Introduction
Ultrasound-targeted microbubble destruction (UTMD)
technology is as a novel method of non-viral vector gene
transfection, that is increasingly favored by researchers. UTMD
technology uses ultrasound microbubbles as nuclei to exert a
cavitation effect, which increases the permeability of cell
membranes during ultrasound exposure. The exogenous DNA can then
enter and transfect a cell through the cell membrane (1). New methods are constantly explored in
biomedical research in order to improve targeted gene transfection
efficiency at levels comparable to viral vectors (1–3);
these methods include the biotin-avidin system (4), the antigen-antibody targeting system
(5,6), and the novel ultrasound microbubbles
system (7,8). At present, these methods have limited
success penetrating the barrier of cell membranes. In addition, for
successful gene delivery, the nuclear membrane is a second barrier
in eukaryotic cells as all materials must go through nuclear pore
complexes to enter the nucleus. Micromolecules with molecular
weight <60 kDa and diameter <9 nm pass through nuclear pore
complexes into the nucleus by passive diffusion, but macromolecule
materials, such as plasmid DNA (pDNA), must contain nuclear
localization signal (NLS) peptides, which mediate the active
transport into the nucleus (9,10).
NLS peptides are composed of functional amino acid
sequences and they are frequently used in the field of gene
transfer research, as a method to enhance nuclear entry (11). Classic NLS (cNLS) peptides are rich
in basic amino acids, such as arginine and lysine, and are a
necessary signal for exogenous DNA or protein to enter into the
nucleus. The nuclear transfer functions of the various NLS peptides
are closely associated with their structure and sequence. The
correct NLS sequence and structure is recognized by nuclear import
proteins and is transported through the nuclear pore complex into
the nucleus in the form of a cargo-import protein complex. cNLSs
should be at the C-terminus of the cargo protein; if the position
of the NLS is wrong, or if the sequence is reversed, the sequence
is not recognized by the nuclear import protein, and thus the NLS
loses its function (12).
Furthermore, when a basic amino acid mutation is introduced in the
cNLS sequence (e.g. 126PKKKRKV132), so that
the lysine (K) at position 128 is mutated to threonine (T), the
resulting mutated NLS (mNLS) peptide loses its nucleus transport
function (13,14). Finally, exogenous lectin wheat germ
agglutinin (WGA) has been reported to act as a nuclear blocker,
sealing the nuclear pore and preventing the NLS peptides from
interacting with the nuclear transport receptor (15,16).
Thus, in the presence of WGA, a targeted protein or gene is not
able to enter into the nucleus, even if it contains a functional
cNLS (15,16).
The present study aimed to investigate novel methods
to improve UMTD-mediated gene transfection. Based on the property
of NLS peptides to enhance nuclear entry, the hypothesis that cNLS
may act as a mediator to increase pDNA entering the nucleus during
UMTD was examined. The present findings suggested that cNLS may be
a potential strategy to improve the transfection efficiency of the
novel UTMD gene transfection technology.
Materials and methods
Plasmid labeling
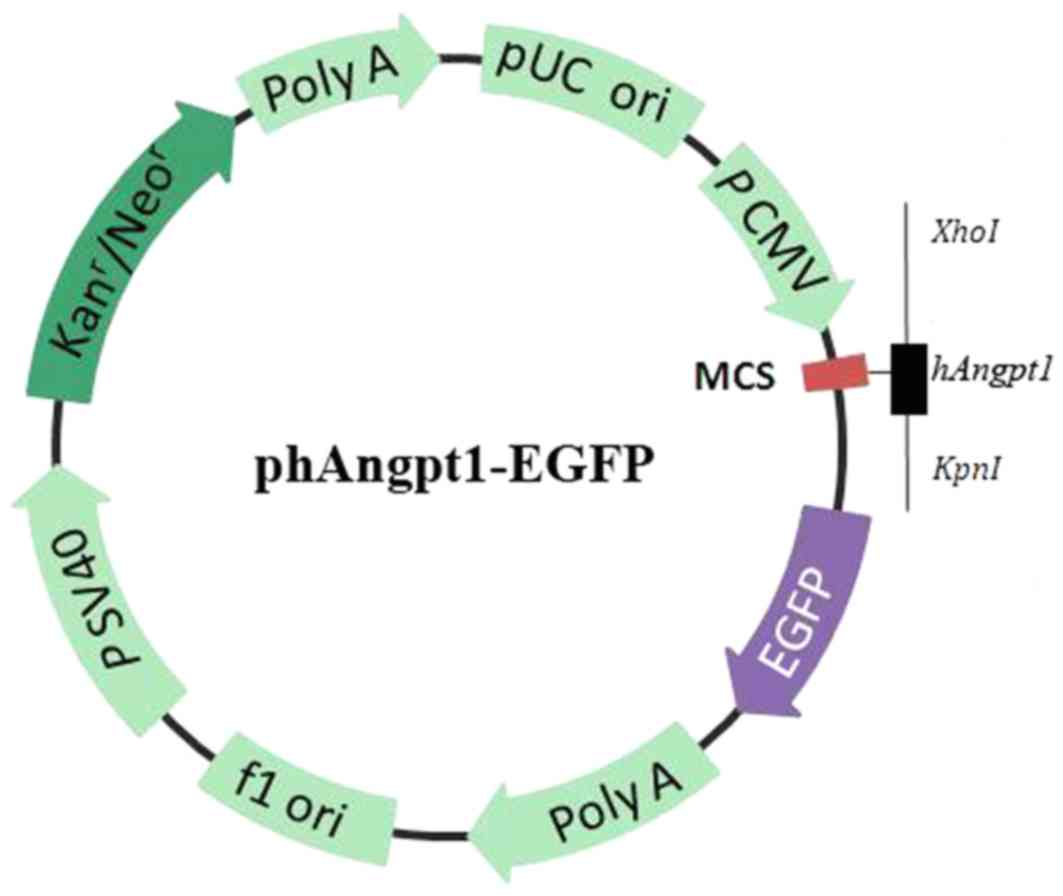
The plasmid hANGPT1-EGFP (cat. no. GOSE63322;
element sequence, CMV-MCS-EGFP-SV40-Neomycin) was constructed by
GeneChem Technology Co., Ltd. (Shanghai, China), amplified in
Escherichia coli, and then purified with the Wizard™
Maxiprep DNA Purification System (Promega Corporation, Madison, WI,
USA). In hANGPT1-EGFP, the angiopoietin 1 (ANGPT1) coding region
was cloned in the vector GV230, under the control of the human
cytomegalovirus promoter/enhancer and followed by the enhanced
green fluorescent protein (EGFP) protein, for visualization of
translation efficiency (Fig. 1).
In addition, pDNA was labeled with Cy3, using a Label IT Tracker
Intracellular Nucleic Acid Localization kit (Mirus Bio, LLC,
Madison, WI, USA), according to the manufacturer's protocol. The
reagent to plasmid weight ratio was 1:2, and the labeled Cy3-pDNA
was purified by ethanol precipitation and resuspended to a
concentration of 1 µg/µl.
Combination of NLS and plasmid
hANGPT1-EGFP
The SV40 T antigen NLS peptide (PKKKRKV; molecular
weight, 883 Da; cat. no. PO15071402) and mutated NLS peptide
(PKTKRKV; molecular weight, 856 Da; cat. no. PO16022402) were
synthesized and high-performance liquid chromatography
(HPLC)-purified by Multiple Peptide Systems (San Diego, CA, USA).
NLS peptides were non-covalently bound to supercoiled phANGPT1-EGFP
plasmid by incubation in 4X phosphate buffer saline (PBS; 547 mM
NaCl, 10.7 mM KCl, 40.6 mM Na2HPO4, 7.1 mM
KH2PO4, pH 7.1) for 30 min at room
temperature. Different molar ratios were tested as follows: 0:1,
5×102:1, 103:1, 5×103:1,
104:1, and 5×104:1 (NLS:phANGPT1-EGFP) The
efficiency of binding was tested by agarose gel electrophoresis
(Geliance 200; Perkin Elmer, Inc., Waltham, MA, USA), and the
lowest molar ratio that resulted in a bound complex was determined
as the optimal binding condition.
The NLS peptide was labeled with fluorescein
isothiocyanate (FITC; Wuhan Boster Biological Technology, Ltd.,
Wuhan, China). A total of 1 M NLS, 40 mM FITC and 2 M DIEA (Suihou
Hongqi Chemical Co., Ltd., Suizhou, Hubei, China) were mixed in the
reactor and then placed on a shaking bed at 30°C for 2 h. pDNA was
labeled with Cy3 as above. The bound complex was observed by
overlapping the two types of fluorescence under an inverted
fluorescence microscope (IX51; Olympus Corporation, Tokyo,
Japan).
Cell culture
Human umbilical vein endothelial cells (HUVECs) were
supplied by the medical center of Renmin Hospital, Wuhan University
(Wuhan, China). HUVECs were cultured in endothelial cell medium
(ECM; ScienCell Research Laboratories, Inc., Carlsbad, CA, USA).
The cells were maintained in 10 cm culture dishes at 37°C in a
humidified and 5% CO2 atmosphere. For transfection
experiments, cells were trypsinized and seeded at a density of
1×106 cells/well in two 6-well plates 24 h prior to
transfection. One of the 6-well plates was transfected using UTMD
with FITC-NLS and Cy3-labeled pDNA, to observe the subcellular
localization following 6 h of transfection, using a fluorescent
microscope and laser confocal microscope (FV1200; Olympus
Corporation), respectively. The other 6-well plate was transfected
with NLS and pDNA, for evaluation of transfection efficiency and
cell survival.
Microbubble preparation
The contrast agent Sulphur hexafluoride
microbubbles, named SonoVue (Bracco Imaging SPA, Milan, Italy), was
prepared as per the manufacturer's instructions. Lyophilized powder
was reconstituted with 5 ml 0.9% sodium chloride w/v solution
following vigorous oscillation, then a suspension containing
~2×108 microbubbles/ml was obtained. The microbubbles
were filled with sulfur hexafluoride gas and encapsulated by a thin
and flexible monolayer of phospholipids. The diameters of the
majority of microbubbles were 2–5 µm.
UTMD-mediated plasmid
transfection
The samples in the present study were divided into 4
groups as follows: Control group, UTMD + pDNA; cNLS group, UTMD +
pDNA + cNLS; mutated NLS (mNLS) group, UTMD + pDNA + mNLS; nuclear
import blocker WGA group, UTMD + pDNA +cNLS + WGA. Gene
transfection was performed by UTMD when the cells in the 6-well
plates were 70–90% confluent. The culture medium was replaced with
4 ml OptiMEM® medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 400 µl microbubbles per well and
60 µg pDNA, with or without NLS. Subsequently, ultrasound
irradiation was performed on the cells and the plasmid-microbubble
suspension, using the UGT2007 ultrasound irradiation system
(Ultrasonic Research Institute of Chongqing Medical University,
Chongqing, China). The parameters of ultrasonic irradiation were
selected based on optimization experiments reported in a previous
study from our group (17), as
follows: Probe was placed upper the cells, and irradiation was
performed with intensity of 1.5 W/cm2, frequency of 1
MHz, irradiation time of 30 sec, duty cycle of 30%, microbubble
concentration of 1×107/ml and plasmid concentration of
15 µg/ml. Each experiment was repeated three times. The cell
culture medium was replaced with ECM medium at 24 h
post-transfection.
Cellular and nuclear uptake of
pDNA
HUVECs were transfected with Cy3-labeled pDNA and
then incubated at 37°C and 5% CO2 atmosphere for 6 h.
HUVECs were then washed with PBS and harvested with trypsin
(Beyotime Institute of Biotechnology, Nantong, China). The number
of fluorescent cells was detected by flow cytometry, and the
percent of Cy3-positive cells over total represented the cellular
uptake efficiency of pDNA.
In order to calculate the nuclear uptake of pDNA,
the cells were seeded on sterile coverslips placed on the bottom of
a 6-well plate and transfected as described above. Cells were then
fixed with 4% paraformaldehyde (Beyotime Institute of
Biotechnology) for 10 min at room temperature. The membrane was
permeabilized by 0.2% Triton X-100 diluted in PBS for 8–10 min at
room temperature. Finally, DAPI was added to the cells in the dark
as a nuclear counterstain, and the cells were washed with PBS three
times for 3–5 min prior to observation. The subcellular
localization of the Cy3-labeled pDNA was observed under a laser
confocal microscope. The fluorescence intensity (FL) of the whole
cell (FLcell) and the nucleus (FLnucleus) was
quantified using Image J, version 1.48 software (http://rsb.info.nih.gov/ij; National Institutes of
Health, Bethesda, MD, USA). The percentage of
FLnucleus/FLcell represented the nuclear
uptake efficiency of the pDNA.
Cell viability
Cell viability was measured using the Cell Counting
Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan), as per the manufacturer's instructions. Briefly,
at 48 h post-transfection, HUVECs were seeded in a 96-well plate at
~5×103 cells/well in 90 µl culture medium. Untransfected
HUVECs were used as a control, and equal volume of ECM medium
without any cells was used as a blank measurement. Following
incubation at 37°C and 5% CO2 atmosphere for 24 h, 10 µl
CCK-8 was added to each well for 4 h, and then absorbance was
measured at 450 nm using an automatic microplate reader (Lambda
1050; Perkin Elmer, Inc., Houston, Texas, USA). Six duplicate
experiments were performed for each sample. Cell viability was
calculated using the optical density (OD) as follows: cell
viability (%)=(ODsample
-ODblank/ODcontrol
-ODblank)x100.
Transfection efficiency
At 48 h post-transfection, up to 500 µl of cell
suspension from each group was analyzed by flow cytometry
(FACSCalibur; BD Biosciences, Franklin Lakes, NJ, USA). CELLQuest
software version 3.0 was used for analysis (BD Biosciences). HUVEC
transfection efficiency was calculated as follows: Transfection
efficiency=the ratio of fluorescent-positive cells per total × cell
viability.
Reverse transcription-polymerase chain
reaction (RT-PCR)
At 48 h post-transfection, total RNA was extracted
using the TRIzol reagent (Aidlab Biotechnologies Co., Ltd, Beijing,
China), cDNA was obtained by reverse transcription and the mRNA
expression of the hANGPT1 gene was semi-quantitatively analyzed
using the Applied Biosystems 7500FAST RT-PCR system and SYBR Green
(Thermo Fisher Scientific, Inc.). The primers were as follows:
hANGPT1, forward 5′-CGTGGAACCGGATTTCTCTT-3′ and reverse
5′-GTACTGCCTCTGACTGGTAATG-3′; and GAPDH, forward
5′-CAAGGTCATCCATGACAACTTTG-3′ and reverse
5′-GTCCACCACCCTGTTGCTGTAG-3′. The reaction conditions were 50°C for
2 min, 95°C for 10 min and then 40 cycles of 95°C for 30 sec and
60°C for 30 sec. PCR products were electrophoresed on a 1% agarose
gel and stained with ethidium bromide. GAPDH was used to normalize
cDNA from different samples. The relative expression of hANGPT1 to
GAPDH was semi-quantified by a gel imaging analysis system
(Geliance 200; Perkin Elmer, Inc.).
Western blot
At 48 h post-transfection, analysis of the hANGPT1
protein expression was performed by western blotting. Cells
(1×106/sample) were collected and
radioimmunoprecipitation (RIPA) buffer (Beyotime Institute of
Biotechnology) was used as the lysis buffer. A total of 200 µl RIPA
was added per well for 3–5 min at room temperature and then placed
on ice for 30 min. Samples were repeatedly agitated to ensure full
cell pyrolysis. Samples were then centrifuged at 4°C and 12,900 × g
for 5 min. The supernatant was collected as the total protein
solution. A bicinchoninic protein concentration determination kit
(Beyotime Institute of Biotechnology) was used to quantify the
concentration of protein according to the manufacturer's
instructions. Total protein (50 µg) was loaded into a 10% sodium
dodecyl sulfate-polyacrylamide gel to separate the protein. The
separated proteins were transferred to polyvinylidene fluoride
membranes (EMD Millipore, Billerica, MA, USA) and blocked with 5%
non-fat dry milk (Aspen Bio, Inc., Wuhan, Hubei, China) for 1 h at
room temperature. The membranes were washed with TBST (TBS with
0.1% Tween-20) then incubated with anti-hANGPT1 antibody (1:1,000;
cat. no. ab133425; Abcam, Cambridge, MA, USA) at 4°C overnight.
Following incubation with horseradish peroxidase-coupled secondary
antibodies (1:10,000; cat. no. BA1054; Boster Biotechnology, Wuhan,
China) for 1 h at room temperature. The membranes were washed in
TBST buffer containing 0.1% Tween-20 (Aspen Bio, Inc.) and protein
expression was detected using a electrochemiluminescence kit (cat.
no. NCI5079; Thermo Fisher Scientific, Inc.). The X-ray film was
scanned and the AlphaEaseFC software (version 4.0.0; Alpha Innotech
Corporation; ProteinSimple; Bio-Techne, Minneapolis, MN, USA) was
used to analyze the optical density. The relative expression of
hANGPT1 to GAPDH was measured.
Statistical analysis
Quantitative data are presented as mean ± standard
error, and statistical analysis was performed using SPSS 19.0
software package (IBM SPSS, Armonk, NY, USA). The parameters of the
four groups were compared using a completely randomized design
analysis of variance. Results with significant differences were
further analyzed by post hoc tests to compare the difference
between two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Combination of NLS and plasmid
hANGPT1-EGFP
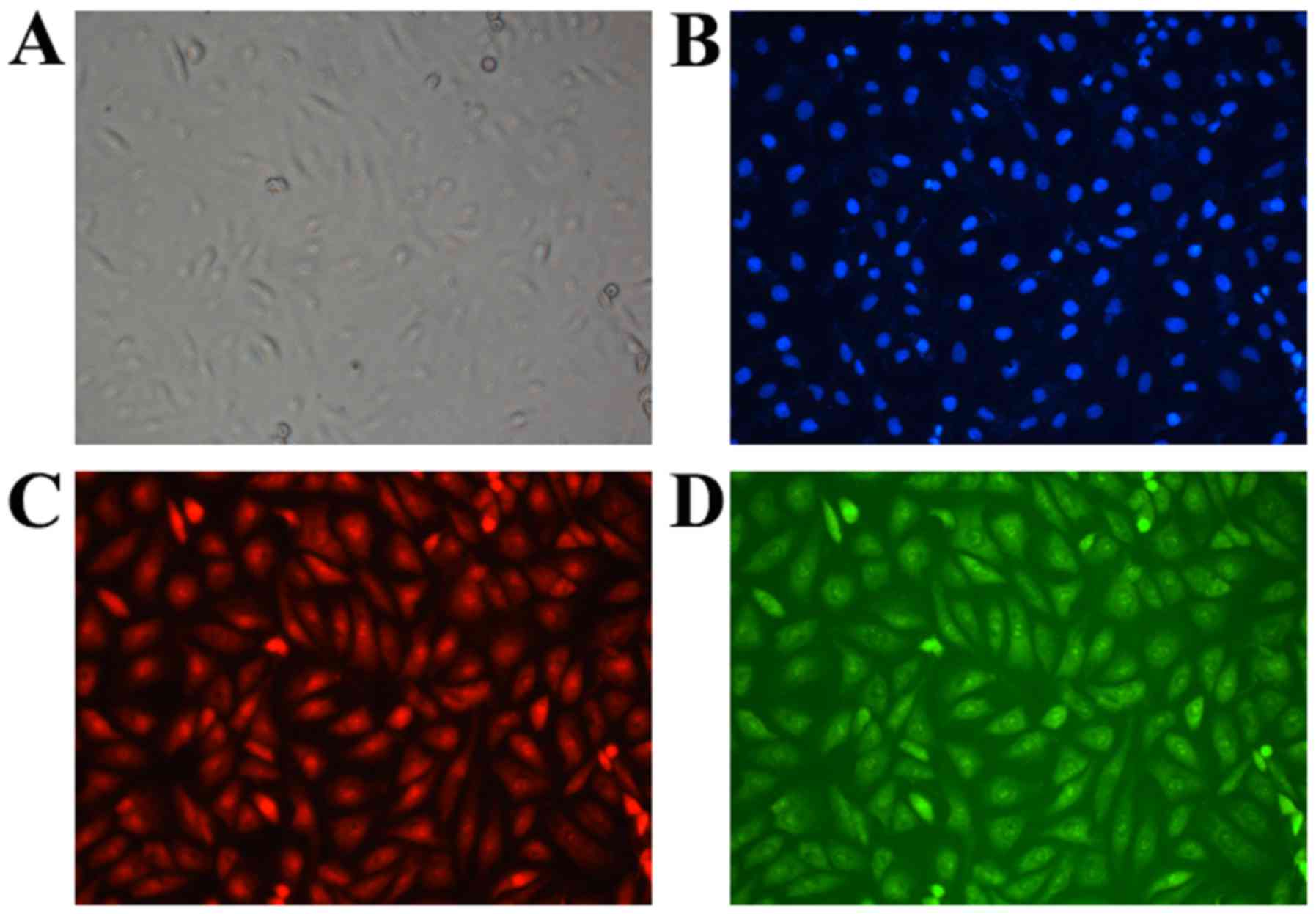
At 6 h post-transfection, green fluorescence-labeled
NLS and red fluorescence-labeled pDNA were detected at the same
locations and with similar signal intensity within the cell, and
this finding suggested a potential combination between them
(Fig. 2). The molecular weight of
a potential NLS and plasmid complex should be increased compared
with the plasmid alone. In addition, the NLS is positively charged
while DNA is negatively charged, therefore their interaction should
partially neutralize electrostatic charge. Both these factors, i.e.
neutralized electrostatic charge and increase in molecular weight,
lead to slower band migration in gel electrophoresis, until
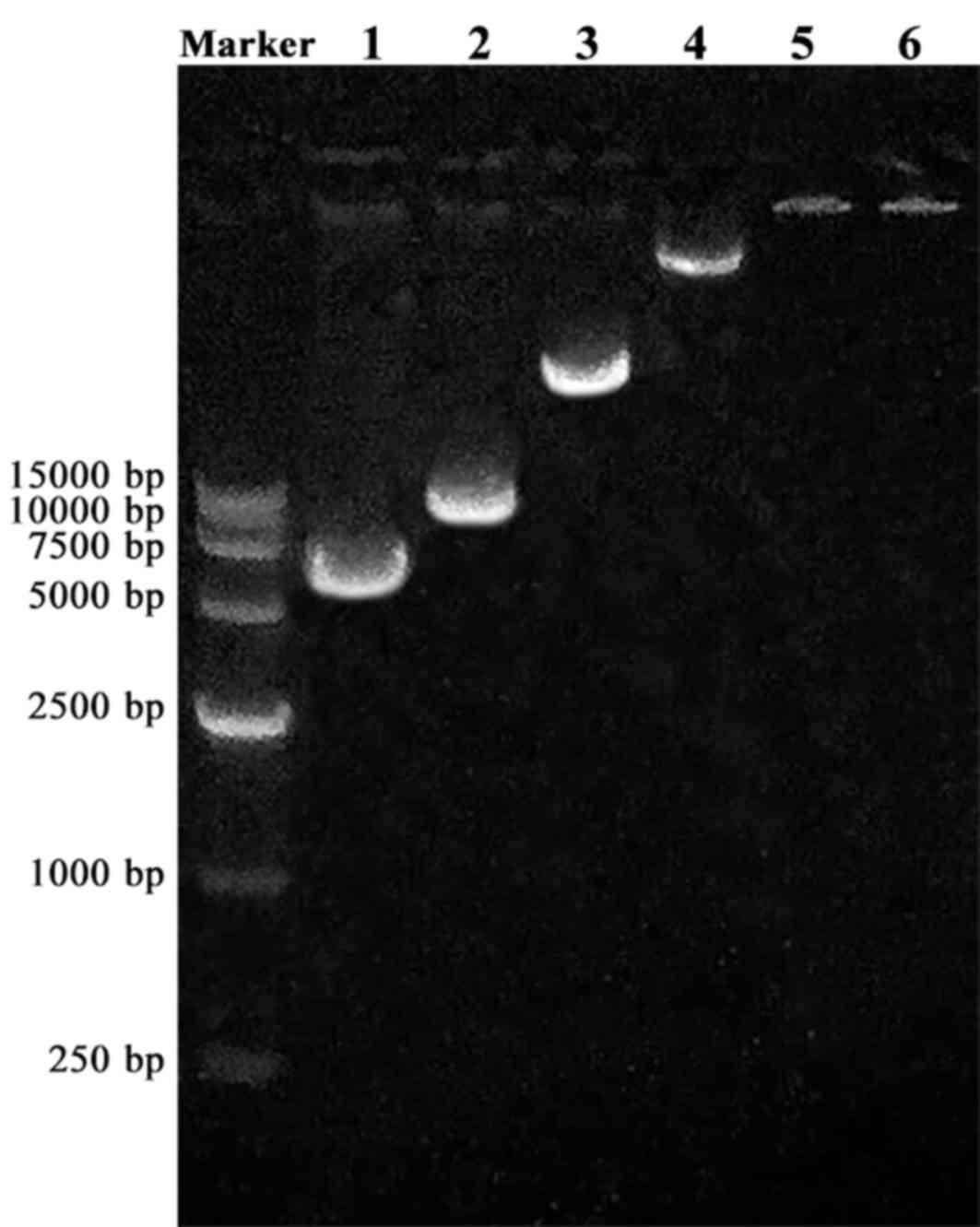
complete blocking is achieved. Agarose gel electrophoresis analysis
demonstrated that the optimum molar ratio of NLS/plasmid
combination was 104:1 (Fig.
3). When the molar ratio was 104:1 or greater, the
NLS and plasmid complex was completely immobilized in the sample
well (Fig. 3).
Distribution of FITC-NLS in cells
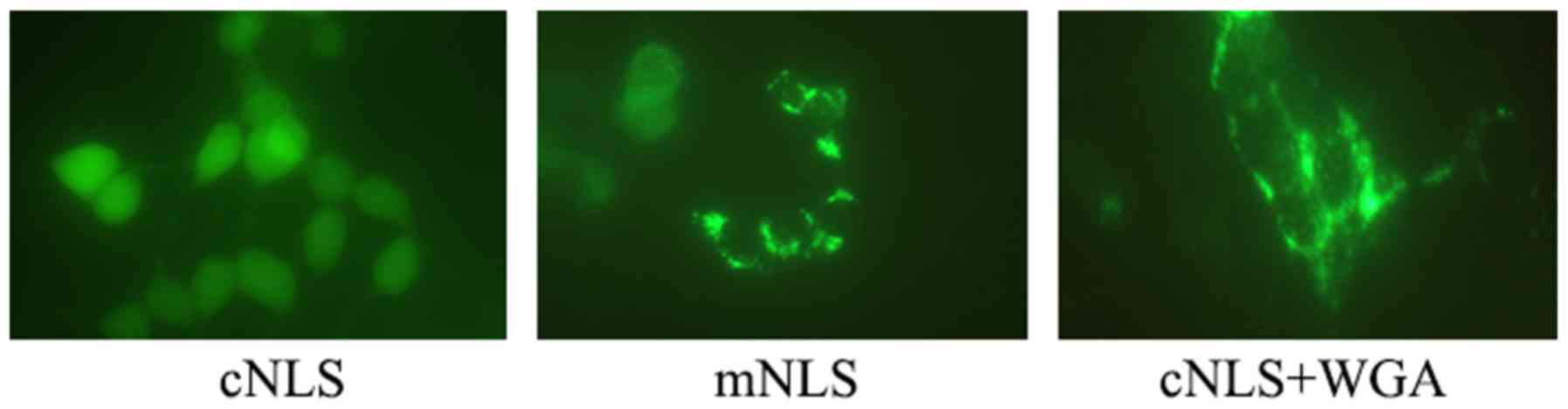
At 6 h post-transfection, the distribution of green
fluorescence FITC-labeled NLS in the cells was observed in the
cNLS, mNLS and WGA groups by fluorescence microscopy. Most green
fluorescence signal was located in the nuclei in the cNLS group
(Fig. 4). Green fluorescence
signal was located in both the cytoplasm and the nuclei in the mNLS
group, while it was almost exclusively in the cytoplasm in the WGA
group (Fig. 4). These findings
revealed that the cNLS peptide successfully entered into the
nucleus following UTMD-mediated transfection, while mNLS only
partly entered into the nucleus and WGA effectively inhibited cNLS
nuclear entry.
cNLS promotes the cellular and nuclear
uptake of pDNA
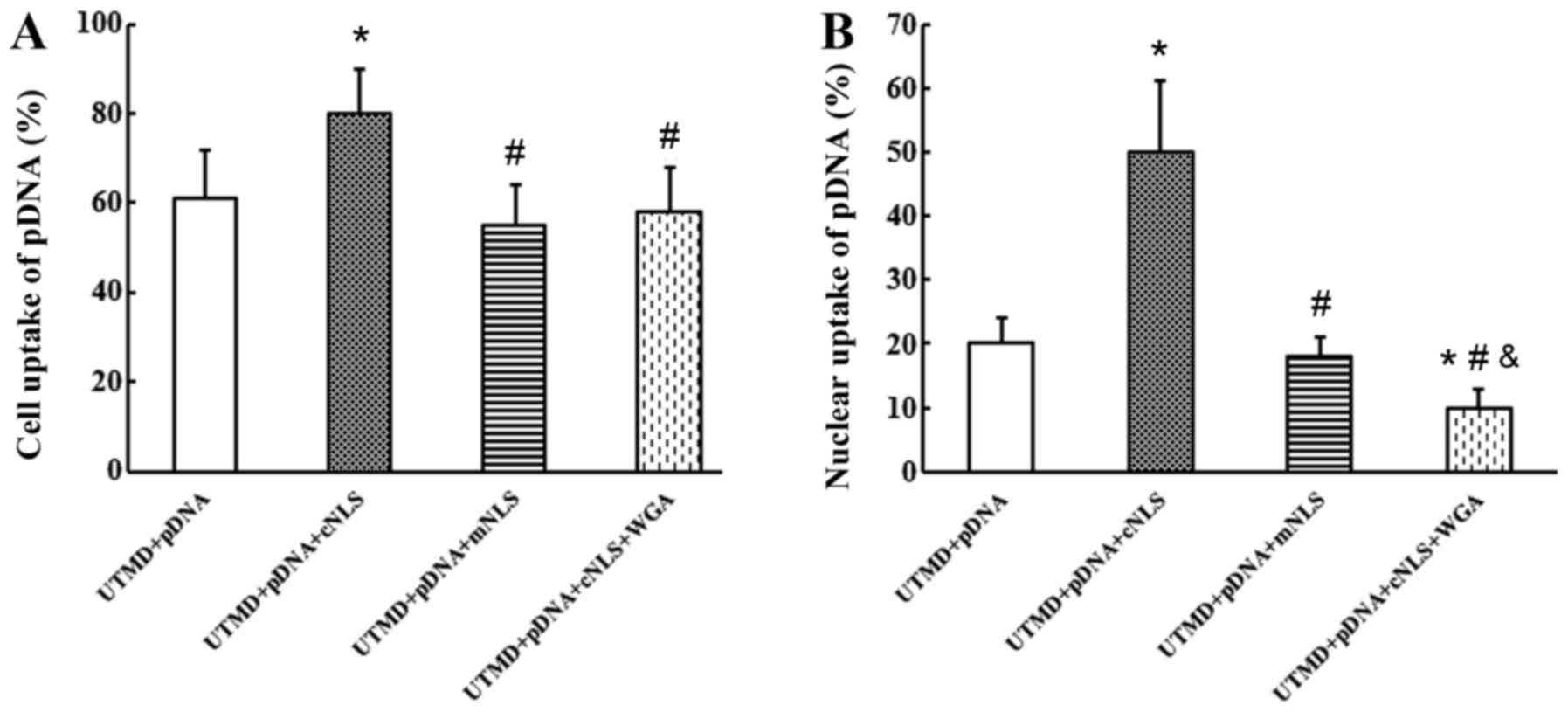
At 6 h post-transfection, the rates of pDNA uptake
into the cells were 61±11, 80±10, 55±9 and 58±10% for the control,
cNLS, mNLS and WGA groups, respectively (Fig. 5A). The rates of pDNA uptake into
the nucleus were 20±4, 50±11, 18±3 and 10±3% for the control, cNLS,
mNLS and WGA groups, respectively (Fig. 5B). These results indicated that the
cNLS significantly enhanced the cellular and nuclear uptake of pDNA
during UTMD-mediated gene transfection.
Effect of cNLS on cell viability and
transfection efficiency
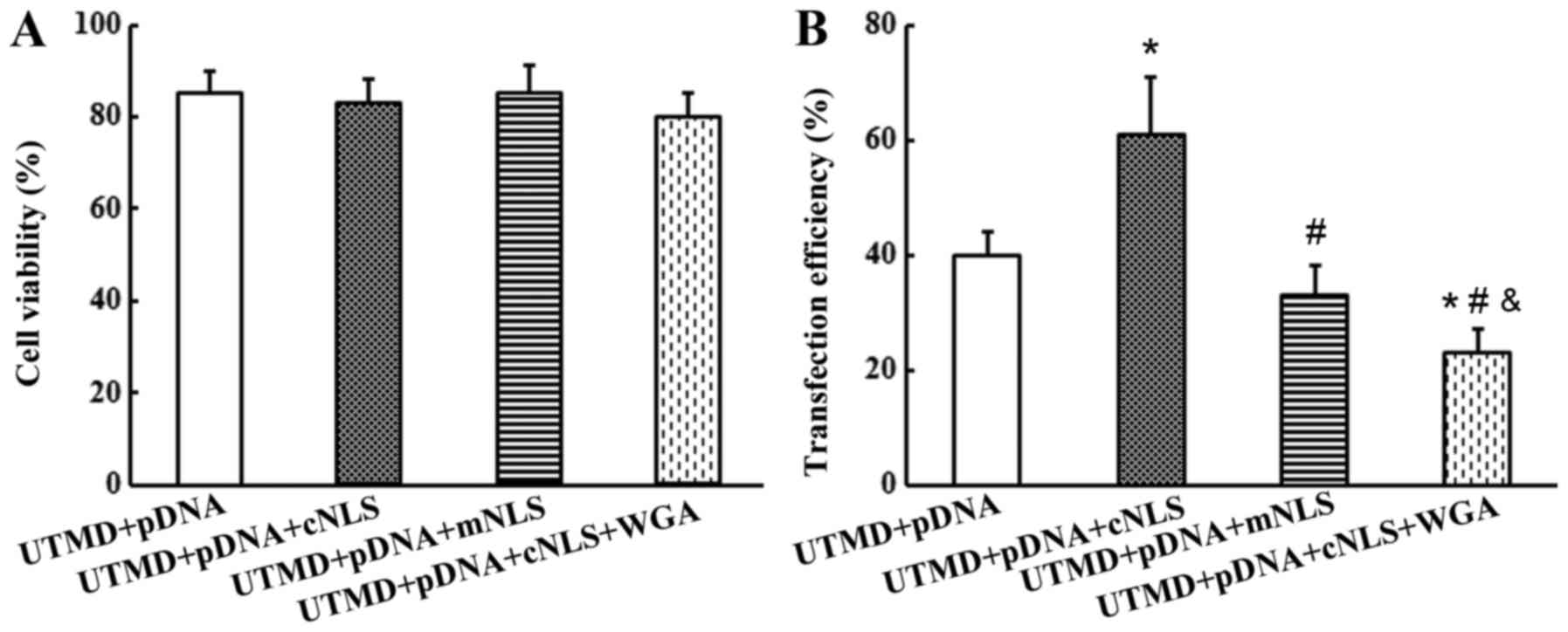
At 48 h post-transfection, the cell viability was
>80% in all 4 experimental groups, and no significant difference
was observed among the groups (P>0.05; Fig. 6A). The transfection efficiency was
40±4, 61±10, 37±5 and 23±4% in the control, cNLS, mNLS and WGA
groups, respectively. The transfection efficiency in the cNLS group
was significantly increased compared with the control (~57%
increase), mNLS and WGA groups (P<0.05; Fig. 6B). This finding indicated that the
cNLS promoted UTMD-mediated transfection efficiency of pDNA.
Transfection efficiency was also determined by expression of EGFP,
encoded on the pDNA vector, via flow cytometry and fluorescence
microscopy. The number of cells expressing EGFP markedly increased
in the cNLS group compared with the other three groups, confirming
enhanced transfection efficiency (Fig.
7).
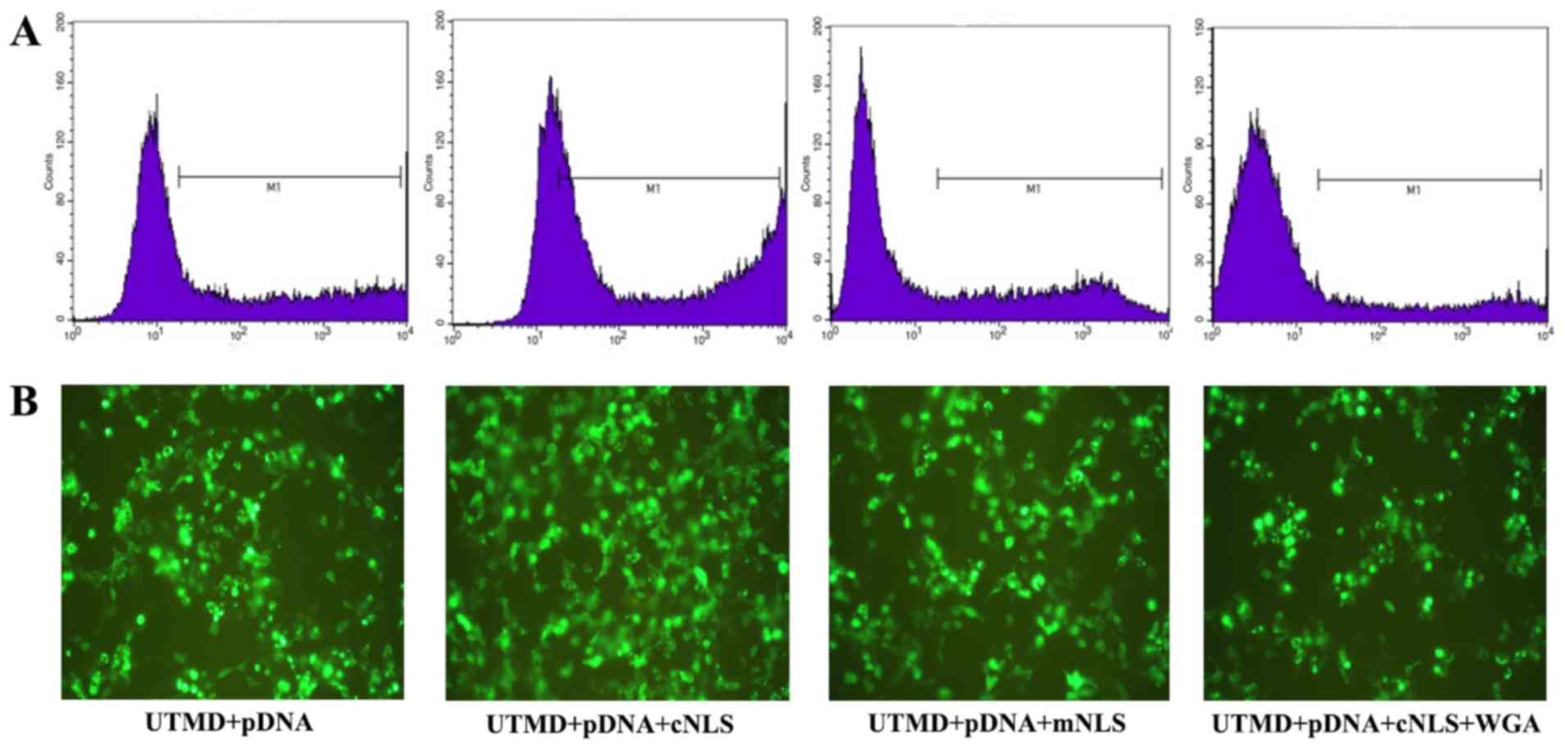 | Figure 7.Evaluation of transfection efficiency
by (A) flow cytometry and (B) fluorescence microscopy in control
group (UTMD + pDNA), cNLS group (UTMD + pDNA + cNLS), mNLS group
(UTMD + pDNA + mNLS), and WGA (UTMD + pDNA + cNLS + WGA) group.
Positive fluorescence signal is evident in the M1 portion of the
flow cytometry plots, and by green signal in the microscopy images
(magnification, ×100). UTMD, ultrasound-targeted microbubble
destruction; pDNA, plasmid DNA; NLS, nuclear localization signal;
c, classic; m, mutated; WGA, wheat germ agglutinin. |
cNLS enhances the expression of
hANGPT1 mRNA and protein
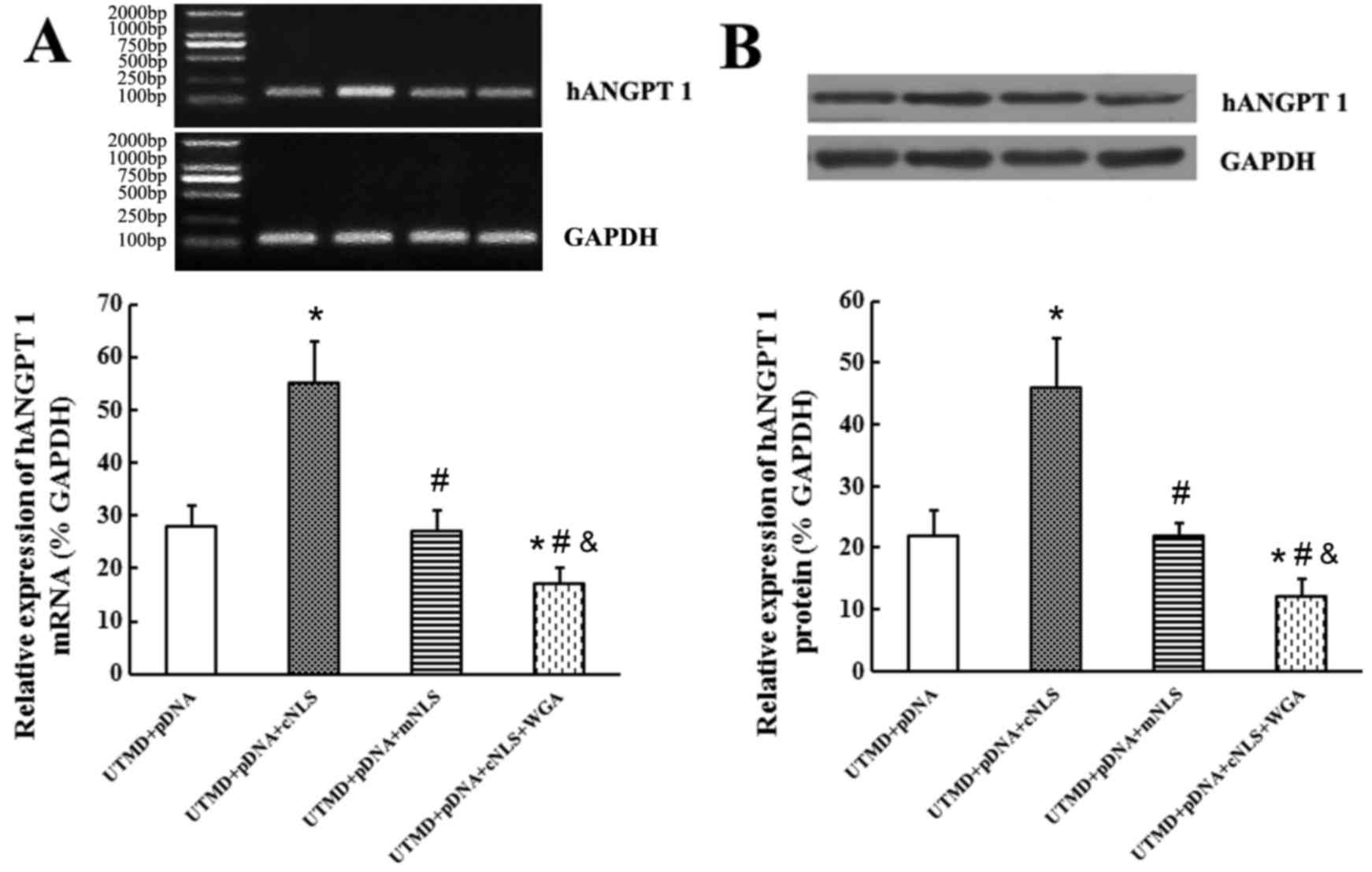
The relative expression levels of hANGPT1 mRNA were
28±4, 55±8, 27±4 and 17±3% in the control, cNLS, mNLS and WGA
groups, respectively, with the cNLS group displaying the highest
mRNA expression levels compared with the other three groups
(P<0.05; Fig. 8A). The relative
expression levels of the hANGPT1 protein were 22±4, 46±8, 22±5 and
12±3% in the control, cNLS, mNLS and WGA groups, respectively, with
the cNLS group again displaying the highest protein expression
levels compared with the other three groups (P<0.05; Fig. 8B). Expression of hANGPT1 mRNA and
protein in the mNLS group was not significantly different to the
control group, whereas the WGA group displayed decreased expression
compared with the control group (P<0.05; Fig. 8).
Discussion
As a new type of non-viral vector gene transfection,
UTMD technology uses ultrasound microbubbles as nuclei to exert a
cavitation effect, which increases the permeability of cell
membranes during ultrasound exposure (1,18–21).
Exogenous DNA can enter a cell through the cell membrane and
transfect the cell under the appropriate conditions. The barrier
function of the cell and nuclear membranes, and nuclease
degradation, are some of the limiting factors of transfection
efficiency by UTMD-mediated exogenous gene in eukaryotic cells.
Several methods have been tested in order to improve transfection
efficiencies: The biotin-avidin system can gather large amounts of
exogenous DNA around the microbubbles; the antigen-targeted
antibody system can identify the damaged endothelial cells
specifically; a new type of ultrasound microbubbles can change
their physical and chemical properties; and addition of cationic
polymers can improve the transfection efficiency (22,23).
These methods have some effect in penetrating the cell membrane
barrier, but have provided little benefit to cross the nuclear
membrane barrier. Macromolecules, such as pDNA, must contain an NLS
in order to be actively transported into the nucleus, by specific
interaction of the NLS with the nuclear import proteins (9,10).
NLS peptides are positively charged because their
sequence is rich in arginines and lysines, and therefore can bind
negatively charged pDNA by electrostatic interactions (24). The present study confirmed that the
NLS and pDNA combined in a complex and the optimum molar ratio for
this binding was 104:1 NLS/pDNA, as detected by agarose
gel electrophoresis, performed similarly to a previous study by
Duvshani-Eshet et al (25).
For different exogenous DNAs, the differences in molecular weight
and electrostatic charge result in different molar ratios for
binding. Part of the positively charged NLS can also interact with
the negatively charged SonoVue microbubbles, thereby increasing
pDNA uptake into cells indirectly during UTMD; this phenomenon may
also partly explain the higher transfection efficiency in the cNLS
group compared with the control group.
The nuclear import of NLS-mediated exogenous DNA is
divided into at least two steps. The first step is the process of
NLS identification by the NLS receptor, which then forms a nuclear
pore positioning complex gathered on the cytoplasmic side of the
nuclear membrane. The second step is the process of nuclear
transfer, which is dependent on temperature and energy, and results
in the NLS peptide passing through the nuclear pores and into the
nucleus (26). When the ultrasound
cavitation effect is generated by UTMD, the absorption or
dissipation of the ultrasonic energy can cause a local and
transient temperature increase, which may be advantageous to the
exogenous DNA to get into the cytoplasm and may provide energy to
promote the NLS-mediated pDNA entry into the nucleus (2–3,27).
This phenomenon may also explain the NLS-mediated exogenous DNA
entry into the nucleus observed in the present study; the rates of
cell and nuclear pDNA uptake increased significantly when the
transfection was conducted by UTMD combined with cNLS.
In the present study, the rates of cell and nuclear
uptake of pDNA both increased following addition of the optimal
proportion of cNLS/pDNA. The transfection efficiency also improved
significantly, as well as expression of mRNA and protein of the
target gene. The present finding indicated that NLS may be
important in transfection efficiency by UTMD method.
Previously, methods of exogenous gene transfection
have had great progress using the function of NLS peptides
(28,29). For example, in nerve cells that are
difficult to transfect, previous studies have performed fusion
expression of an NLS peptide with a target gene, and reported that
recombinant transcription factors were efficiently transferred into
the neural stem cells and successfully induced nerve cell
differentiation (30). In addition
to vertebrates, NLS peptides can improve the transfection
efficiency in crustaceans; compared with DNA alone, the
non-covalent bound DNA/NLS group exhibited >2-fold greater
transfection efficiency, faster nuclear input rate as detected by
β-galactose activity determination (the progress is 20 min vs. 90
min in the control group), and decreased degradation by DNA
hydrolytic enzymes (31). The use
of cNLS in the present study may have enhanced the transfection
efficiency by to promoting the entry of the exogenous DNA into the
nucleus in great quantities, as well as avoiding exogenous DNA
degradation in the cytoplasm. Thus, the combination of the two
kinds of effects may significantly improve transfection
efficiency.
However, various studies have adopted different
experimental methods; the transfection efficiency was not always
improved by combining an NLS peptide with the target gene. As such,
the characteristics and advantages of the NLS peptide need be
thoroughly investigated in order to explore how to optimally use
NLS as a nuclear guide to enhance the rate of exogenous DNA entry
into the nucleus and to thus improve the transfection efficiency in
the future.
A limitation of the present study was the method of
nuclei staining. Although the nuclei are displayed under a
fluorescent microscope, the location of the cell nucleus is visibly
more apparent if cells are co-stained with DAPI. This methodology
will be further improved in the group's future investigations.
Nevertheless, the present study demonstrated that the UTMD method
increased cell membrane permeability for exogenous DNA entry into
the cytoplasm, and addition of NLS promoted pDNA entry into the
nucleus, thereby improving the overall transfection efficiency. The
findings suggest that NLS combination with UTMD method may be a
novel efficient strategy of non-viral vector gene transfection.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81471674 and
81501495) and the Fundamental Research Funds for the Central
Universities (grant no. 2015301020203).
References
|
1
|
Noble ML, Kuhr CS, Graves SS, Loeb KR, Sun
SS, Keilman GW, Morrison KP, Paun M, Storb RF and Miao CH:
Ultrasound-targeted microbubble destruction-mediated gene delivery
into canine livers. Mol Ther. 21:1687–1694. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hernot S and Klibanov AL: Microbubbles in
ultrasound-triggered drug and gene delivery. Adv Drug Deliv Rev.
60:1153–1166. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan Z, Chen D and Deng CX: Improving
ultrasound gene transfection efficiency by controlling ultrasound
excitation of microbubbles. J Control Release. 170:401–413. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Teulon JM, Delcuze Y, Odorico M, Chen SW,
Parot P and Pellequer JL: Single and multiple bonds in
(strept)avidin-biotin interactions. J Mol Recognit. 24:490–502.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kocbek P, Obermajer N, Cegnar M, Kos J and
Kristl J: Targeting cancer cells using PLGA nanoparticles surface
modified with monoclonal antibody. J Control Release. 120:18–26.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Laing ST and McPherson DD: Cardiovascular
therapeutic uses of targeted ultrasound contrast agents. Cardiovasc
Res. 83:626–635. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Suzuki R, Oda Y, Utoguchi N and Maruyama
K: Progress in the development of ultrasound-mediated gene delivery
systems utilizing nano- and microbubbles. J Control Release.
149:36–41. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mannell H, Pircher J, Fochler F, Stampnik
Y, Räthel T, Gleich B, Plank C, Mykhaylyk O, Dahmani C, Wörnle M,
et al: Site directed vascular gene delivery in vivo by ultrasonic
destruction of magnetic nanoparticle coated microbubbles.
Nanomedicine. 8:1309–1318. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meinema AC, Poolman B and Veenhoff LM: The
transport of integral membrane proteins across the nuclear pore
complex. Nucleus. 3:322–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marfori M, Mynott A, Ellis JJ, Mehdi AM,
Saunders NFW, Curmi PM, Forwood JK, Bodén M and Kobe B: Molecular
basis for specificity of nuclear import and prediction of nuclear
localization. Biochim Biophys Acta. 1813:1562–1577. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lupberger J, Schaedler S, Peiran A and
Hildt E: Identification and characterization of a novel bipartite
nuclear localization signal in the hepatitis B virus polymerase.
World J Gastroenterol. 19:8000–8010. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Adam SA, Lobl TJ, Mitchell MA and Gerace
L: Identification of specific binding proteins for a nuclear
location sequence. Nature. 337:276–279. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Collins E, Birchall JC, Williams JL and
Gumbleton M: Nuclear localisation and pDNA condensation in
non-viral gene delivery. J Gene Med. 9:265–274. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yoo HS and Jeong SY: Nuclear targeting of
non-viral gene carriers using psoralen-nuclear localization signal
(NLS) conjugates. Eur J Pharm Biopharm. 66:28–33. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Finlay DR, Newmeyer DD, Price TM and
Forbes DJ: Inhibition of in vitro nuclear transport by a lectin
that binds to nuclear pores. J Cell Biol. 104:189–200. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng P, Gong X and Jiang Y: A novel method
for studying nuclear localization signal-mediated nuclear
translocation. Nan Fang Yi Ke Da Xue Xue Bao. 32:1148–1150.
2012.(In Chinese). PubMed/NCBI
|
|
17
|
Deng Q, Chen JL, Zhou Q, Hu B, Chen Q,
Huang J and Guo RQ: Ultrasound microbubbles combined with the NFκB
binding motif increase transfection efficiency by enhancing the
cytoplasmic and nuclear import of plasmid DNA. Mol Med Rep.
8:1439–1445. 2013.PubMed/NCBI
|
|
18
|
Villanueva FS: Getting good vibes: The
therapeutic power of microbubbles and ultrasound. JACC Cardiovasc
Imaging. 5:1263–1266. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kooiman K, Emmer M, Foppen-Harteveld M,
van Wamel A and de Jong N: Increasing the endothelial layer
permeability through ultrasound-activated microbubbles. IEEE Trans
Biomed Eng. 57:29–32. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meijering BDM, Juffermans LJ, van Wamel A,
Henning RH, Zuhorn IS, Emmer M, Versteilen AM, Paulus WJ, van Gilst
WH, Kooiman K, et al: Ultrasound and microbubble targeted delivery
of macromolecules is regulated by induction of endocytosis and pore
formation. Circ Res. 104:679–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bekeredjian R, Kroll RD, Fein E, Tinkov S,
Coester C, Winter G, Katus HA and Kulaksiz H: Ultrasound targeted
microbubble destruction increases capillary permeability in
hepatomas. Ultrasound Med Biol. 33:1592–1598. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li P, Gao Y, Liu Z, Tan K, Zuo Z, Xia H,
Yang D, Zhang Y and Lu D: DNA transfection of bone marrow stromal
cells using microbubble-mediated ultrasound and polyethylenimine:
An in vitro study. Cell Biochem Biophys. 66:775–786. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Werth S, Urban-Klein B, Dai L, Höbel S,
Grzelinski M, Bakowsky U, Czubayko F and Aigner A: A low molecular
weight fraction of polyethylenimine (PEI) displays increased
transfection efficiency of DNA and siRNA in fresh or lyophilized
complexes. J Control Release. 112:257–270. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim BK, Kang H, Doh KO, Lee SH, Park JW,
Lee SJ and Lee TJ: Homodimeric SV40 NLS peptide formed by disulfide
bond as enhancer for gene delivery. Bioorg Med Chem Lett.
22:5415–5418. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Duvshan-Eshet M, Keren H, Oz S,
Radzishevsky IS, Mor A and Machluf M: Effect of peptides bearing
nuclear localization signals on therapeutic ultrasound mediated
gene delivery. J Gene Med. 10:1150–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yi WJ, Yang J, Li C, Wang HY, Liu CW, Tao
L, Cheng SX, Zhuo RX and Zhang XZ: Enhanced nuclear import and
transfection efficiency of TAT peptide-based gene delivery systems
modified by additional nuclear localization signals. Bioconjug
Chem. 23:125–134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duvshan-Eshet M, Haber T and Machluf M:
Insight concerning the mechanism of therapeutic ultrasound
facilitating gene delivery: Increasing cell membrane permeability
or interfering with intracellular pathways? Hum Gene Ther.
25:156–164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Opanasopit P, Rojanarata T, Apirakaramwong
A, Ngawhirunpat T and Ruktanonchai U: Nuclear localization signal
peptides enhance transfection efficiency of chitosan/DNA complexes.
Int J Pharm. 382:291–295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kawazu T, Kanzaki H, Uno A, Azuma H and
Nagasaki T: HVJ-E/importin-β hybrid vector for overcoming
cytoplasmic and nuclear membranes as double barrier for non-viral
gene delivery. Biomed Pharmacother. 66:519–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stock K, Nolden L, Edenhofer F, Quandel T
and Brüstle O: Transcription factor-based modulation of neural stem
cell differentiation using direct protein transduction. Cell Mol
Life Sci. 67:2439–2449. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Arenal A, Pimentel R, García C, Pimentel E
and Aleström P: The SV40 T antigen nuclear localization sequence
enhances nuclear import of vector DNA in embryos of a crustacean
(Litopenaeus schmitti). Gene. 337:71–77. 2004. View Article : Google Scholar : PubMed/NCBI
|