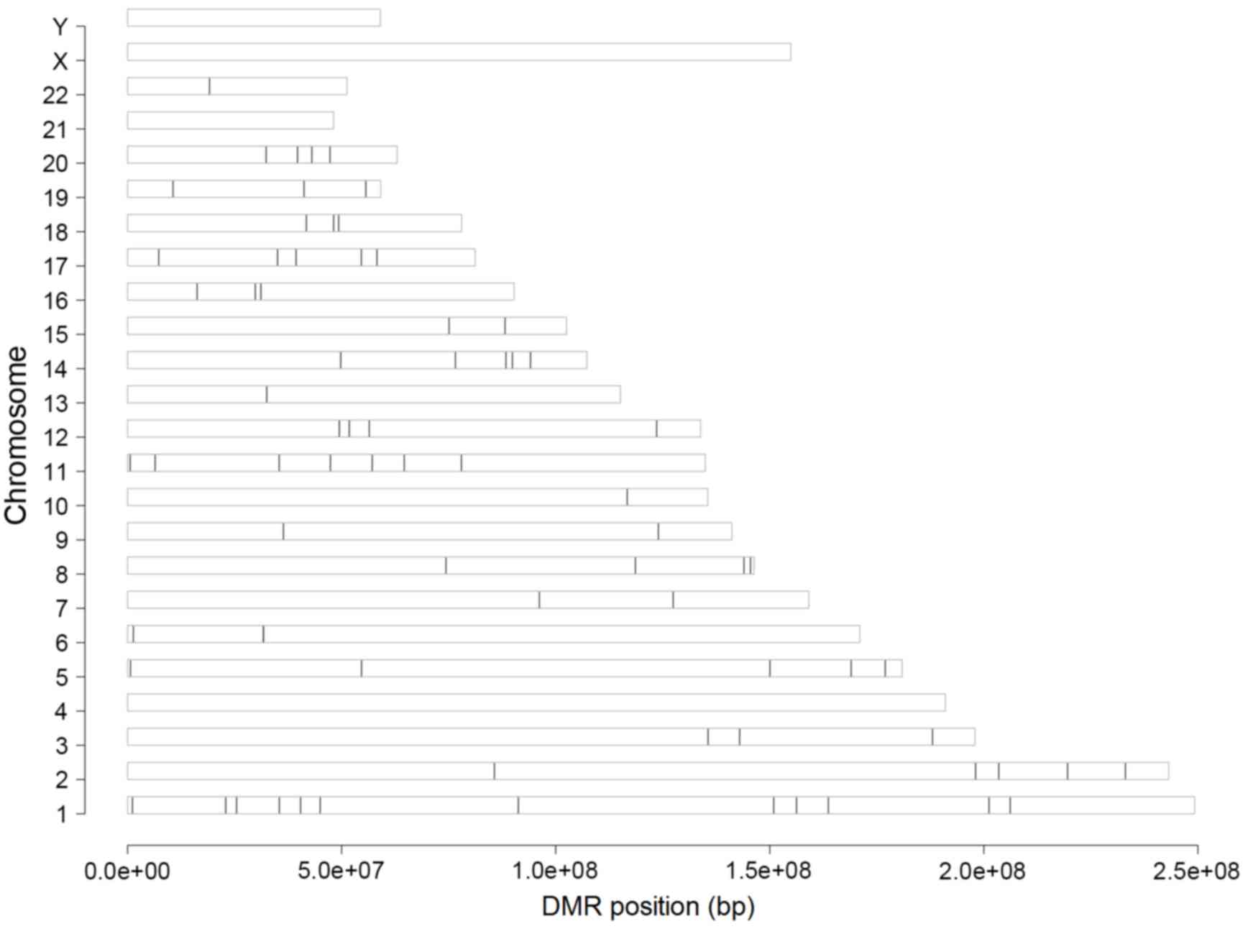
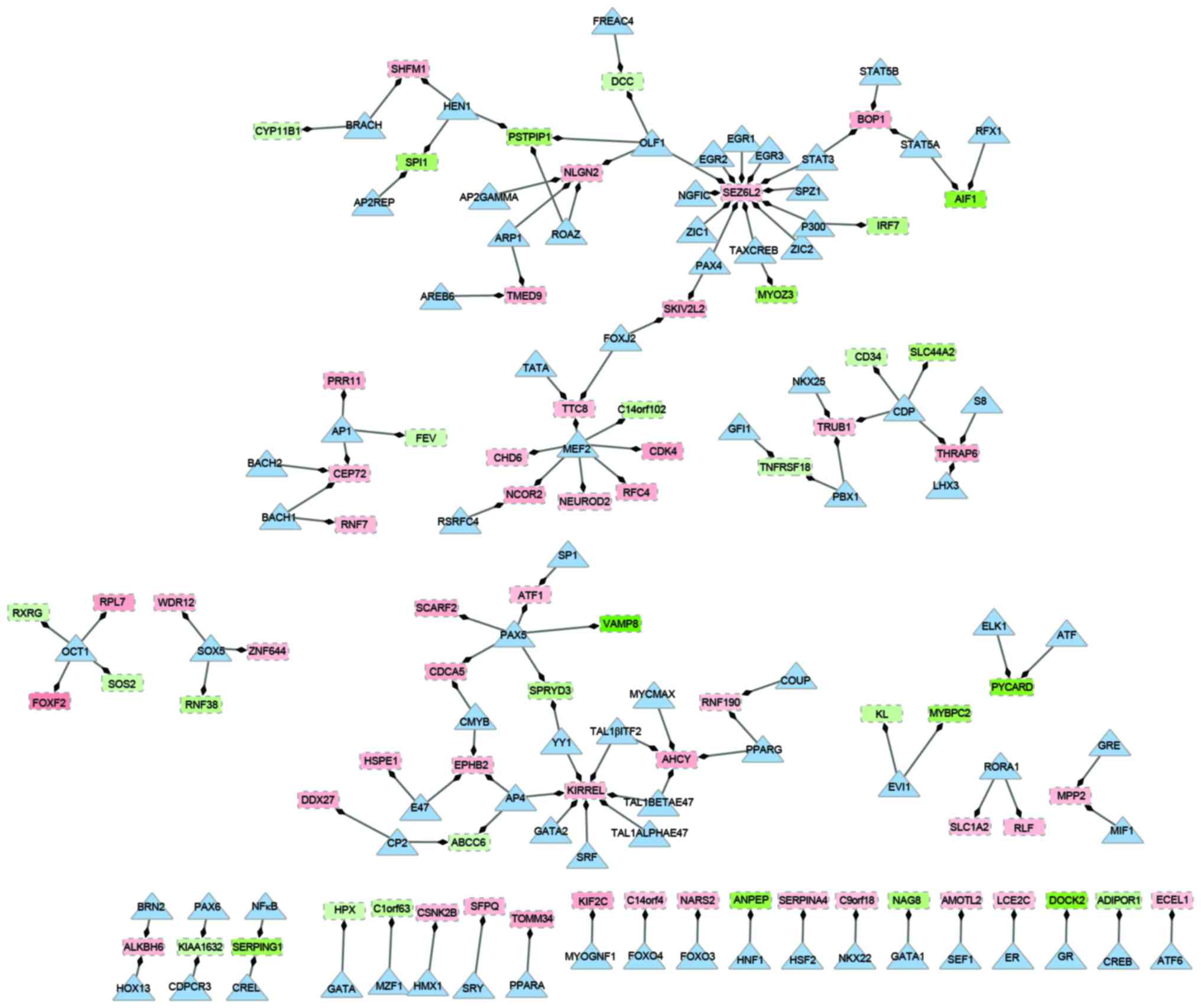
|
1
|
Luetke A, Meyers PA, Lewis I and Juergens
H: Osteosarcoma treatment-where do we stand? A state of the art
review. Cancer Treat Rev. 40:523–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jaffe N, Bruland OS and Bielack S:
Pediatric and adolescent osteosarcoma. Springer Science &
Business Media; 2010, View Article : Google Scholar
|
|
3
|
Buddingh EP, Anninga JK, Versteegh MI,
Taminiau AH, Egeler RM, van Rijswijk CS, Hogendoorn PC, Lankester
AC and Gelderblom H: Prognostic factors in pulmonary metastasized
high-grade osteosarcoma. Pediatr Blood Cancer. 54:216–221.
2010.PubMed/NCBI
|
|
4
|
Lewis IJ, Nooij MA, Whelan J, Sydes MR,
Grimer R, Hogendoorn PC, Memon MA, Weeden S, Uscinska BM, van
Glabbeke M, et al: Improvement in histologic response but not
survival in osteosarcoma patients treated with intensified
chemotherapy: A randomized phase III trial of the European
Osteosarcoma Intergroup. J Natl Cancer Inst. 99:112–128. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Berman SD, Calo E, Landman AS, Danielian
PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein
ML, et al: Metastatic osteosarcoma induced by inactivation of Rb
and p53 in the osteoblast lineage. Proc Natl Acad Sci USA.
105:11851–11856. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scott MC, Sarver AL, Phan F, Gupta R,
Thayanithy V, Subramanian S and Modiano F: Abstract B19: RB
function as a central component of osteosarcoma behavior: A
comparative assessment in dogs and humans. Molecular Cancer Res.
12:B192014. View Article : Google Scholar
|
|
7
|
Ternovoi VV, Curiel DT, Smith BF and
Siegal GP: Adenovirus-mediated p53 tumor suppressor gene therapy of
osteosarcoma. Lab Invest. 86:748–766. 2006.PubMed/NCBI
|
|
8
|
Scott MC, Sarver AL, Tomiyasu H, Cornax I,
Van Etten J, Varshney J, O'Sullivan MG, Subramanian S and Modiano
JF: Aberrant retinoblastoma (RB)-E2F transcriptional regulation
defines molecular phenotypes of osteosarcoma. J Biol Chem.
290:28070–28083. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Patiño-García A, Piñeiro ES, Díez MZ,
Iturriagagoitia LG, Klüssmann FA and Ariznabarreta LS: Genetic and
epigenetic alterations of the cell cycle regulators and tumor
suppressor genes in pediatric osteosarcomas. J Pediatr Hematol
Oncol. 25:362–367. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cui J, Wang W, Li Z, Zhang Z, Wu B and
Zeng L: Epigenetic changes in osteosarcoma. Bull Cancer.
98:E62–E68. 2011.PubMed/NCBI
|
|
11
|
Skarn M, Namlos HM, Ahmed D, Lind GE,
Meza-Zepeda LA and Myklebost O: Epigenetic regulation of miRNA
expression in osteosarcoma. Cancer Res. 72:1972012. View Article : Google Scholar
|
|
12
|
Kresse SH, Rydbeck H, Skarn M, Skårn M,
Namløs HM, Barragan-Polania AH, Cleton-Jansen AM, Serra M, Liestøl
K, Hogendoorn PC, et al: Integrative analysis reveals relationships
of genetic and epigenetic alterations in osteosarcoma. PLoS One.
7:e482622012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Xie G, Singh M, Ghanbarian AT,
Raskó T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al:
Primate-specific endogenous retrovirus-driven transcription defines
naive-like stem cells. Nature. 516:405–409. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Altman NS: An introduction to kernel and
nearest-neighbor nonparametric regression. The American
Statistician. 46:175–185. 1992. View
Article : Google Scholar
|
|
16
|
Hastie T, Tibshirani R, Narasimhan B and
Chu G: Impute: imputation for microarray data. Bioinformatics.
17:520–525. 2001.PubMed/NCBI
|
|
17
|
Barfield RT, Kilaru V, Smith AK and
Conneely KN: CpGassoc: An R function for analysis of DNA
methylation microarray data. Bioinformatics. 28:1280–1281. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meyer LR, Zweig AS, Hinrichs AS, Karolchik
D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al:
The UCSC Genome Browser database: Extensions and updates 2013.
Nucleic Acids Res. 41:D64–D69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Data Mining in Proteomics. Springer; pp. 291–303.
2011
|
|
20
|
Csardi G and Nepusz T: The igraph software
package for complex network research. Inter J Complex Systems.
1695:2006.
|
|
21
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei D, Le X, Zheng L, Wang L, Frey JA, Gao
AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL and Xie K: Stat3
activation regulates the expression of vascular endothelial growth
factor and human pancreatic cancer angiogenesis and metastasis.
Oncogene. 22:319–329. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ryu K, Choy E, Yang C, Susa M, Hornicek
FJ, Mankin H and Duan Z: Activation of signal transducer and
activator of transcription 3 (Stat3) pathway in osteosarcoma cells
and overexpression of phosphorylated-Stat3 correlates with poor
prognosis. J Orthop Res. 28:971–978. 2010.PubMed/NCBI
|
|
24
|
Liu LH, Li H, Li JP, Zhong H, Zhang HC,
Chen J and Xiao T: miR-125b suppresses the proliferation and
migration of osteosarcoma cells through down-regulation of STAT3.
Biochem Biophys Res Commun. 416:31–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Matsunoshita Y, Ijiri K, Ishidou Y, Nagano
S, Yamamoto T, Nagao H, Komiya S and Setoguchi T: Suppression of
osteosarcoma cell invasion by chemotherapy is mediated by urokinase
plasminogen activator activity via up-regulation of EGR1. PLoS One.
6:e162342011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhuo W, Ge W, Meng G, Jia S, Zhou X and
Liu J: MicroRNA-20a promotes the proliferation and cell cycle of
human osteosarcoma cells by suppressing early growth response 2
expression. Mol Med Rep. 12:4989–4994. 2015.PubMed/NCBI
|
|
27
|
Sellin L, Huber TB, Gerke P, Quack I,
Pavenstädt H and Walz G: NEPH1 defines a novel family of podocin
interacting proteins. FASEB J. 17:115–117. 2003.PubMed/NCBI
|
|
28
|
Durcan PJ, Al-Shanti N and Stewart CE:
Identification and characterization of novel Kirrel isoform during
myogenesis. Physiol Rep. 1:e000442013. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duelli D and Lazebnik Y: Cell fusion: A
hidden enemy? Cancer Cell. 3:445–448. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pawelek JM and Chakraborty AK: The cancer
cell-leukocyte fusion theory of metastasis. Adv Cancer Res.
101:397–444. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rechache NS, Wang Y, Stevenson HS, Killian
JK, Edelman DC, Merino M, Zhang L, Nilubol N, Stratakis CA, Meltzer
PS and Kebebew E: DNA methylation profiling identifies global
methylation differences and markers of adrenocortical tumors. J
Clin Endocrinol Metab. 97:E1004–E1013. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jackstadt R, Röh S, Neumann J, Jung P,
Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A
and Hermeking H: AP4 is a mediator of epithelial-mesenchymal
transition and metastasis in colorectal cancer. J Exp Med.
210:1331–1350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bébien M, Salinas S, Becamel C, Richard V,
Linares L and Hipskind RA: Immediate-early gene induction by the
stresses anisomycin and arsenite in human osteosarcoma cells
involves MAPK cascade signaling to Elk-1, CREB and SRF. Oncogene.
22:1836–1847. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsai FY and Orkin SH: Transcription factor
GATA-2 is required for proliferation/survival of early
hematopoietic cells and mast cell formation, but not for erythroid
and myeloid terminal differentiation. Blood. 89:3636–3643.
1997.PubMed/NCBI
|
|
35
|
Kang JU, Koo SH, Kwon KC, Park JW and Kim
JM: Gain at chromosomal region 5p15.33, containing TERT, is the
most frequent genetic event in early stages of non-small cell lung
cancer. Cancer Genet Cytogenet. 182:1–11. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cho HJ, Kang JH, Kwak JY, Lee TS, Lee IS,
Park NG, Nakajima H, Magae J and Chang YC: Ascofuranone suppresses
PMA-mediated matrix metalloproteinase-9 gene activation through the
Ras/Raf/MEK/ERK-and Ap1-dependent mechanisms. Carcinogenesis.
28:1104–1110. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Trovesi C, Manfrini N, Falcettoni M and
Longhese MP: Regulation of the DNA damage response by
cyclin-dependent kinases. J Mol Biol. 425:4756–4766. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Finn RS, Crown JP, Lang I, Boer K,
Bondarenko IM, Kulyk SO, Etti J, Patel R, Pinter T, Schmidt M, et
al: Results of a randomized phase 2 study of PD 0332991, a
cyclin-dependent kinase (CDK) 4/6 inhibitor, in combination with
letrozole vs. letrozole alone for first-line treatment of
ER+/HER2-advanced breast cancer (BC). Cancer Res. 72:S1–S6. 2012.
View Article : Google Scholar
|
|
39
|
Grivennikov SI and Karin M: Dangerous
liaisons: STAT3 and NF-kappaB collaboration and crosstalk in
cancer. Cytokine Growth Factor Rev. 21:11–19. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ishdorj G, Johnston JB and Gibson SB:
Cucurbitacin-I (JSI-124) activates the JNK/c-Jun signaling pathway
independent of apoptosis and cell cycle arrest in B leukemic cells.
BMC Cancer. 11:2682011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oshimori N, Li X, Ohsugi M and Yamamoto T:
Cep72 regulates the localization of key centrosomal proteins and
proper bipolar spindle formation. EMBO J. 28:2066–2076. 2009.
View Article : Google Scholar : PubMed/NCBI
|