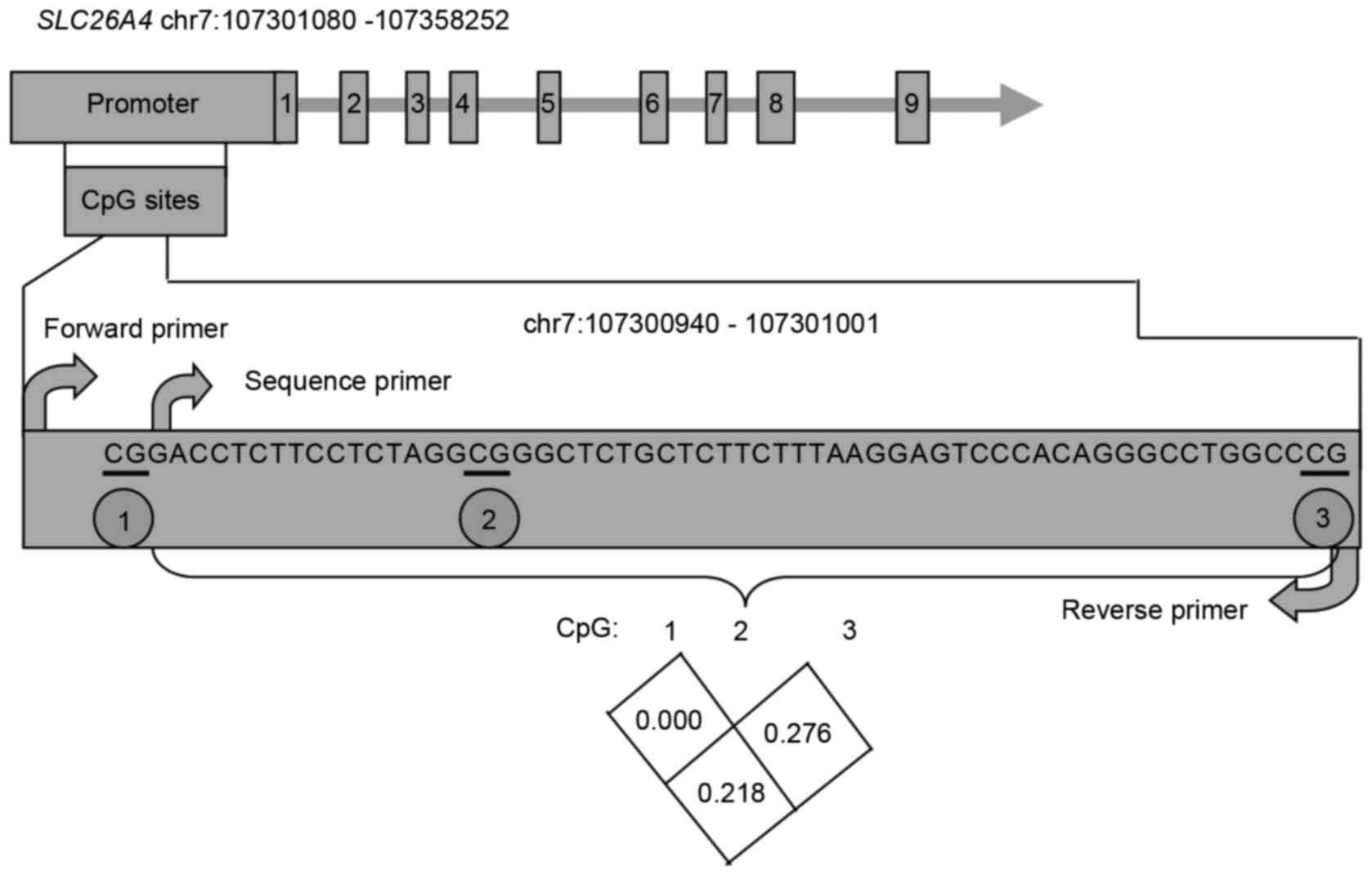
In general, the 1-kb region upstream of a
transcription start site (TSS) of a given gene is considered to be
an important promoter region, therefore, this region of
SLC26A4 was selected for analysis. Based on the PyroMark
Assay Design software, which automatically selects the appropriate
CpG sites with high scores within 70-nt fragments for primer
design, the best-scoring primers harbored three CpG sites and were
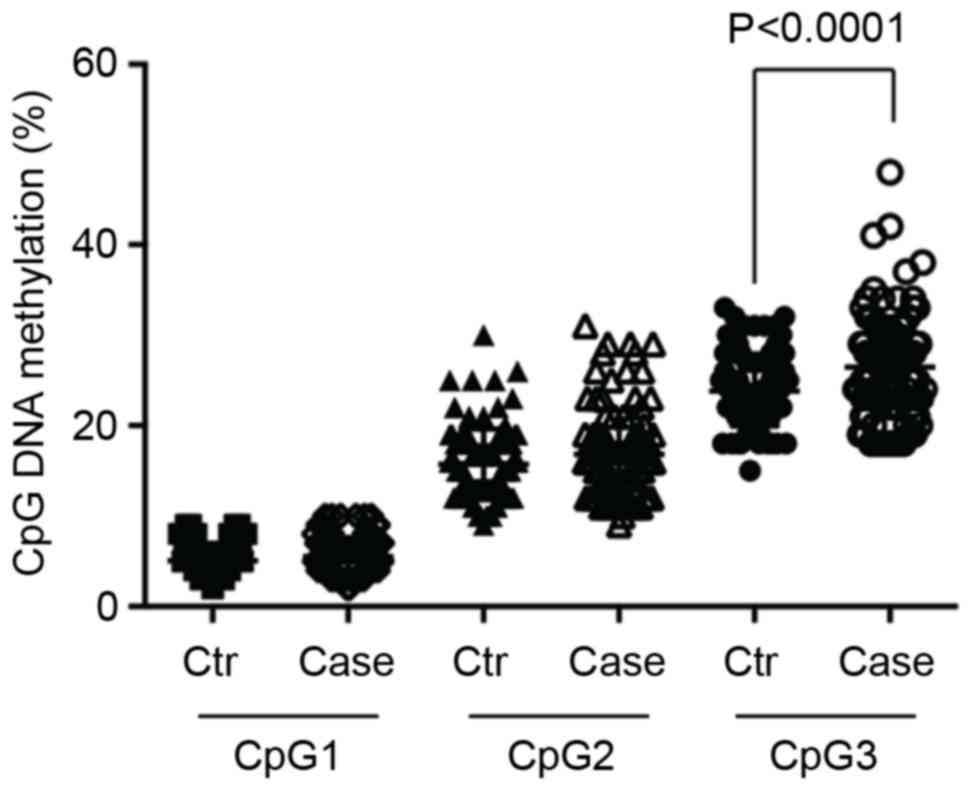
selected for methylation level evaluation (Fig. 1). As illustrated in Table I and Fig. 2, the methylation of the CpG3 site
was significantly elevated (P<0.0001) in the presbycusis cases
(26.5±5.56%) compared with the controls (23.8±3.85%). However, no
association was identified between CpG1 and CpG2 with presbycusis
(CpG1, P=0.106; and CpG2, P=0.076; Table I).
 | Table I.Comparison of the SLC26A4
methylation levels between the controls and presbycusis cases. |
Table I.
Comparison of the SLC26A4
methylation levels between the controls and presbycusis cases.
| Characteristic | Control (mean ±
SD) | Case (mean ± SD) | P-value |
|---|
| All | (n=104) | (n=102) |
| CpG1 (%) | 5.1±1.85 | 5.5±2.07 | 0.106 |
| CpG2 (%) | 15.7±3.91 | 16.8±4.97 | 0.076 |
| CpG3 (%) | 23.8±3.85 | 26.5±5.56 |
<0.0001a |
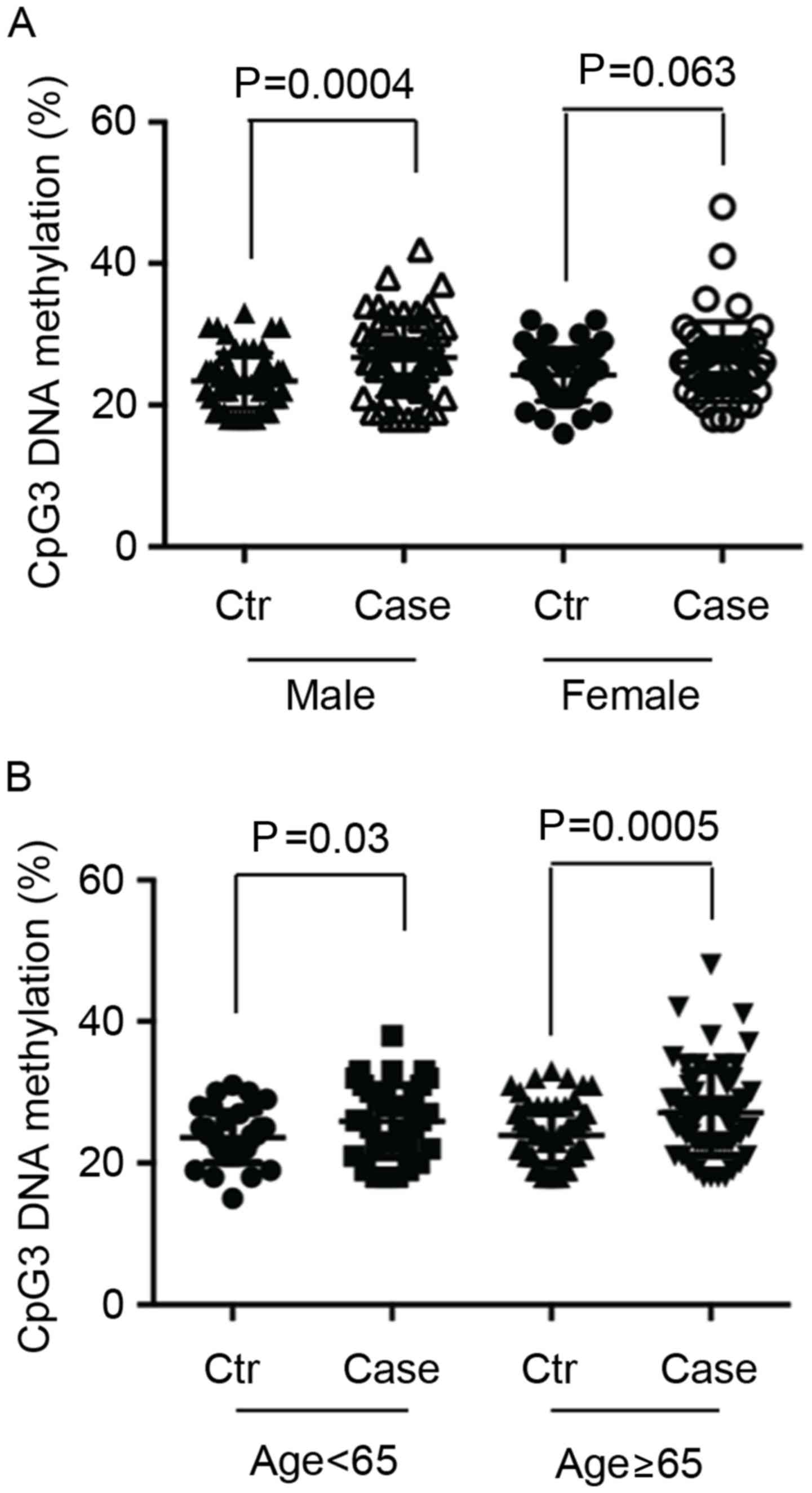
To further investigate the association between
SLC26A4 methylation and presbycusis, subgroup analyses were
conducted according gender and age (Fig. 3). Notably, a significant difference
was observed between the patients with presbycusis (28.6±5.54%) and
the controls (23.4±3.92%) only among the male participants
(P=0.0004; Fig. 3A), and no
difference was observed among the female subgroup (P=0.063;
Fig. 3A). Regarding the age
subgroups, a significant difference was observed between the
patients with presbycusis (27.1±5.92%) and controls (23.9±4.02%) in
those ≥65 years old (P=0.0005; Fig.
3B) than in those <65 years old (P=0.03; Fig. 3B).
SLC26A4 transcription was markedly
decreased in the patients with presbycusis with elevated SLC26A4
methylation levels
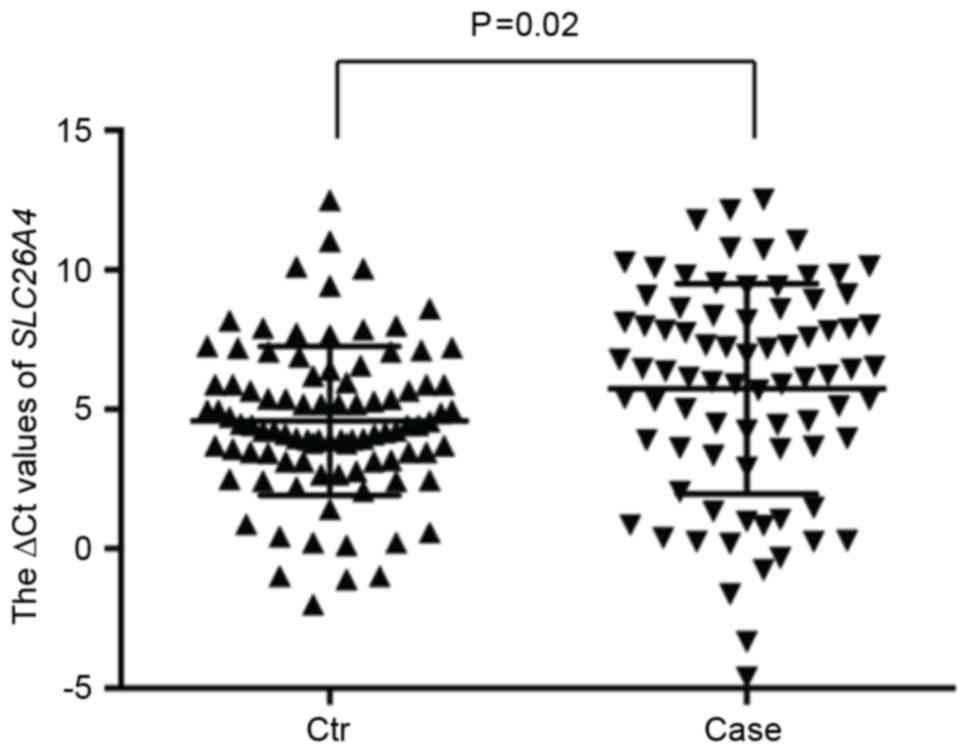
RT-qPCR analyses were performed to investigate
whether the evaluated methylation affected the transcription level
of SLC26A4 using the total RNAs extracted from the
peripheral blood samples of 93 controls and 80 patients with
presbycusis. As illustrated in Fig.
4, a significant difference in the SLC26A4 transcription
levels between the cases and controls was observed (P=0.02).
CpG3 methylation in SLC26A4 predicted
the risk of presbycusis in the males
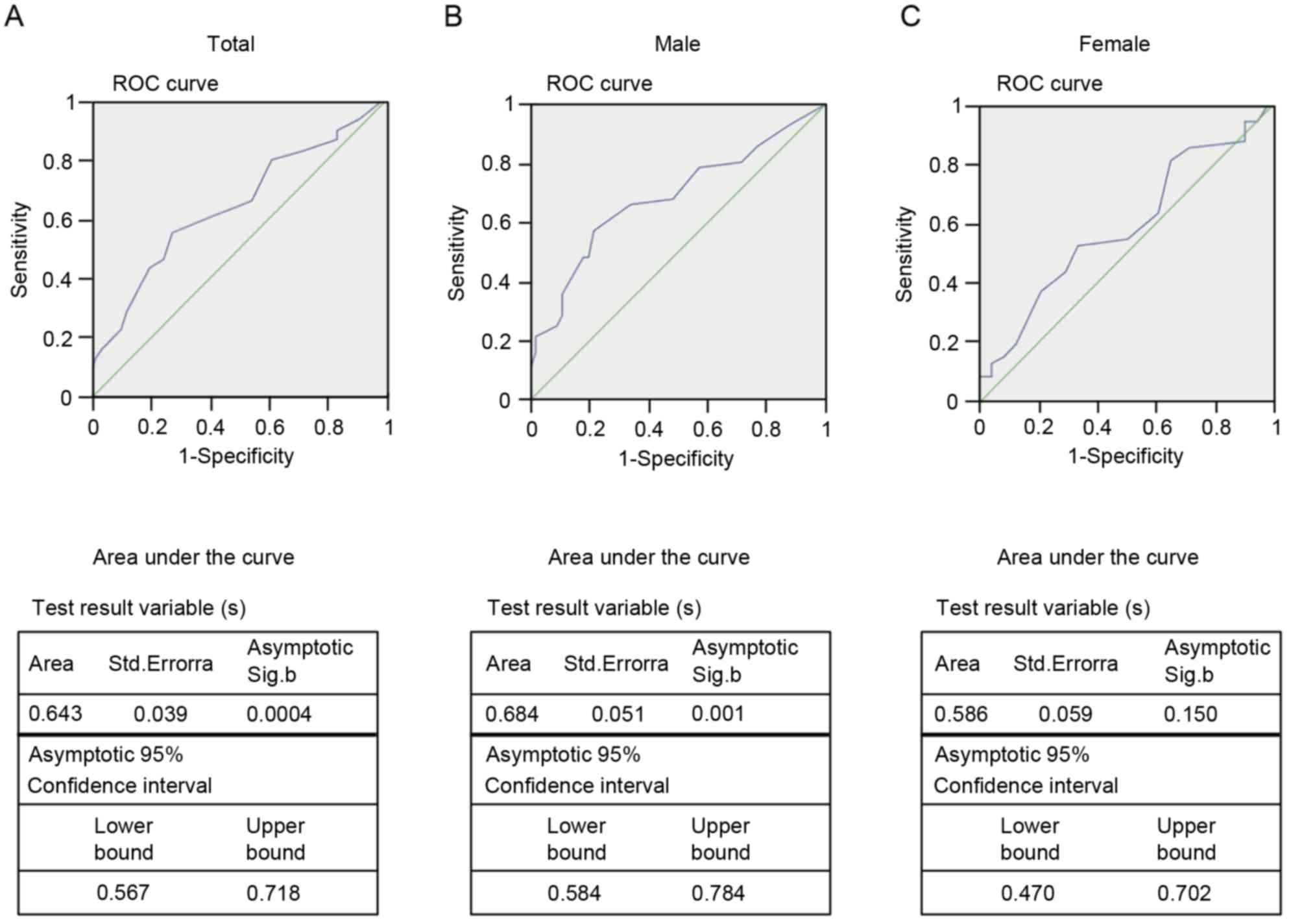
As illustrated in Fig.
5, we compared the differences between the presbycusis cases
and controls based on the cut-off value (25.5%) were obtained from
the ROC curve. The areas under the ROC curves (AUCs) reached 0.643
[95% confidence interval (CI) from 0.567–0.718; P=0.0004; Fig. 5A], and the further subgroup
analyses demonstrated a male-dependent effect of SLC26A4
methylation (AUC=0.684; 95% CI=0.584–0.784; P=0.001; Fig. 5B) and no correlation among the
female participants (AUC=0.570; 95% CI=0.470–0.702; P=0.150;
Fig. 5C). These results suggest
that the CpG3 methylation of SLC26A4 predicted the risk of
presbycusis among the male participants.
Discussion
In addition to being one of the most important
epigenetic alterations, the methylation of DNA CpG sites is closely
associated with environmental factors (6). Numerous studies have verified the
contribution of CpG site methylations to various disorders. In the
present study, 102 presbycusis cases and 104 controls were analyzed
to test the association between the DNA methylation level of the
SLC26A4 gene and the risk of presbycusis. In the presbycusis
cases, it was observed that the CpG3 methylation of SLC26A4
in the peripheral blood was significantly elevated. Additionally,
the male gender was positively correlated with the CpG3 methylation
level. Widespread CpG island methylation has been previously
identified to cause the silencing of the tumor suppressor gene in
colorectal cancer (16), and in
cases of hepatocellular carcinoma, the normal tissues exhibit lower
CpG island methylation levels than those of the tumor tissue, and
the promoters in the tumor tissues possesses higher CpG island
methylation levels (17). In
addition, the majority of cholangiocarcinomas (91%) exhibit
aberrant methylation in at least one locus, which suggests that CpG
island methylation is a common event (18). Specific mutation loci in
SLC26A4, including IVS7-2A>G (c.919-2A>G) and p.H723R
(c.2168A>G), are recognized as risk factors for non-syndromic
hearing loss in Asian populations, in particular in Chinese
populations (5). The results of
the current study indicated that CpG3 methylation of the
SLC26A4 gene was significantly increased in the male
presbycusis patients compared with the male controls. In addition,
the area under the ROC curve reached 0.684, which indicates the
importance of the CpG methylation of SLC26A4. Methylation at
this site can result in moderate to severe hearing loss in a manner
similar to mutations at specific loci (19), thus could serve as a potential
diagnostic biomarker.
Pendrin is the protein encoded by the SLC26A4
gene and is known to mediate Cl−/HCO3−
exchange in the inner ear and to serve a role in the maintenance of
the higher pH of the cochlear endolymph compared with the
perilymph. Abnormal pendrin expression could disrupt the pH balance
and lead to the acidification and enlargement of the membranous
labyrinth, which would directly affect the cochlea and cause
deafness (7–9,20).
As indicated in a previous study (21), a novel c.-103T→C mutation in the
key transcriptional regulatory element of the SLC26A4
promoter can interfere with the binding of forkhead box I1 (FOXI1),
which is a transcriptional activator of the SLC26A4 gene,
and can completely abolish FOXI1-mediated transcriptional
activation (22). However,
inactivating SLC26A4 mutations that cause profound deafness
can also be involved in the etiology of moderate to severe hearing
loss, which indicates that the severity of hearing loss cannot be
predicted by analyzing the type of mutation in all cases (19). In the current study, the
significant difference in SLC26A4 transcription between the
presbycusis cases and controls strongly suggested a correlation
between elevated CpG site methylation in SLC26A4 and
presbycusis.
However, there are several limitations to the
present study that need to be taken into account. Firstly, due to a
limitation of the PyroMark Assay Design software, which
automatically selects the appropriate CpG sites with high scores
within a 70-nt fragment for primer design, the best-scoring primers
harbored only three CpG sites that may not fully represent the
overall contribution of SLC26A4 to presbycusis. Further
analysis of the far upstream region and gene body sequence may be
necessary to reduce the deviation. In addition, the current study
involved only a moderate number of samples. Although positive
results were detected, future studies with larger pools of
individuals are required to provide a more reliable conclusion,
particularly in terms of those with high levels of CpG site
methylation. Due to the fact that ear tissues were not obtained,
the SLC26A4 DNA methylation and gene transcription levels
were tested only in the peripheral blood. Additional comprehensive
studies are required to test the concordances of SLC26A4
methylation and gene transcription levels between ear tissues and
peripheral blood.
In conclusion, the present study indicated that the
evaluation of SLC26A4 CpG site methylation reflected an
increased risk of presbycusis among the male participants. This
association may aid in the clarification of the molecular
mechanisms that underlie the pathogenesis of presbycusis, and
provide a potential clinical diagnostic marker.
Acknowledgements
The present study was supported in part by the
Ningbo Social Development Research Project (grant no. 2014C50066),
the Natural Science Foundation of Ningbo (grant nos. 2013A610207
and 2012A610217), and the K.C. Wong Magna Fund of Ningbo
University.
References
|
1
|
Kidd Iii AR and Bao J: Recent advances in
the study of age-related hearing loss: A mini-review. Gerontology.
58:490–496. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dai P, Yuan Y, Huang D, Zhu X, Yu F, Kang
D, Yuan H, Wu B, Han D and Wong LJ: Molecular etiology of hearing
impairment in Inner Mongolia: Mutations in SLC26A4 gene and
relevant phenotype analysis. J Transl Med. 6:742008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fransen E, Lemkens N, Van Laer L and Van
Camp G: Age-related hearing impairment (ARHI): Environmental risk
factors and genetic prospects. Exp Gerontol. 38:353–359. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohlemiller KK: Mechanisms and genes in
human strial presbycusis from animal models. Brain Res. 1277:70–83.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Du W, Wang Q, Zhu Y, Wang Y and Guo Y:
Associations between GJB2, mitochondrial 12S rRNA, SLC26A4
mutations, and hearing loss among three ethnicities. Biomed Res
Int. 2014:7468382014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamasoba T, Lin FR, Someya S, Kashio A,
Sakamoto T and Kondo K: Current concepts in age-related hearing
loss: Epidemiology and mechanistic pathways. Hear Res. 303:30–38.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lo Nigro C, Monteverde M, Lee S, Lattanzio
L, Vivenza D, Comino A, Syed N, McHugh A, Wang H, Proby C, et al:
NT5E CpG island methylation is a favourable breast cancer
biomarker. Br J Cancer. 107:75–83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang L, Ye H, Hong Q, Wang L, Wang Q, Wang
H, Xu L, Bu S, Zhang L, Cheng J, et al: Elevated CpG island
methylation of GCK gene predicts the risk of type 2 diabetes in
Chinese males. Gene. 547:329–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Joo MK, Kim KH, Park JJ, Yoo HS, Choe J,
Kim HJ, Lee BJ, Kim JS and Bak YT: CpG island promoter
hypermethylation of Ras association domain family 1A gene
contributes to gastric carcinogenesis. Mol Med Rep. 11:3039–3046.
2015.PubMed/NCBI
|
|
10
|
Yoshino T, Sato E, Nakashima T, Teranishi
M, Yamamoto H, Otake H and Mizuno T: Distribution of pendrin in the
organ of Corti of mice observed by electron immunomicroscopy. Eur
Arch Otorhinolaryngol. 263:699–704. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ledford H: Language: Disputed definitions.
Nature. 455:1023–1028. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo HJ, Yang T and Wu H: Genetic research
of age-related hearing impairment. Zhonghua Er Bi Yan Hou Tou Jing
Wai Ke Za Zhi. 48:78–81. 2013.(In Chinese). PubMed/NCBI
|
|
13
|
Wangemann P, Nakaya K, Wu T, Maganti RJ,
Itza EM, Sanneman JD, Harbidge DG, Billings S and Marcus DC: Loss
of cochlear HCO3- secretion causes deafness via endolymphatic
acidification and inhibition of Ca2+ reabsorption in a
Pendred syndrome mouse model. Am J Physiol Renal Physiol.
292:F1345–F1353. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim HM and Wangemann P: Epithelial cell
stretching and luminal acidification lead to a retarded development
of stria vascularis and deafness in mice lacking pendrin. PLoS One.
6:e179492011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song H, Sun W, Ye G, Ding X, Liu Z, Zhang
S, Xia T, Xiao B, Xi Y and Guo J: Long non-coding RNA expression
profile in human gastric cancer and its clinical significances. J
Transl Med. 11:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tanaka N, Huttenhower C, Nosho K, Baba Y,
Shima K, Quackenbush J, Haigis KM, Giovannucci E, Fuchs CS and
Ogino S: Novel application of structural equation modeling to
correlation structure analysis of CpG island methylation in
colorectal cancer. Am J Pathol. 177:2731–2740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song MA, Tiirikainen M, Kwee S, Okimoto G,
Yu H and Wong LL: Elucidating the landscape of aberrant DNA
methylation in hepatocellular carcinoma. PLoS One. 8:e557612013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sriraksa R, Zeller C, El-Bahrawy MA, Dai
W, Daduang J, Jearanaikoon P, Chau-In S, Brown R and Limpaiboon T:
CpG-island methylation study of liver fluke-related
cholangiocarcinoma. Br J Cancer. 104:1313–1318. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Khan MR, Bashir R and Naz S: SLC26A4
mutations in patients with moderate to severe hearing loss. Biochem
Genet. 51:514–523. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Uchida Y, Sugiura S, Ando F, Nakashima T
and Shimokata H: Hearing impairment risk and interaction of folate
metabolism related gene polymorphisms in an aging study. BMC Med
Genet. 12:352011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang T, Vidarsson H, Rodrigo-Blomqvist S,
Rosengren SS, Enerback S and Smith RJ: Transcriptional control of
SLC26A4 is involved in Pendred syndrome and nonsyndromic
enlargement of vestibular aqueduct (DFNB4). Am J Hum Genet.
80:1055–1063. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rozenfeld J, Efrati E, Adler L, Tal O,
Carrithers SL, Alper SL and Zelikovic I: Transcriptional regulation
of the pendrin gene. Cell Physiol Biochem. 28:385–396. 2011.
View Article : Google Scholar : PubMed/NCBI
|