Introduction
Coronary artery disease (CAD) has been a leading
cause of mortality and disability worldwide for the past decades,
and is likely to remain so for a number of years to come (1). Acute coronary syndromes (ACS) is a
high-risk clinical type of CAD which occurs as a result of
myocardial ischaemia, and includes acute myocardial infarction and
unstable angina (UA). Effective prevention and treatment strategies
are important for reducing the morbidity and mortality of CAD.
Statins, 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase
inhibitors, are the foundation of medical therapy in primary and
secondary prevention of cardiovascular diseases. Lipid-lowering
therapy uses statins to reduce cardiovascular risk in patients with
stable CAD (2) and ACS (3,4).
Statin therapy is also recommended (Level of Evidence 1A) by the
American College of Cardiology/American Heart Association (ACC/AHA)
guidelines for all patients with ACS, regardless of baseline
low-density lipoprotein (LDL) levels prior to hospital discharge
(5). Although statins were first
developed to lower total serum cholesterol and improve the lipid
profile, a number of studies have suggested that statins may exert
atheroprotective effects beyond cholesterol lowering (6,7),
such as improving endothelial function, increasing nitric oxide
(NO) activity, reducing oxidative stress, alleviating inflammation,
and inhibiting platelet adhesion and the coagulation cascade. Our
previous research also demonstrated that statins could improve
endothelial function independent of LDL cholesterol reduction
(8). All of these results
indicated that the clinical benefit of statins in ACS was
independent of lipid-reducing effects, but the potential mechanism
remains unclear.
microRNAs (miRNAs) are small non-coding RNAs that
negatively regulate gene expression at the post-transcription level
by combining with target mRNA 3′ untranslated region (3′UTR)
(9). Single miRNA species can
regulate multiple mRNA targets, and single mRNAs may contain
several miRNA recognition sites on their 3′UTR, which forms a
complex regulatory network and controls important biological
functions (10,11). Alterations in miRNA levels are
associated with numerous human pathologies, including cancer
(12,13), and metabolic (14,15)
and cardiovascular diseases (16,17).
miRNAs have also been investigated in the blood, where they have
been detected in plasma, platelets, erythrocytes and nucleated
blood cells, and serve as novel diagnostic markers (18). It has also been identified that
miRNAs are capable of mediating cell-cell communication transferred
by microvesicles, and serve an important regulatory role in a
number of diseases (19).
It has been reported that statins are able to serve
their biological role by regulating miRNA expression in
CAD-associated cells, including platelets (20), endothelial cells (21), endothelial progenitor cells
(22,23) and monocytes (24). Statins may enhance the stability of
atherosclerotic plaques mediated by miRNAs in UA patients;
therefore, the present study aimed to investigate the influence of
statins on the circulating miRNA profile in UA patients, and
analyzed the miRNA-mediated regulatory network in these
patients.
Materials and methods
Patients
The present study was performed in accordance with
the Helsinki Declaration and was approved by the Ethics Review
Board of Peking University People's Hospital (Beijing, China). The
patients were recruited from Peking University People's Hospital
and were as follows: 8 non-statin controls without CAD, as assessed
by coronary angiography (group 1: Control group); 8 UA patients
with non-statin medication (group 2: UA group, also designated
non-statin group); and 8 UA patients with statin treatment (group
3: statin group). All subjects gave their written informed consent.
Criteria for the diagnosis of UA were according to the ACC/AHA 2011
guidelines (1). Patients
presenting elevated troponin I (≥ 0.04 ng/ml) and/or creatine
kinase (≥ 5 ng/ml) or with myocarditis, cardiogenic shock, a
history of severe hepatic or renal dysfunction, leukemia, ongoing
inflammation and malignant disease, were excluded.
Blood collection and RNA
extraction
Blood was collected from each patient via
venipuncture into PAXgene Blood RNA tubes (BD Diagnostics, Inc.,
Sparks, MD, USA) prior to coronary angiography. A PAXgene Blood
miRNA kit (Qiagen, Inc., Valencia, CA, USA) was used for RNA
isolation according to manufacturer's protocol.
miRNA taqman low density array
(TLDA)
TLDA was used to determine differentially expressed
miRNAs in whole blood from subjects (n=8/group). Approximately 15
ng of total RNA was reverse-transcribed with a Taqman miRNA reverse
transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and Taqman miRNA Multiplex RT assays (Human
Pool A; Applied Biosystems; Thermo Fisher Scientific, Inc.). The
reverse transcription products were analyzed using Human MicroRNA
TLDA card A version 3.0 (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), which can detect the
expression of 372 miRNAs simultaneously. miRNAs levels were
normalized to the levels of U6. DataAssist software version 3.01
(www.lifetechnologies.com/us/en/home/technical-resources/software-downloads/dataassist-software.html)
was used to calculate the relative levels of miRNAs, using the
quantitation threshold (Cq) method (25). Significance analysis of microarrays
was used to analyze differentially expressed miRNAs between two
groups. The criteria for the differentially expressed miRNAs were a
fold change ≥2 or ≤0.5, q-value <0.05 and a false discovery rate
<0.05 in comparison between two groups.
Bioinformatic analysis
The target genes of miRNAs were predicted in
TargetScan (www.targetscan.org/) and miRanda (miracle.igib.res.in/miracle/) databases, and the
target genes simultaneously predicted by the two databases were
selected for the next step of signaling pathway analysis. The
Database for Annotation, Visualization and Integrated Discovery
(DAVID; david.abcc.ncifcrf.gov/) 6.7 platform was used to
input target genes, and the enriched pathways of these genes were
obtained in PANTHER (www.pantherdb.org/). Clustering of target genes
according to different cell types was based on CGAP SAGE (for
monocytes and endothelium; cgap.nci.nih.gov/SAGE) and a UP tissue-specific
library (for platelets) via the DAVID platform. To visualize the
putative target genes or functional pathways of miRNAs, the network
dataset was entered in Cytoscape version 3.0.0 beta 1 (www.cytoscape.org).
Statistical analysis
Quantitative data are presented as the mean ±
standard deviation. For continuous variables, statistical
significance was calculated using Student's t-test for the
comparison of two groups. For categorical variables, statistical
significance was calculated using the chi-square test for the
comparison of two groups. All tests were two-sided. SPSS version
17.0 (SPSS, Inc., Chicago, IL, USA) was used for all statistical
analyses. P<0.05 was considered to indicate a statistically
significant difference.
Results
Regulatory effect of statins on
blood-borne miRNA expression profiling in UA patients
To determine the role of statins in UA patients
mediated by circulating miRNA, the characteristic of circulating
miRNA profiling in non-statin patients with UA (n=8) compared with
non-CAD controls (n=8) was first studied (Table I). Under the pathological condition
of UA without statin therapy, there were 21 differentially
expressed miRNAs; 20 miRNAs were downregulated and 1 miRNA was
upregulated (Fig. 1A; Table II). However, 33 upregulated miRNAs
were identified in UA patients treated with statin (n=8) compared
with non-statin patients (Fig. 1B;
Table III). Among the 33
upregulated miRNAs, there were 20 nascent miRNAs and 13 initially
downregulated miRNAs in non-statin-treated patients with UA
(Fig. 1C), which indicated that
statins may alter the circulating miRNA expression profiles of UA
patients.
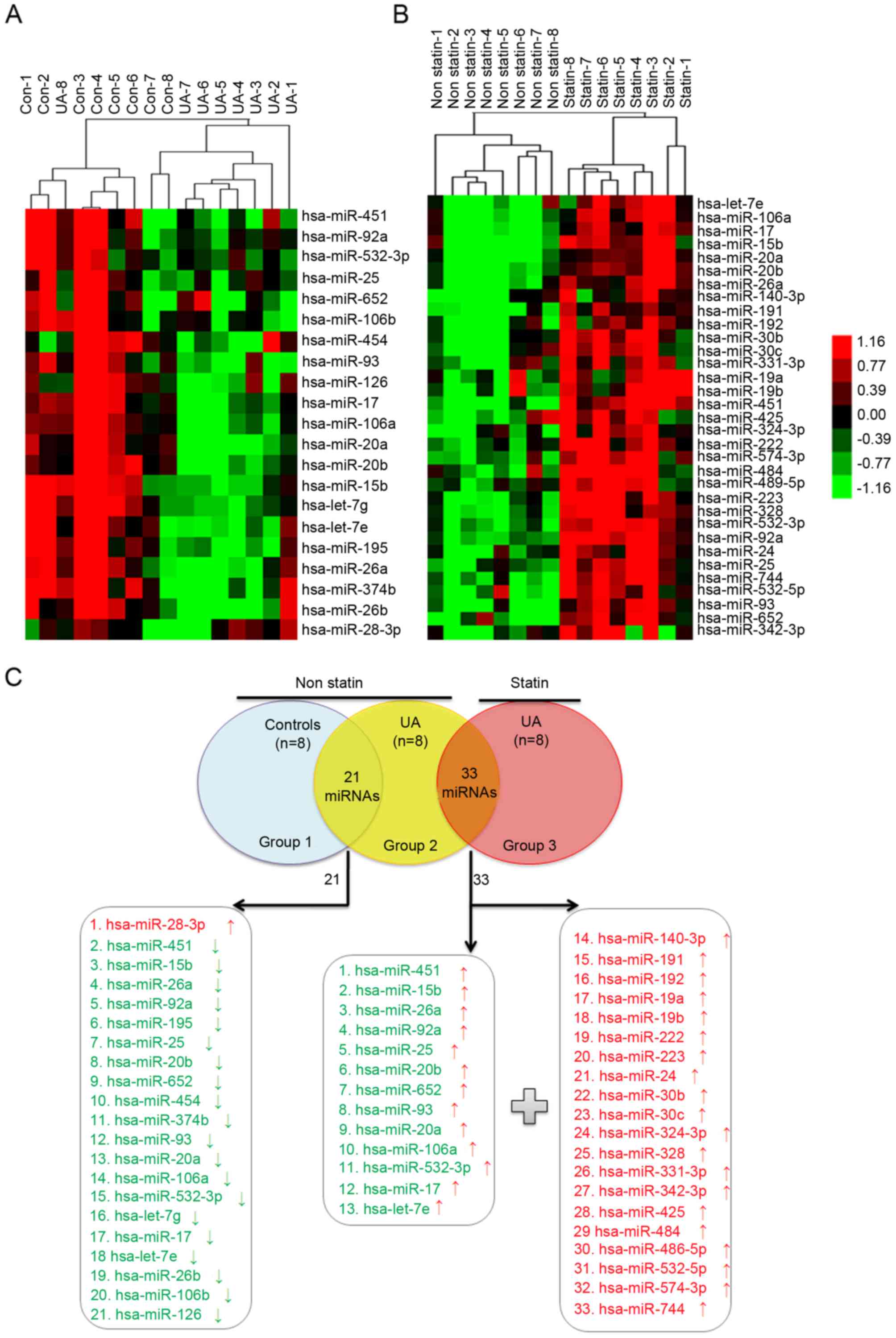 | Figure 1.Blood-borne miRNA differential
expression profiling in (A) UA patients without statin treatment
compared with non-CAD controls and (B) in UA patients with statin
treatment compared with non-statin-treated UA patients, as detected
by TLDA. (A) RNA was isolated from whole blood of subjects with no
statin treatment (UA, n=8 vs. Con, n=8). (B) RNA was isolated from
whole blood of UA patients with and with no statin therapy (statin,
n=8 vs. non-statin, n=8). The heat map illustrates levels of
significantly altered miRNAs. Color intensity is scaled within each
row so that the highest expression value corresponds to bright red
and the lowest to bright green. (C) There were 33 upregulated
miRNAs in the statin group compared with the non-statin group, of
which 13 miRNAs were downregulated in the non-statin group compared
with the non-CAD group, and 20 nascent miRNAs. miRNA/miR, microRNA;
UA, unstable angina; CAD, coronary artery disease; TLDA, taqman low
density array; Con, control. |
 | Table I.Clinical characteristics of
patients. |
Table I.
Clinical characteristics of
patients.
|
| Non-statin | Statin | P-value |
|---|
|
|
|
|
|
|---|
|
| Group 1 controls
(n=8) | Group 2 UA
(n=8) | Group 3 UA
(n=8) | Group 2 vs. Group
1 | Group 3 vs. Group
2 |
|---|
| General data |
| Age
(years) | 62±9 | 66±9 | 58±8 | 0.42 | 0.08 |
| Sex
(male/female) | 5/3 | 4/4 | 3/5 | 0.61 | 0.61 |
|
SBP | 133±11 | 136±15 | 132±18 | 0.76 | 0.67 |
|
DBP | 82±4 | 79±6 | 82±14 | 0.24 | 0.53 |
| Medical history,
% |
|
Hypertension | 62.5 | 87.5 | 62.5 | 0.25 | 0.25 |
|
Diabetes | 0 | 12.5 | 12.5 | 0.30 | >0.99 |
|
Hyperlipaemia | 12.5 | 25.0 | 50.0 | 0.52 | 0.30 |
| Laboratory test,
mmol/l |
|
LDL-C | 2.13±1.06 | 2.61±0.97 | 2.41±0.97 | 0.36 | 0.68 |
|
HDL-C | 1.26±0.72 | 1.04±0.10 | 1.08±0.24 | 0.40 | 0.67 |
| TC | 4.07±0.90 | 4.22±1.07 | 4.07±0.82 | 0.77 | 0.77 |
| TG | 1.35±0.61 | 1.18±0.38 | 1.27±0.78 | 0.51 | 0.76 |
|
Glucose | 6.28±1.57 | 4.99±0.83 | 5.25±0.77 | 0.06 | 0.52 |
|
Creatinine | 91.25±29.20 | 85.75±33.04 | 78.13±40.75 | 0.73 | 0.69 |
| Medication, % |
|
Aspirin | 12.5 | 50.0 | 75.0 | 0.11 | 0.30 |
|
Clopidogrel | 12.5 | 25.0 | 50.0 | 0.52 | 0.30 |
| Calcium
antagonist | 12.5 | 50.0 | 25.0 | 0.11 | 0.30 |
|
ACEI | 0 | 25.0 | 12.5 | 0.13 | 0.52 |
|
ARB | 0 | 37.5 | 12.5 | 0.06 | 0.25 |
|
β-blocker | 37.5 | 50.0 | 37.5 | 0.61 | 0.61 |
 | Table II.Circulating miRNA differential
expression profiling in non-statin-treated patients with UA
compared with non-CAD controls. |
Table II.
Circulating miRNA differential
expression profiling in non-statin-treated patients with UA
compared with non-CAD controls.
| No. | Gene ID | Score (d) | Fold change
(UA/Control) | q-value, % |
|---|
| 1 | hsa-miR-28-3p | 1.71 | 3.77 | <0.01 |
| 2 | hsa-miR-451 | −2.54 | 0.32 | <0.01 |
| 3 | hsa-miR-15b | −2.47 | 0.21 | <0.01 |
| 4 | hsa-miR-26a | −2.21 | 0.28 | <0.01 |
| 5 | hsa-miR-92a | −2.07 | 0.41 | <0.01 |
| 6 | hsa-miR-195 | −2.02 | 0.09 | <0.01 |
| 7 | hsa-miR-25 | −1.92 | 0.47 | <1.35 |
| 8 | hsa-miR-20b | −1.89 | 0.08 | <1.35 |
| 9 | hsa-miR-652 | −1.88 | 0.39 | <1.35 |
| 10 | hsa-miR-454 | −1.86 | 0.18 | <1.35 |
| 11 | hsa-miR-374b | −1.80 | 0.23 | <1.35 |
| 12 | hsa-miR-93 | −1.78 | 0.21 | <1.35 |
| 13 | hsa-miR-20a | −1.76 | 0.13 | <1.35 |
| 14 | hsa-miR-106a | −1.75 | 0.20 | <1.35 |
| 15 | hsa-miR-532-3p | −1.74 | 0.49 | <1.35 |
| 16 | hsa-let-7g | −1.69 | 0.09 | <1.35 |
| 17 | hsa-miR-17 | −1.68 | 0.20 | <1.35 |
| 18 | hsa-let-7e | −1.67 | 0.16 | <1.35 |
| 19 | hsa-miR-26b | −1.64 | 0.16 | <1.35 |
| 20 | hsa-miR-106b | −1.63 | 0.32 | <1.35 |
| 21 | hsa-miR-126 | −1.62 | 0.17 | <1.35 |
 | Table III.Circulating miRNAs differential
expression profiling in UA patients treated with statin compared
with non-statin patients. |
Table III.
Circulating miRNAs differential
expression profiling in UA patients treated with statin compared
with non-statin patients.
| No | Gene ID | Score (d) | Fold change
(statin/non statin) | q-value, % |
|---|
| 1 | hsa-miR-191 | 5.52 | 2.28 | <0.01 |
| 2 | hsa-miR- 92a | 5.15 | 3.28 | <0.01 |
| 3 | hsa-miR-223 | 4.92 | 3.17 | <0.01 |
| 4 | hsa-miR-532-3p | 4.46 | 2.69 | <0.01 |
| 5 | hsa-miR-451 | 4.31 | 4.77 | <0.01 |
| 6 | hsa-miR-30b | 4.22 | 2.80 | <0.01 |
| 7 | hsa-miR-15b | 4.15 | 3.57 | <0.01 |
| 8 | hsa-miR-26a | 3.77 | 3.38 | <0.01 |
| 9 | hsa-miR-30c | 3.65 | 2.50 | <0.01 |
| 10 | hsa-miR-19b | 3.41 | 2.38 | <0.01 |
| 11 | hsa-miR- 25 | 3.39 | 2.92 | <0.01 |
| 12 | hsa-miR-222 | 3.34 | 2.54 | <0.01 |
| 13 | hsa-miR-574-3p | 3.33 | 2.98 | <0.01 |
| 14 | hsa-miR-484 | 3.31 | 2.88 | <0.01 |
| 15 | hsa-miR-24 | 3.31 | 2.02 | <0.01 |
| 16 | hsa-miR-652 | 3.30 | 3.06 | <0.01 |
| 17 | hsa-miR-486-5p | 3.27 | 2.53 | <0.01 |
| 18 | hsa-miR-324-3p | 3.20 | 2.22 | <0.01 |
| 19 | hsa-miR-331-3p | 3.10 | 2.07 | <0.01 |
| 20 | hsa-miR-106a | 3.06 | 3.46 | <0.01 |
| 21 | hsa-miR-328 | 3.05 | 2.70 | <0.01 |
| 22 | hsa-miR-20a | 3.02 | 4.15 | <0.01 |
| 23 | hsa-miR-140-3p | 2.77 | 2.23 | <0.01 |
| 24 | hsa-miR-93 | 2.71 | 3.61 | <0.01 |
| 25 | hsa-miR-17 | 2.67 | 3.61 | <0.01 |
| 26 | hsa-miR-192 | 2.67 | 2.72 | <0.01 |
| 27 | hsa-miR-20b | 2.61 | 4.13 | <0.01 |
| 28 | hsa-miR-532-5p | 2.55 | 2.74 | <0.01 |
| 29 | hsa-miR-744 | 2.54 | 3.68 | <0.01 |
| 30 | hsa-miR-342-3p | 2.49 | 2.21 | <0.01 |
| 31 | hsa-miR-19a | 2.49 | 2.04 | <0.01 |
| 32 | hsa-miR-425 | 2.39 | 2.05 | <0.01 |
| 33 | hsa-let-7e | 2.28 | 3.17 | <0.01 |
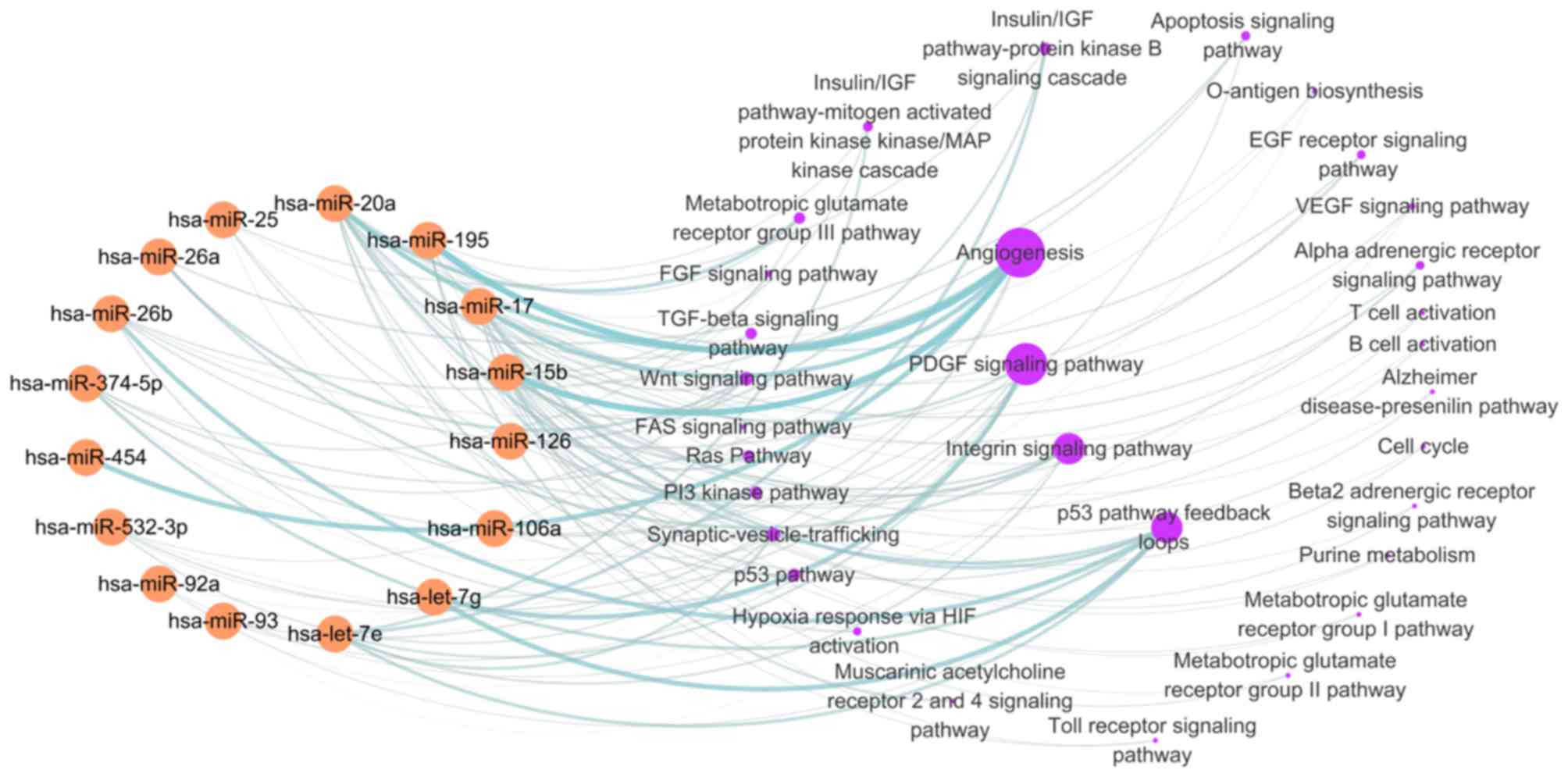
Signaling pathways analysis targeted
by differentially expressed miRNAs induced by statin in UA
patients
In order to understand the biological role of
statins in UA patients, the signaling pathways targeted by 21
differentially expressed miRNAs in non-statin patients with UA were
first analyzed. Each miRNA target was entered into the DAVID
platform and the signaling pathways referring to these targets were
obtained from the PANTHER database. Bioinformatics analysis results
demonstrated that the target genes were mainly enriched in the
following pathways: Angiogenesis (regulated by miR-15b, 17, 20a,
93, 195, 374-5p, 454), platelet derived growth factor (PDGF)
signaling pathway (regulated by let-7 g, 7e, miR-17, 20a, 532-3p),
integrin signaling pathway (regulated by let-7e, miR-25, 26a,
532-3p) and p53 pathway feedback loops (regulated by let-7 g,
miR-25, 26b, 92a; Fig. 2). There
were 16 out of 21 miRNAs involved in the regulation of signaling
pathways in non-statin-treated patients with UA. The effects of
these pathways were enhanced in unstable coronary heart disease for
the downregulation of associated miRNAs, which may contribute to
plaque destabilization.
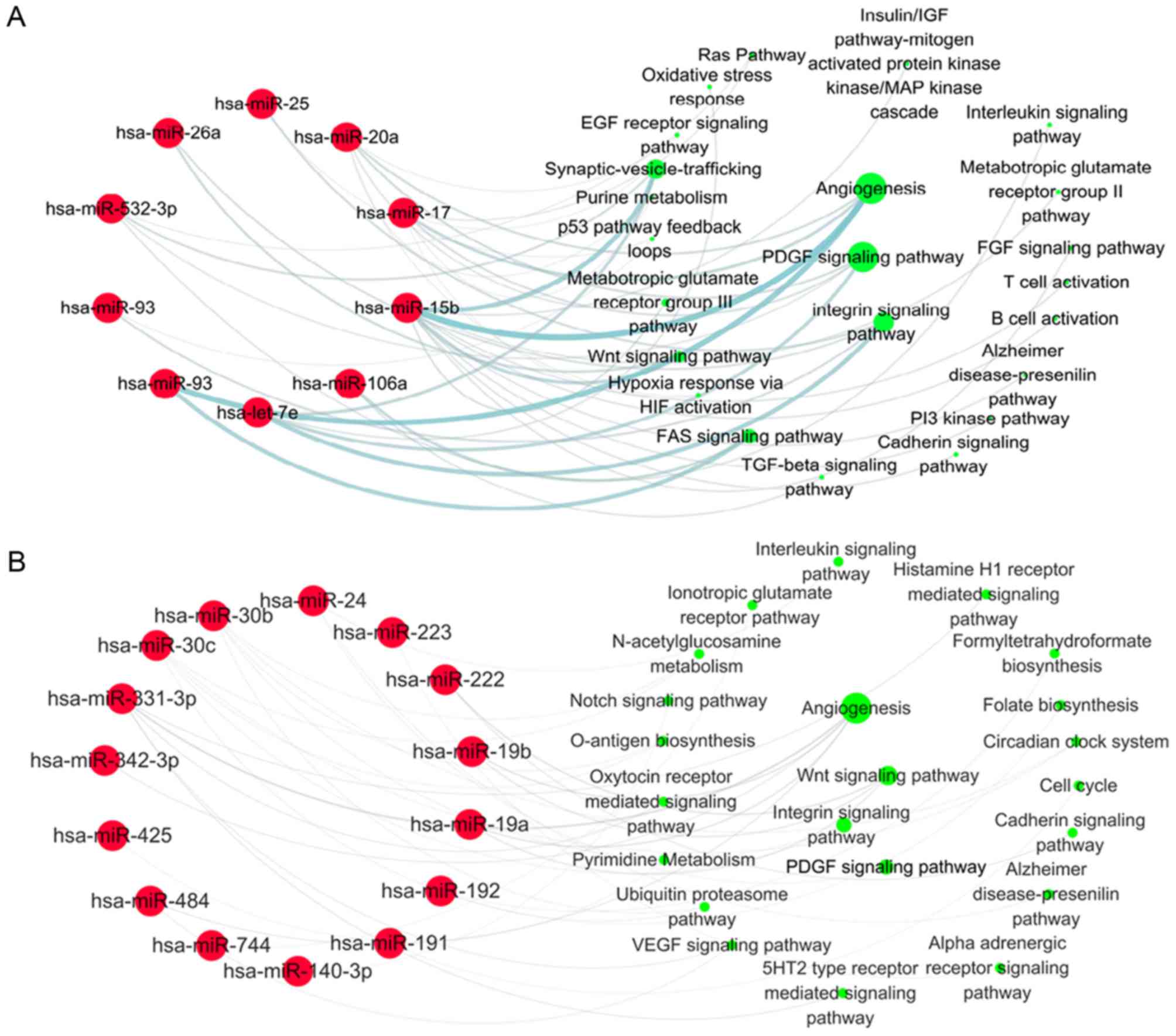
Subsequently, the target pathways of 33 upregulated
miRNAs in statin-treated UA patients were analyzed by the same
bioinformatic method. The results demonstrated that the target
genes of 13 initially downregulated miRNAs were primarily involved
in angiogenesis (regulated by miR-15b, 17, 20a, 93), the PDGF
signaling pathway (regulated by let-7e, miR-17, 20a, 532-3p) and
the integrin signaling pathway (regulated by let-7e, miR-25, 26a,
92a; Fig. 3A). Although 20 nascent
miRNAs were primarily involved in angiogenesis (regulated by
miR-19a, 19b, 331-3p, 342-3p, 484) and the Wnt signaling pathway
(regulated by miR-19a, 19b, 222; Fig.
3B), the first three signaling pathways targeted by 33
statin-induced miRNAs were still the angiogenesis, integrin and
PDGF signaling pathways. A total of 25 of 33 miRNAs were involved
in regulating these biological pathways in UA patients treated with
statins (Fig. 3). The effects of
these signaling pathways were inhibited by statins in UA patients
by upregulation of associated miRNAs, which suggested the
atheroprotective effects of statins in UA patients.
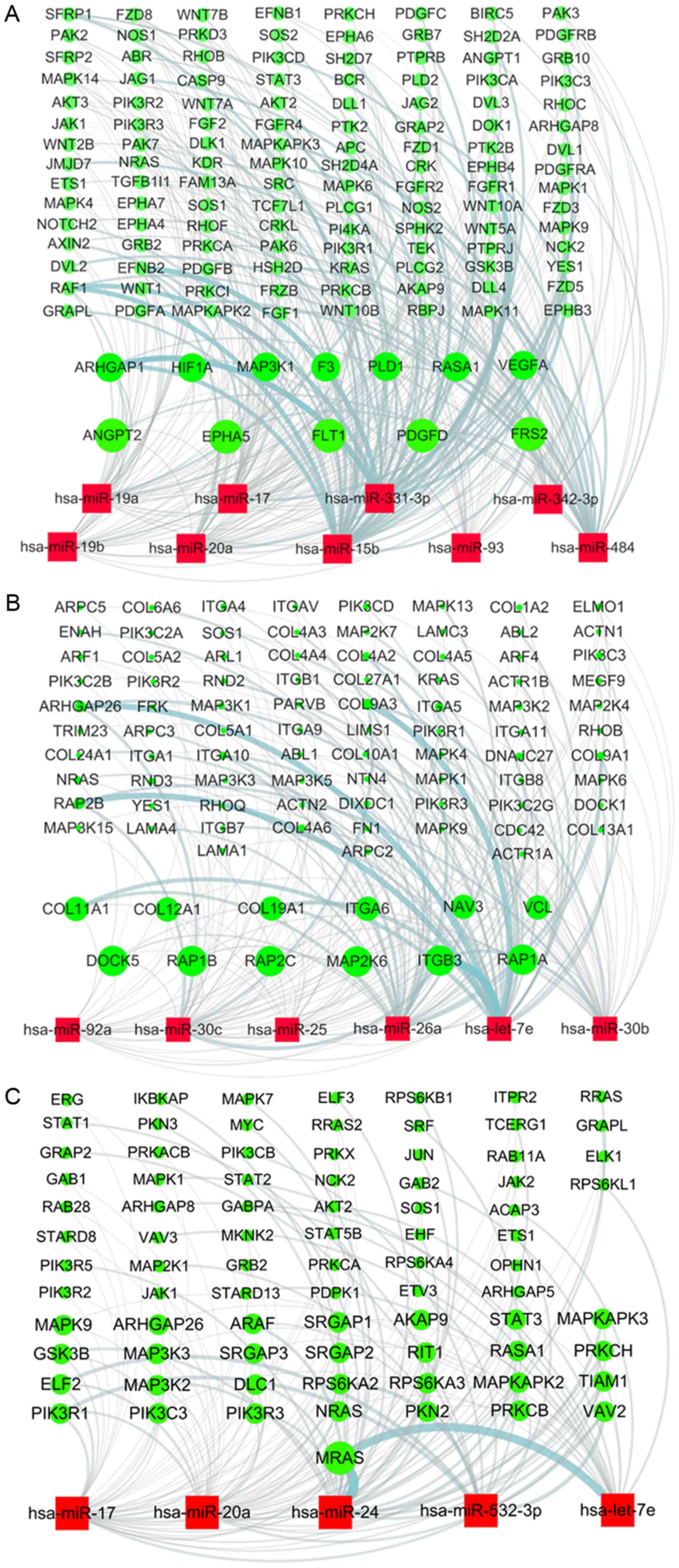
Target genes of statin-induced
differentially expressed miRNAs involved in the angiogenesis,
integrin and PDGF signaling pathways
Since the angiogenesis, integrin and PDGF signaling
pathways are three of the most important pathways targeted by
upregulated miRNAs induced by statins in UA patients, the miRNAs
and their target genes involved were analyzed. Each group of target
genes of 33 miRNAs was entered into the DAVID platform, and then
the angiogenesis, integrin and PDGF signaling pathways were
obtained as well as the relevant genes and miRNAs in the PANTHER
database. Bioinformatic analysis revealed that 9 of 33 miRNAs were
involved in the angiogenesis pathway referring to 132 target genes
(Fig. 4A), 6 of 33 miRNAs in the
integrin signaling pathway by targeting 95 genes (Fig. 4B), and 5 of 33 miRNAs in the PDGF
signaling pathway including 81 target genes (Fig. 4C). By upregulating miRNA levels,
statins may suppress the expression of relevant genes to inhibit
the angiogenesis, integrin and PDGF signaling pathways in UA
patients.
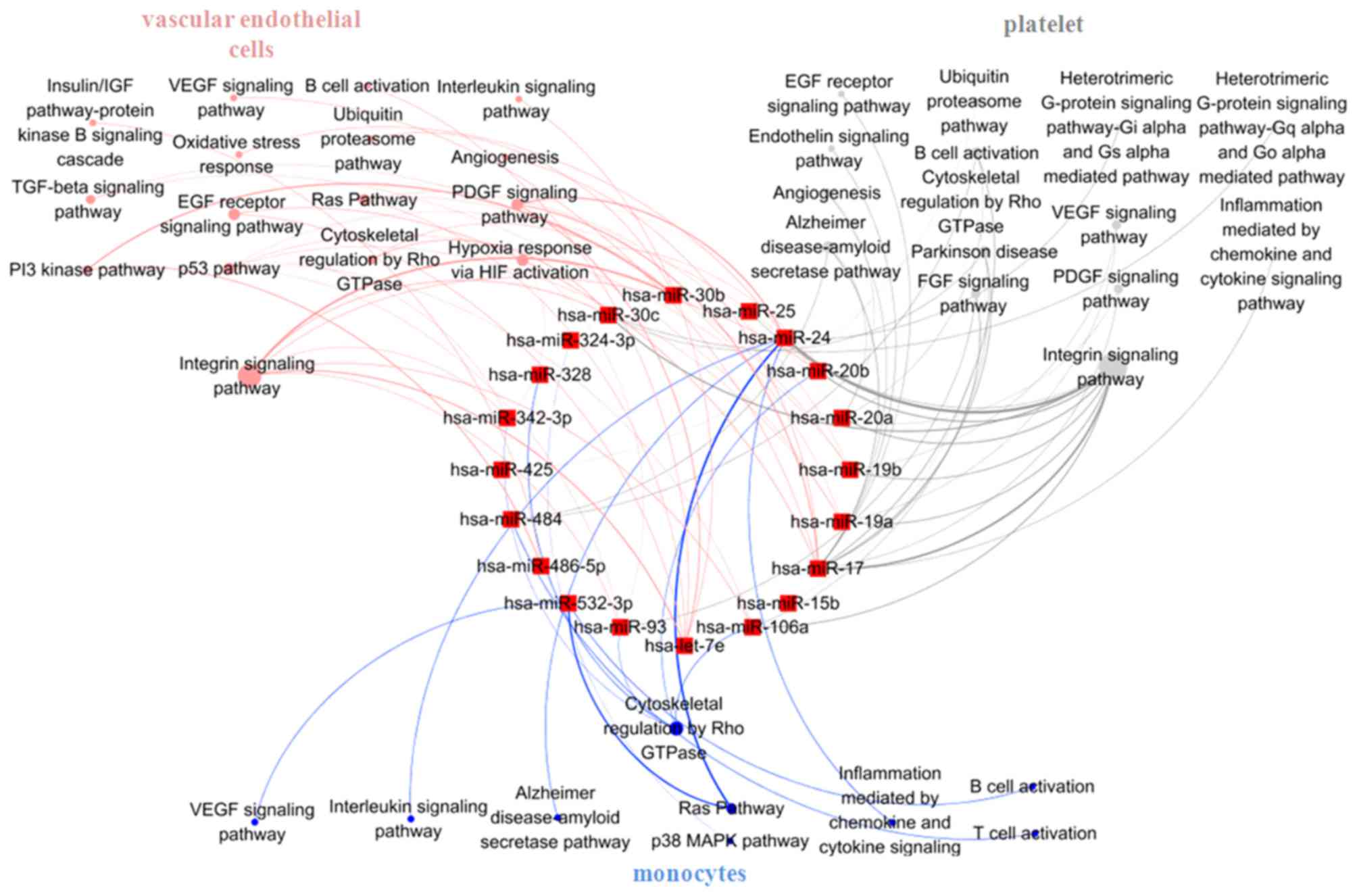
Signaling pathways analysis in
unstable plaque-associated cell types of UA patients treated with
statins
Vascular endothelial cells, monocytes and platelets
are the main sources of circulating miRNAs in CAD patients
(26) and these three cell types
are predominantly involved in the formation of unstable plaque and
plaque rupture. Therefore, to clarify the role of statin in the
formation of unstable plaque, the target pathways of 33
statin-induced miRNAs were analyzed in vascular endothelial cells,
monocytes and platelets, separately. Target genes of the 33 miRNAs
in the three different cell types were obtained from the CGAP SAGE
database (for monocytes and endothelial cells) and a UP
tissue-specific database (for platelets). These genes were then
entered into the DAVID platform and the signaling pathways in the
PANTHER database. The results indicated that differentially
expressed miRNAs induced by statins mainly targeted integrin
signaling pathways both in vascular endothelial cells (regulated by
let-7e, miR-17, 19a, 19b, 20a, 20b, 24, 30b, 30c, 93, 106a, 342-3p,
486-5p) and platelets (regulated by miR-15b, 17, 19a, 19b, 20a,
20b, 24, 25, 93, 30c, 106a, 425, 484; Fig. 5), and cytoskeletal regulation by
Rho GTPase pathway in monocytes (regulated by miR-20b, 24, 93,
106a, 324-3p, 328, 342-3p, 484, 532-3p; Fig. 5). Statins may inhibit
atherosclerosis progression by influencing the effects of different
signaling pathways in unstable plaque-related cells mediated by
miRNAs.
Discussion
Statins serve an important role in the prevention
and treatment of cardiovascular diseases due to their pleiotropic
effects. It has been previously reported that statins improve
endothelial function in patients with CAD (8), alleviating inflammation in the aorta
of hypercholesterolaemic atherosclerotic rabbits (27) and increasing NO synthesis in rat
vascular smooth muscle cells (28). Previous studies have demonstrated
that statins can influence cellular biological activity by
regulating the expression of particular miRNAs. For example,
simvastatin can decrease miR-155 expression through interfering
with the mevalonate-geranylgeranyl-pyrophosphate-RhoA signaling
pathway, and then increasing endothelial nitric oxide synthase
expression and endothelium-dependent vasodilation (21). Atorvastatin treatment increased
angiogenesis-associated miR-221, miR-222 and miR-92a expression in
endothelial progenitor cells (23)
and inhibited immune response by downregulating toll-like receptor
4 signaling by inducing let-7i expression in monocytes from CAD
patients (24). However, whether
statins serve systematic biological roles in CAD patients by
regulating the miRNAs network remains to be elucidated. The present
study demonstrated that in UA patients, statins may exert
pleiotropic effects in endothelial cells, platelets and monocytes
by influencing the blood-borne miRNA regulatory network.
The present study first examined the miRNA
expression profile in the whole blood of non-statin-treated UA
patients and non-CAD controls. The TLDA results demonstrated that
there were 21 differentially expressed miRNAs in non-statin-treated
patients compared with controls. The majority of the miRNAs were
downregulated and mainly targeted angiogenesis, p53 pathway
feedback loops, integrin and PDGF signaling pathways, which
suggested the pathological states of UA patients at the molecular
level. The function enhancement of the four signaling pathways may
partially explain atherosclerotic plaque progression in UA patients
(29–35). Nevertheless, compared with the UA
patients without statin treatment, there were 33 upregulated miRNAs
in statin-treated UA patients. The 33 upregulated miRNAs were
composed of 13 initially downregulated miRNAs in non-statin-treated
UA patients and 20 nascent miRNAs.
In order to understand the biological role of statin
in UA patients, the signaling pathways mediated by the
differentially expressed miRNAs were next analyzed. Bioinformatic
analysis revealed that the 33 upregulated miRNAs induced by statin
were primarily involved in angiogenesis, integrin and PDGF
signaling pathways. Consistent with these observations, statins
were demonstrated to inhibit inflammation/hypoxia-induced
angiogenesis in endothelial cells or mice which may protect against
plaque inflammatory angiogenesis and rupture (36–39),
to reduce monocytes or hepatocellular carcinoma adhering to
endothelium by interfering in the integrin signaling pathway
(40,41) and to suppress PDGF-mediated
vascular smooth muscle proliferation and migration (42–44).
A further target signaling pathways analysis of 33 upregulated
miRNAs in atherosclerosis-associated vascular endothelial cells,
platelets and monocytes demonstrated that statins primarily
regulate the integrin signaling pathway in vascular endothelial
cells and platelets, and mediate cytoskeletal regulation by the Rho
GTPase pathway in monocytes, which was also confirmed to be
associated with statins (45). The
above results suggested that statins may facilitate atherosclerotic
plaque stability through inhibiting angiogenesis,
atherosclerosis-associated cell proliferation, monocyte migration,
platelet adhesion and the coagulation cascade, mediated by
circulating miRNAs.
The target genes of 33 upregulated miRNAs involved
in the angiogenesis, integrin and PDGF signaling pathways were
extracted. Bioinformatic analysis demonstrated that the targets in
angiogenesis mainly included PDGF D, fms related tyrosine kinase 1,
vascular endothelial growth factor receptor 1, fibroblast growth
factor receptor substrate 2, ephrin type-a receptor 5 and
angiopoietin-2. In the integrin signaling pathway they mainly
included dedicator of cytokinesis protein 5, dual specificity
mitogen-activated protein kinase kinase 6, integrin β-3,
ras-related protein Rap-1A and ras-related protein Rap-2C. The PDGF
signaling pathway included ras-related protein M-Ras,
mitogen-activated protein kinase kinase kinase 2/3, protein kinase
C η type, mitogen-activated protein kinase 9, rho GTPase-activating
protein 26, rho GTPase-activating protein1/2/3, rho
GTPase-activating protein 7, ras GTPase- activating protein 1,
GTP-binding protein Rit 1, ribosomal protein S6 kinase α-2/3 and
signal transducer and activator of transcription 3. These targets
are all essential genes in the pathological process of plaque
progression. Statins may reduce the expression of these genes by
directly affecting miRNA levels in blood vessel cells and blood
cells. Statins may also affect the release of miRNAs from the above
cells, and subsequently enter into recipient cells and regulate
their bioactivities by acting on their target genes via cell-cell
communication (19).
In conclusion, the findings of the present study
suggested that statins may exert protective effects on plaque
stability by regulating the blood-borne miRNA network in UA
patients. The definite role of statins in miRNA regulatory networks
requires further validation through further biological
experiments.
Acknowledgements
The present study was supported by National Natural
Science Foundation of China (grant nos. 81600340, 81270274,
81470473, 81400265 and 81400264) and Beijing Municipal Science and
Technology Project (grant no. D141100000114002).
References
|
1
|
Heidenreich PA, Trogdon JG, Khavjou OA,
Butler J, Dracup K, Ezekowitz MD, Finkelstein EA, Hong Y, Johnston
SC, Khera A, et al: Forecasting the future of cardiovascular
disease in the United States: A policy statement from the American
Heart Association. Circulation. 123:933–944. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
LaRosa JC, Grundy SM, Waters DD, Shear C,
Barter P, Fruchart JC, Gotto AM, Greten H, Kastelein JJ, Shepherd
J, et al: Treating to New Targets (TNT) Investigators: Intensive
lipid lowering with atorvastatin in patients with stable coronary
disease. N Engl J Med. 352:1425–1435. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cannon CP, Braunwald E, McCabe CH, Rader
DJ, Rouleau JL, Belder R, Joyal SV, Hill KA, Pfeffer MA and Skene
AM: Pravastatin or Atorvastatin Evaluation and Infection
Therapy-Thrombolysis in Myocardial Infarction 22 Investigators:
Intensive versus moderate lipid lowering with statins after acute
coronary syndromes. N Engl J Med. 350:1495–1504. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schwartz GG, Olsson AG, Ezekowitz MD, Ganz
P, Oliver MF, Waters D, Zeiher A, Chaitman BR, Leslie S and Stern
T: Myocardial Ischemia Reduction with Aggressive Cholesterol
Lowering (MIRACL) Study Investigators: Effects of atorvastatin on
early recurrent ischemic events in acute coronary syndromes: The
MIRACL study: A randomized controlled trial. JAMA. 285:1711–1718.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wright RS, Anderson JL, Adams CD, Bridges
CR, Casey DE Jr, Ettinger SM, Fesmire FM, Ganiats TG, Jneid H,
Lincoff AM, et al: 2011 ACCF/AHA focused update incorporated into
the ACC/AHA 2007 Guidelines for the Management of Patients with
Unstable Angina/Non-ST-Elevation Myocardial Infarction: A report of
the American College of Cardiology Foundation/American Heart
Association Task Force on Practice Guidelines developed in
collaboration with the American Academy of Family Physicians,
Society for Cardiovascular Angiography and Interventions, and the
Society of Thoracic Surgeons. J Am Coll Cardiol. 57:e215–e367.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ray KK and Cannon CP: The potential
relevance of the multiple lipid-independent (pleiotropic) effects
of statins in the management of acute coronary syndromes. J Am Coll
Cardiol. 46:1425–1433. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Angeli F, Reboldi G, Mazzotta G, Garofoli
M, Cerasa MF and Verdecchia P: Statins in acute coronary syndrome:
Very early initiation and benefits. Ther Adv Cardiovasc Dis.
6:163–174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen H, Ren JY, Xing Y, Zhang WL, Liu X,
Wu P, Wang RJ and Luo Y: Short-term withdrawal of simvastatin
induces endothelial dysfunction in patients with coronary artery
disease: A dose-response effect dependent on endothelial nitric
oxide synthase. Int J Cardiol. 131:313–320. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu N and Olson EN: MicroRNA regulatory
networks in cardiovascular development. Dev Cell. 18:510–525. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jacobsen A, Silber J, Harinath G, Huse JT,
Schultz N and Sander C: Analysis of microRNA-target interactions
across diverse cancer types. Nat Struct Mol Biol. 20:1325–1332.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Price NL, Ramírez CM and
Fernández-Hernando C: Relevance of microRNA in metabolic diseases.
Crit Rev Clin Lab Sci. 51:305–320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De Rosa S, Curcio A and Indolfi C:
Emerging role of microRNAs in cardiovascular diseases. Circ J.
78:567–575. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hagiwara S, Kantharidis P and Cooper ME:
MicroRNA as biomarkers and regulator of cardiovascular development
and disease. Curr Pharm Des. 20:2347–2370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Creemers EE, Tijsen AJ and Pinto YM:
Circulating microRNAs: Novel biomarkers and extracellular
communicators in cardiovascular disease? Circ Res. 110:483–495.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loyer X, Vion AC, Tedgui A and Boulanger
CM: Microvesicles as cell-cell messengers in cardiovascular
diseases. Circ Res. 114:345–353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tabuchi T, Satoh M, Itoh T and Nakamura M:
MicroRNA-34a regulates the longevity-associated protein SIRT1 in
coronary artery disease: Effect of statins on SIRT1 and
microRNA-34a expression. Clin Sci (Lond). 123:161–171. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun HX, Zeng DY, Li RT, Pang RP, Yang H,
Hu YL, Zhang Q, Jiang Y, Huang LY, Tang YB, et al: Essential role
of microRNA-155 in regulating endothelium-dependent vasorelaxation
by targeting endothelial nitric oxide synthase. Hypertension.
60:1407–1414. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Minami Y, Satoh M, Maesawa C, Takahashi Y,
Tabuchi T, Itoh T and Nakamura M: Effect of atorvastatin on
microRNA 221/222 expression in endothelial progenitor cells
obtained from patients with coronary artery disease. Eur J Clin
Invest. 39:359–367. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Q, Kandic I and Kutryk MJ:
Dysregulation of angiogenesis-related microRNAs in endothelial
progenitor cells from patients with coronary artery disease.
Biochem Biophys Res Commun. 405:42–46. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Satoh M, Tabuchi T, Minami Y, Takahashi Y,
Itoh T and Nakamura M: Expression of let-7i is associated with
Toll-like receptor 4 signal in coronary artery disease: Effect of
statins on let-7i and Toll-like receptor 4 signal. Immunobiology.
217:533–539. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Leroyer AS, Isobe H, Lesèche G, Castier Y,
Wassef M, Mallat Z, Binder BR, Tedgui A and Boulanger CM: Cellular
origins and thrombogenic activity of microparticles isolated from
human atherosclerotic plaques. J Am Coll Cardiol. 49:772–777. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qiao Z, Ren J and Chen H: Simvastatin
reduces expression and activity of lipoprotein-associated
phospholipase A(2) in the aorta of hypercholesterolaemic
atherosclerotic rabbits. J Int Med Res. 37:1029–1037. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen H, Ikeda U, Shimpo M, Ikeda M, Minota
S and Shimada K: Fluvastatin upregulates inducible nitric oxide
synthase expression in cytokine-stimulated rat vascular smooth
muscle cells. Hypertension. 36:923–928. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yahagi K, Kolodgie FD, Otsuka F, Finn AV,
Davis HR, Joner M and Virmani R: Pathophysiology of native
coronary, vein graft, and in-stent atherosclerosis. Nat Rev
Cardiol. 13:79–98. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen
C, Cai Y, Huang H, Yang Y, Liu Y, et al: LincRNA-p21 regulates
neointima formation, vascular smooth muscle cell proliferation,
apoptosis, and atherosclerosis by enhancing p53 activity.
Circulation. 130:1452–1465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sayin VI, Khan OM, Pehlivanoglu LE,
Staffas A, Ibrahim MX, Asplund A, Agren P, Nilton A, Bergström G,
Bergo MO, et al: Loss of one copy of Zfp148 reduces lesional
macrophage proliferation and atherosclerosis in mice by activating
p53. Circ Res. 115:781–789. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang CK, Pang H, Wang L, Niu Y, Luo J,
Chang E, Sparks JD, Lee SO and Chang C: New therapy via targeting
androgen receptor in monocytes/macrophages to
battleatherosclerosis. Hypertension. 63:1345–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karshovska E, Zhao Z, Blanchet X, Schmitt
MM, Bidzhekov K, Soehnlein O, von Hundelshausen P, Mattheij NJ,
Cosemans JM, Megens RT, et al: Hyperreactivity of junctional
adhesion molecule A-deficient platelets acceleratesatherosclerosis
in hyperlipidemic mice. Circ Res. 116:587–599. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ricci C and Ferri N: Naturally occurring
PDGF receptor inhibitors with potential anti-atherosclerotic
properties. Vascul Pharmacol. 70:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Heldin CH: Targeting the PDGF signaling
pathway in the treatment of non-malignant diseases. J Neuroimmune
Pharmacol. 9:69–79. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li X, Liu X, Xu Y, He Y, Liu J and Xie M:
Expression profile of apoptotic and proliferative proteins in
hypoxic HUVEC treated with statins. Int J Oncol. 46:677–684.
2015.PubMed/NCBI
|
|
37
|
Araújo FA, Rocha MA, Mendes JB and Andrade
SP: Atorvastatin inhibits inflammatory angiogenesis in mice through
down regulation of VEGF, TNF-alpha and TGF-beta1. Biomed
Pharmacother. 64:29–34. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Massaro M, Zampolli A, Scoditti E,
Carluccio MA, Storelli C, Distante A and De Caterina R: Statins
inhibit cyclooxygenase-2 and matrix metalloproteinase-9 in human
endothelial cells: Anti-angiogenic actions possibly contributing to
plaque stability. Cardiovasc Res. 86:311–320. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Weis M, Heeschen C, Glassford AJ and Cooke
JP: Statins have biphasic effects on angiogenesis. Circulation.
105:739–745. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Weber C, Erl W, Weber KS and Weber PC:
HMG-CoA reductase inhibitors decrease CD11b expression and CD11b-
dependent adhesion of monocytes to endothelium and reduce increased
adhesiveness of monocytes isolated from patients with
hypercholesterolemia. J Am Coll Cardiol. 30:1212–1217. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Relja B, Meder F, Wang M, Blaheta R,
Henrich D, Marzi I and Lehnert M: Simvastatin modulates the
adhesion and growth of hepatocellular carcinoma cells via decrease
of integrin expression and ROCK. Int J Oncol. 38:879–885. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen S, Liu B, Kong D, Li S, Li C, Wang H
and Sun Y: Atorvastatin calcium inhibits phenotypic modulation of
PDGF-BB-induced VSMCs via down-regulation the Akt signaling
pathway. PLoS One. 10:e01225772015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yoshikawa M, Nakamura K, Nagase S,
Sakuragi S, Kusano KF, Matsubara H and Ohe T: Effects of combined
treatment with angiotensin II type 1 receptor blocker and statin on
stent restenosis. J Cardiovasc Pharmacol. 53:179–186. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang D, Yuan J, Liu G, Ling Z, Zeng H,
Chen Y, Zhang Y, She Q and Zhou X: Angiotensin receptor blockers
and statins could alleviate atrial fibrosis via regulating
platelet-derived growth factor/Rac1/nuclear factor-kappa B Axis.
Int J Med Sci. 10:812–824. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sawada N and Liao JK: Rho/Rho-associated
coiled-coil forming kinase pathway as therapeutic targets for
statins in atherosclerosis. Antioxid Redox Signal. 20:1251–1267.
2014. View Article : Google Scholar : PubMed/NCBI
|