Introduction
The prevalence of diabetes is increasing rapidly,
and type 2 diabetes now accounts for 20–50% of cases of new-onset
diabetes in young people (1,2).
Diabetic peripheral neuropathy (DN) is the most common complication
in both type 1 and type 2 diabetes (3). To explore a more effective treatment
for DN, efforts have been focused on the molecular mechanisms
underlying the etiology of DN. Serine protease inhibitors, termed
serpins, are key regulators of many biological events (4). In blood, they circulate as inactive
proforms or zymogens, and once activated, are quickly and
irreversibly inhibited by circulating inhibitors, in particular by
serine protease inhibitors, termed serpins. The extracellular
serine protease inhibitor E2 (SerpinE2), also called Protease
Nexin-1 (PN-1) belongs to the Serpin gene superfamily (5). Santoro et al (6) demonstrated that SerpinE2 prevents
cartilage catabolism by inhibiting the expression of matrix
metalloproteinase 13, one of the most relevant collagenases,
involved in cartilage breakdown in OA. Another study using
PN-1-deficient mice revealed that SerpinE2 confers antithrombotic
and antifibrinolytic properties that had previously not been
recognized (4). However, the
effect of this factor upon diabetes neuropathy remains unknown.
MicroRNAs (miRNAs or miRs) are a family of
non-coding RNAs that inhibit gene expression by interacting
directly with the 3′-untranslated regions (3′-UTRs) of target mRNAs
(7,8). Through these interactions, they can
inhibit translation of the targeted mRNAs or degrade them (9). miRNAs have multiple functions,
affecting many aspects of all kinds of diseases. Although there is
much evidence linking altered miRNA expression with various cancers
(10,11), the role of miRNAs in diabetes has
not been sufficiently examined. Through suppressing key genes
involved in disease development and progression, miRNAs can affect
many disease-related signaling pathways via local or systemic
regulation (12). The association
between changes in miRNA expression and the development of diabetes
neuropathy presents us with a new angle to explore pathogenesis and
progression of diabetes: In diabetes, several miRNAs including
miR-590-3p, miR-155 and miR-323b-5p have been demonstrated to be
associated with genesis and prognosis of diabetes patients
(13–15). Limited studies have been performed
on miRNA expression profiles in patients with DN, especially in
DN-induced pain, and miRNA expression has not previously been
linked to SerpinE2 (16).
In the present study, miRNA-199a-3p expression was
detected in the plasma of patients with diabetes and healthy
controls in order to study the pathogenesis of diabetes. In
addition, miRNA-199a-3p expression was examined in lower limb
tissue samples isolated from patients with DN and from healthy
volunteers. Finally, the molecular mechanisms underlying the
effects of miRNA-199a-3p in DN were examined using endothelial cell
lines.
Materials and methods
Patients and samples
Plasma was collected from 60 patients who have a
family history of type II diabetes, at the Department of Pain
Management, The Central Hospital of Wuhan (Wuhan, China) between
May 2014 and March 2015. The 60 diabetes patients were aged 35–65
and included 32 female and 28 male patients. All diabetes patients
had at least one parent or sibling who also suffered from diabetes.
Plasma samples isolated from 5 healthy volunteers were used as
controls. Lower limb skin samples from 30 DN patients and 20
volunteer samples were collected between July 2011 and October
2015. All samples were cut into small pieces and stored at −80°C.
Written consent was obtained from all patients and prior approval
for the study was obtained from the Institutional Research Ethics
Committee of Wuhan Central Hospital.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted from plasma obtained from patients
and control volunteers using the mirVana PARIS kit (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). RNA
was then reverse transcribed using the PrimeScript™
RT-PCR kit (Takara Bio, Inc., Otsu, Japan) and qPCR performed using
Real Time PCR Master Mix SYBR-Green PCR (Toyobo Co., Ltd., Osaka,
Japan). Thermocycling conditions were as follows: A total of 45
cycles of denaturation at 95°C for 15 sec, annealing at 55°C for 30
sec, and extension at 72°C for 30 sec. U6 was used as the internal
control. The primers used were as follows: miR-199a-3p, forward
5′-GCGGCGGACAGTAGTCTGCAC-3′, reverse 5′-ATCCAGTGCAGGGTCCGAGG-3′;
U6, forward 5′-CTCGCTTCGGCAGCACA-3′, reverse
5′-AACGCTTCACGAATTTGGT-3′. Relative miRNA expression levels were
calculated using the 2−ΔΔCq method (17). All assays were carried out in
triplicate.
Total RNA from DN lower limb skins and control group
tissues was obtained using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Total RNA (1 µg) was reverse
transcribed and subjected to qPCR using the same method as that
used for plasma microRNA.
miRNA-199a-3p expression was measured using a CFX
Connect™ Real-Time PCR Detection System (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The process was repeated in
the CRL-4025 cell line to measure the transfection efficiency of
miR-199a-3p mimic and inhibitor.
Cell lines, cell culture and transient
transfection
CRL-4025, a human dermal microvascular endothelium
cell line, was purchased from the Typical Training Content
Preservation Committee Cell Bank, Chinese Academy of Sciences
(Shanghai, China). The cells were cultured in Vascular Cell Basal
Medium (cat. no. PCS-100-030; American Type Culture Collection,
Manassas, VA, USA), supplemented with Microvascular Endothelial
Cell Growth Kit-VEGF (cat. no. PCS-110-041; American Type Culture
Collection) and 12.5 µg/ml blasticidine and penicillin/streptomycin
at 37°C in a humidified atmosphere with 5% CO2. To
manipulate miR-199a-3p expression, miRNA mimics
(5′-UACCCCUCCCCCCAUCCCGCCUGCCCACCCCCCCCCCCCCCCCGUGUUCAGACUACCUGUUCAGGAAGUAGUGGUUGUACAGUAGUCUGCACAUUGGUUAGGCUGGUUAGGGAAGUGCG-3′)
and inhibitor
(5′-AAGAACCUGCUCCGUCGCCCCAGUGUUCAGACUACCUGUUCAGGACAAUGCUGUUGUACAGUAGUCUGCACAUUGGUUAGACUGGGCAUGGGACAG-3′;
Thermo Fisher Scientific, Inc.) were used to transiently transfect
CRL-4025 cell lines. Small interfering RNA (siRNA) targeting
different coding regions of human SerpinE2 and its scrambled siRNA
sequence (NC) were synthesized by Thermo Fisher Scientific Inc. The
siRNA sequences were as follows: siRNA1, sense
5′-AAGACATTGTGACAGTGGCTA-3′, antisense 3′-TTCTGTAACACTGTCACCGAT-5′;
siRNA2, sense 5′-AAGACCATAGACAGCTGGATG-3′, antisense
3′-TTCTGGTATCTGTCGACCTAC-5′; and scramble siRNA, sense
5′-AAGACCAACTGACAGTGGCTA-3′ and antisense
3′-TTCTGTAATTAGGTCACCGAT-5′. Transfections were performed using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Cells were plated at a
density of 6×104 per well in a 6-well plate. The
expression of miR-199a-3p was measured 48 h after reaching
confluence; protein expression was measured 72 h after reaching
confluence.
Luc-UTR vectors and tumor protein p53
(P53)-expressing vector
The full-length SerpinE2 3′-UTR was cloned into the
Sac1 and Mlu1 sites of the pMIR-REPORT luciferase
vector (Ambion; Thermo Fisher Scientific, Inc.). A mutated vector
with the first 5 nucleotides complementary to the miR-199a-3p
seed-region mutated was constructed as a control.
Full-length P53 cDNA entirely lacking the 3′-UTR was
purchased from Gene Chem (Shanghai, CHINA) and subcloned into the
pcDNA3.1(+) vector (Invitrogen; Thermo Fisher Scientific, Inc.).
The blank pcDNA3.1(+) vector was applied as a negative control.
Western blot analysis
Cells were lysed using radioimmunoprecipitation
assay lysis buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40,
1% sodium deoxycholate, 0.1% SDS) for 30 min on ice, sonicated for
5–10 sec and then centrifuged at 12,000 × g for 20 min at 4°C.
Protein concentrations were determined using a DC™
Protein Assay (Bio-Rad Laboratories, Inc.). Equal amounts (40 µg)
of extracted protein samples were separated by 10% SDS-PAGE and
transferred onto nitrocellulose membranes. Membranes were blocked
with 2% milk in PBS containing 0.1% Tween-20 at room temperature
for 2 h, and subsequently probed with the following primary
antibodies overnight at 4°C: Anti-SerpinE2 (cat. no. ab154591;
1:1,000; Abcam, Cambridge, MA, USA), anti-tissue plasminogen
activator (tPA; cat. no. ab157469; 1:1,000; Abcam), anti-β-actin
(cat. no. sc-130300; 1:1,000; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and anti-GADPH (cat. no. ab37168; 1:1,000; Abcam).
Membranes were then incubated with the following horseradish
peroxidase-conjugated secondary antibodies for 2 h at room
temperature: Anti-rabbit (cat. no. A0545; 1:5,000; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) and anti-mouse (cat. no. A9044;
1:5,000; Sigma-Aldrich; Merck KGaA). The protein bands were
detected using an enhanced chemiluminescence detection system
(Pierce; Thermo Fisher Scientific, Inc.).
Immunohistochemistry
Immunohistochemical staining was performed on 4-µm
thick paraffin-embedded skin tissue sections with an anti-SerpinE2
primary antibody (cat. no. ab154591; 1:200; Abcam). Briefly,
sections were deparaffinized in xylene and hydrated with graded
ethanol. Antigen retrieval was performed using 0.01 mM citrate
buffer (pH 6.0) in a pressure cooker at 95°C for 20 min, and
endogenous peroxidase activity was blocked with incubation with 3%
hydrogen peroxide for 10 min at room temperature. Sections were
incubated with the primary antibody in a moist chamber overnight at
4°C, washed 3 times in PBS and then incubated with a horseradish
peroxidase-conjugated secondary antibody (cat. no. 31460; 1:5,000;
Thermo Fisher Scientific, Inc.) for 1 h at 37°C. Anibody-antigen
complexes were visualized using a 3,3′-diaminobenzidine detection
kit (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA).
Samples were counterstained with 100% hematoxylin for 10 sec at
room temperature. The mean percentage of SerpinE2-positive tumor
cells was determined under an inverted microscope. At least 5
random fields under ×400 magnification were assessed from each
section.
Luciferase activity of different
promoter constructs and chromatin immunoprecipitation (ChIP)
Transcriptional factor binding sites in the human
miR-199a-3p promoter region were predicted using the JASPAR 2016
server (http://jaspar.genereg.net) and the ECR
browser software (https://ecrbrowser.dcode.org/). Putative P53 binding
site: 5′-GGGCTTT-3′; −1747 bp to −1741 bp. Mutant P53 binding site:
5′-AAATGGG-3′. ChIP assays were performed on CRL-4025 cells
transfected with P53 or empty vector using the Magna ChIP Assay Kit
(EMD Millipore, Billerica, MA, USA). Protein-DNA complexes were
precipitated with normal IgG (cat. no. I5006; 1:1,000;
Sigma-Aldrich; Merck KGaA) and anti-P53 (cat. no. P5813; 1:200;
Sigma-Aldrich) at 4°C overnight with rotation.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Comparisons of clinical-pathologies were performed by
the log-rank test and χ2 test. P<0.05 was considered
to indicate a statistically significant difference. Statistical
analysis was performed using SPSS statistical software (version
15.0; SPSS, Inc., Chicago, IL, USA).
Results
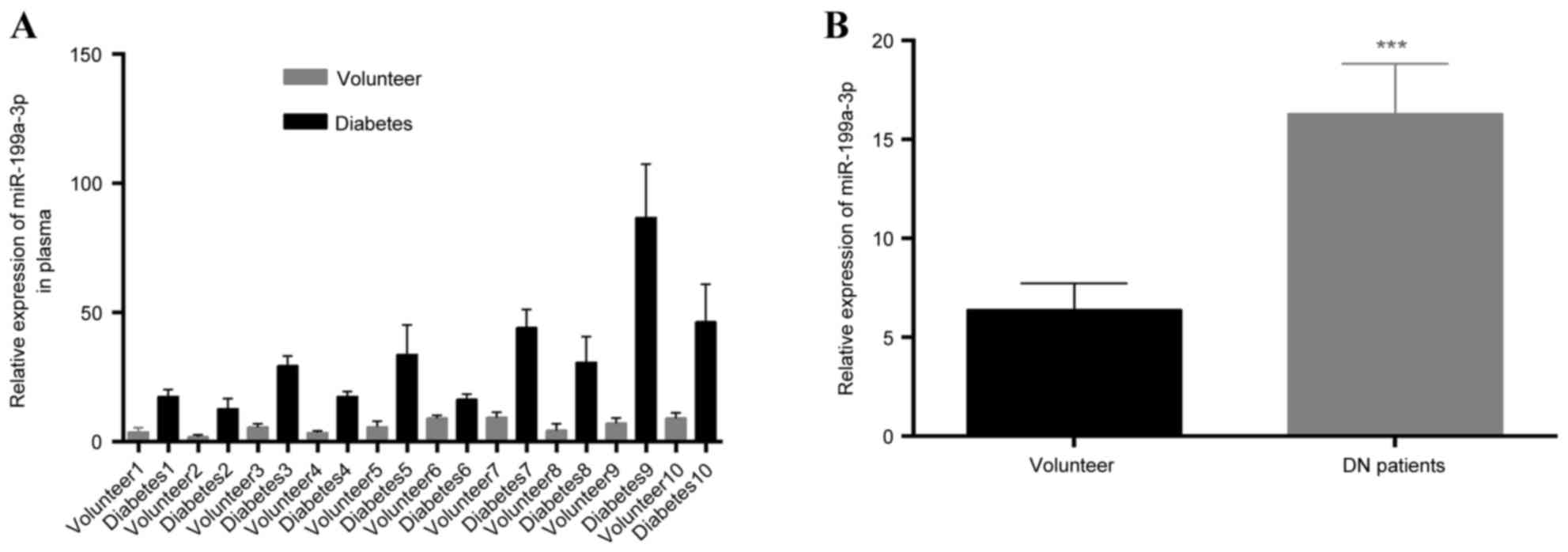
miR-199a-3p is upregulated in the
plasma of patients with a family history of type II diabetes
RT-qPCR was employed to detect miR-199a-3p
expression in the plasma of the 60 patients with diabetes and a
family history of diabetes and healthy volunteers. miR-199a-3p
expression appeared to be increased in patients with diabetes
compared with the controls (Fig.
1A). In addition, the expression of miR-199a-3p was measured in
30 DN lower limb skin samples and 20 volunteer tissues. We found
that miR-199a-3p expression was significantly increased in skin
tissues compared with volunteer tissues (P<0.05; Fig. 1B).
miR-199a-3p expression is positively
associated with clinical features of type 2 diabetes patients,
especially peripheral blood coagulation
To examine the relationships between miR-199a-3p
expression and biological characters of diabetes, the 60 patients
were divided into two groups according to miR-199a-3p expression
(Table I). High disease duration,
glycated hemoglobin (HbA1C) and fibrinogen (FIB) were demonstrated
to be associated with high miR-199a-3p expression (P<0.05;
Table I). This suggests that high
miR-199a-3p expression in diabetes patients may be considered as
indicative of DN progression.
 | Table I.miR-199a-3p expression in type 2
diabetes patients clinical and pathological properties. |
Table I.
miR-199a-3p expression in type 2
diabetes patients clinical and pathological properties.
| Parameters | High miR-199a-3p
expression, N=36 (%) | Low miR-199a-3p
expression, N=24 (%) |
χ2-test |
|---|
| Sex |
|
| P=0.853 |
| Male | (16) (44.4) | (10) (41.7) |
|
|
Female | (20) (55.6) | (14) (58.3) |
|
| Age (years) |
|
| P=0.203 |
|
≥45 | (22) (61.1) | (11) (45.8) |
|
|
<45 | (14) (38.9) | (13) (54.2) |
|
| Disease duration
(years) |
|
|
P=0.041a |
|
≥10 | (16) (44.4) | (3) (12.5) |
|
|
<10 | (20) (55.6) | (21) (87.5) |
|
| HbA1C (%) |
|
|
P=0.033a |
| ≥7 | (22) (61.1) | (9) (37.5) |
|
|
<7 | (14) (38.9) | (15) (62.5) |
|
| BMI |
|
| P=0.971 |
|
≥28 | (25) (69.4) | (17) (70.8) |
|
|
<28 | (11) (30.6) | (7) (29.2) |
|
| APTT |
|
| P=0.179 |
|
≥25 | (11) (30.6) | (11) (45.8) |
|
|
<25 | (25) (69.4) | (13) (54.2) |
|
| PT |
|
| P=0.076 |
|
≥11 | (10) (27.8) | (12) (50.0) |
|
|
<11 | (26) (72.2) | (12) (50.0) |
|
| FIB |
|
|
P=0.003a |
| ≥4 | (27) (75.0) | (5) (20.9) |
|
|
<4 | (9) (25.0) | (19) (79.1) |
|
miR-199a-3p promotes coagulation by
targeting SerpinE2 in dermal microvascular endothelium
To study the mechanism by which miR-199a-3p may act,
several computational algorithms, such as TargetScan version 7.0
(18), and miRBase (19–23),
were used to predict potential miR-199a-3p target sites. The seed
sequences of mature miR-199a-3p and the 3′-UTR of SERPINE2 mRNA
were matched (Fig. 2A). Following
transfection with a miR-199a-3p mimic, the expression of
miR-199a-3p was significantly upregulated, whereas transfection
with an inhibitor effectively suppressed the expression of
miR-199a-3p (Fig. 2B).
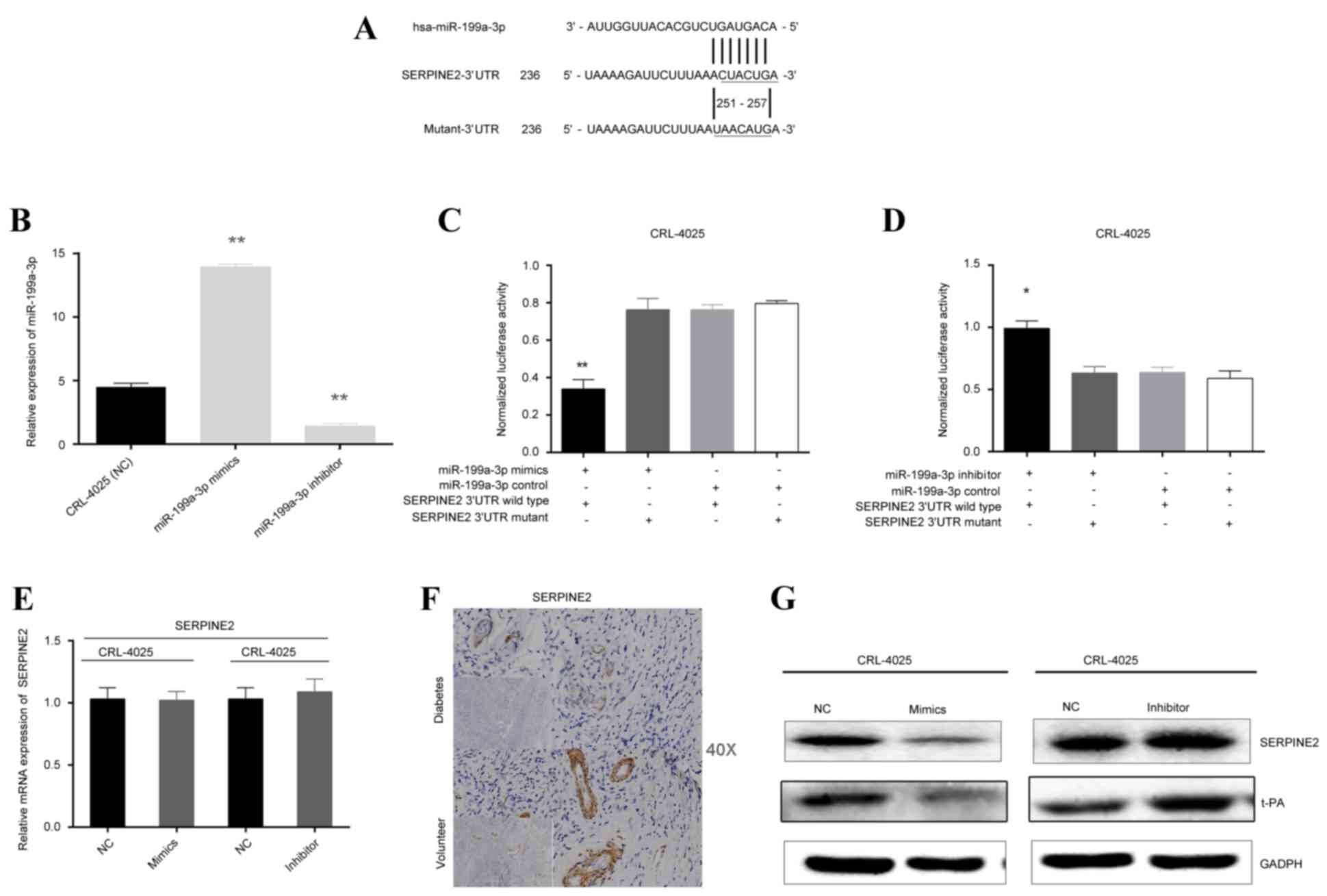 | Figure 2.SERPINE2 is a direct target of
miR-199a-3p. (A) Sequence alignment of miR-199a-3p with the
SERPINE2 3′-UTR. The seed-recognizing sites (underlined) in the
SERPINE2 sequence matched with the seed regions of miR-199a-3p. (B)
Following transfection of CRL-4025 cells with a miR-199a mimic, the
expression of miR-199a was upregulated, whereas transfection with
an inhibitor effectively suppressed the expression of miR-199a-3p.
**P<0.01 vs. NC. (C) Luciferase assay in CRL-4025 cells
demonstrated that miR-199a-3p mimics significantly suppressed
luciferase activity in wild type reporter constructs, compared with
NC. (D) miR-199a-3p inhibitor significantly promoted luciferase
activity in wild type constructs, compared with NC. *P<0.05 vs.
NC. (E) Reverse transcription-quantitative polymerase chain
reaction demonstrated that the level of SERPINE2 mRNA in CRL-4025
cells was not affected by transfection with miR-199a-3p mimics or
miR-199a-3p inhibitor. (F) Immunohistochemistry revealed that
SERPINE2 was downregulated in DN microvasculature compared with
volunteers. (G) Transfection of CRL-4025 cells with miR-199a-3p
mimics or miR-199a-3p inhibitor affects SERPINE2 and tPA protein
expression levels. Protein expression of SERPINE2 was negatively
associated with the expression of miR-199a-3p. All data are
presented as the mean ± standard deviation. SERPINE2, serine
protease inhibitor E2; miR, microRNA; UTR, untranslated region; NC,
negative control; DN, diabetic neuropathy; tPa, tissue plasminogen
activator. |
To confirm SERPINE2 as the target of miR-199a-3p,
the full-length SERPINE2 3′-UTR was subcloned into a luciferase
reporter vector (pMIR-REPORT β-galactosidase control vector).
Following transfection of SERPINE2 3′-UTR pMIR-REPORT into CRL-4025
cells, luciferase activity associated with SERPINE2 expression was
demonstrated to be inhibited by co-transfection with miR-199a-3p
mimics compared with co-transfection with miR-199a-3p control
(P<0.01; Fig. 2C). This
reduction in luciferase activity was also demonstrated to be
dependent on the presence of a wild type, rather than mutated,
SERPINE2 3′-UTR; this inhibition was abolished when the predicted
miR-199a-3p target sequences in the SERPINE2 3′-UTR were mutated
(Fig. 2A and C). Furthermore,
inhibition of endogenous miR-199a-3p by the inhibitor in CRL-4025
cells was able to increase luciferase expression compared with
control (P<0.05; Fig. 2D). The
changes in luciferase activity all occurred in the absence of any
changes in the mRNA expression level of SERPINE2 (Fig. 2E). Immunohistochemical analysis of
skin tissues also demonstrated the difference in SERPINE2
expression in microvascular endothelia cells between diabetic
patients and volunteers (Fig. 2F).
To directly assess the effect of miR-199a-3p on SERPINE2 protein
expression, miR-199a-3p was transfected into CRL-4025 cells the
protein expression levels of SERPINE2 detected by western blotting;
overexpression of miR-199a-3p was demonstrated to reduce SERPINE2
protein levels compared with NC (Fig.
2G). Conversely, transfection with miR-199a-3p inhibitor
increased SERPINE2 protein expression levels compared with NC
(Fig. 2G).
miR-199a-3p promotes coagulation by
targeting SerpinE2, thus inhibiting the tPA pathway
Since SERPINE2 lies upstream of the tPA signaling
pathway, the effect of miR-199a-3p dysregulation on this pathway
was examined in CRL-4025 cells. Increased miR-199a-3p in cells was
associated with decreased expression of tPA compared with NC,
whereas, inhibition of miR-199a-3p resulted in increased expression
of tPA compared with NC (Fig. 2G).
miR-199a-3p was, therefore, posited to affect downstream signaling
by targeting SERPINE2.
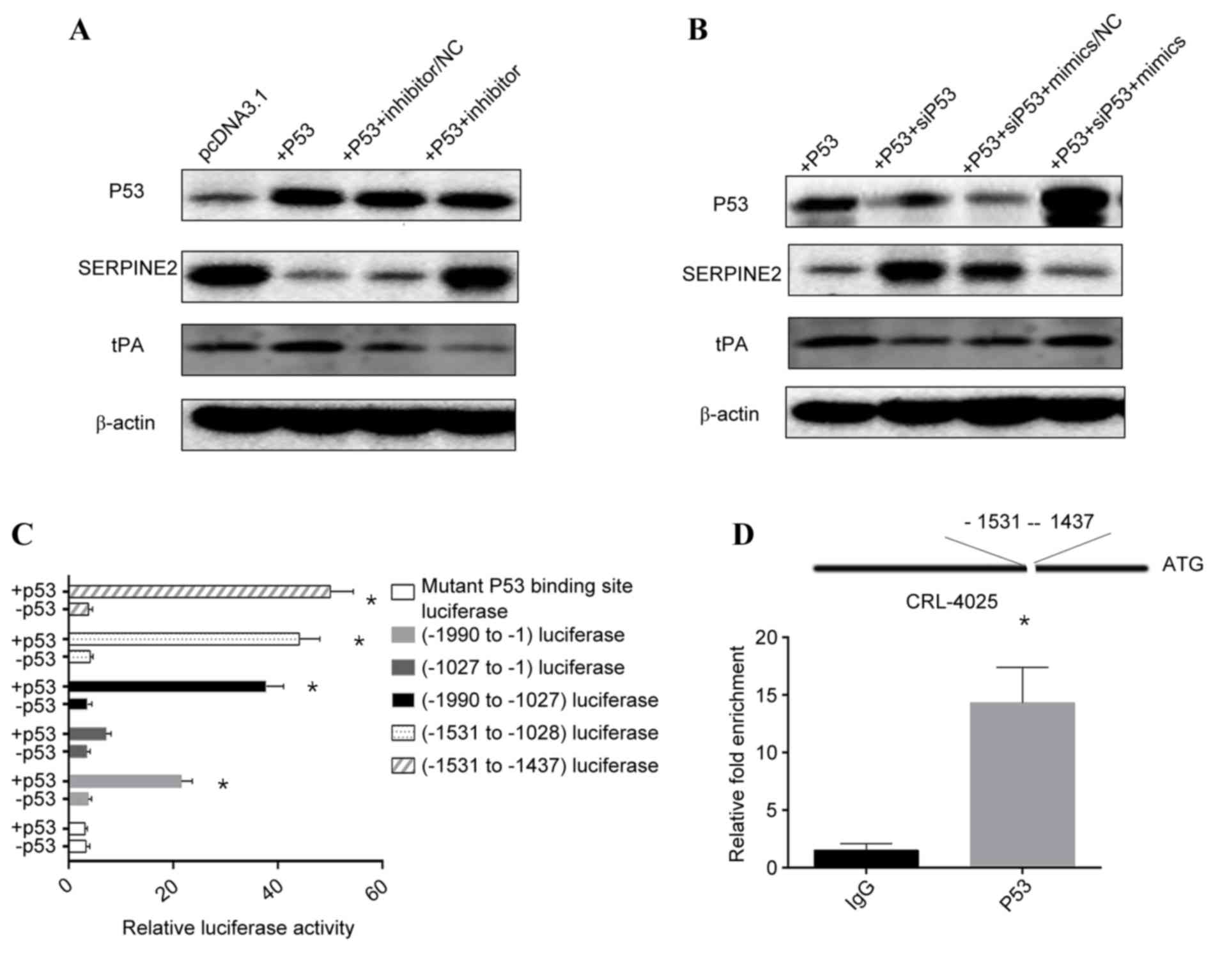
P53 activation of the promoter of
miR-199a-3p and downregulates SERPINE2
P53 was reported to regulate gene expression through
its interaction with transcriptional factors and is downregulated
in diabetes (13). Activation of β
cell glucokinase, initially triggers replication, then results in
apoptosis associated with DNA double-strand breaks and activation
of the tumor suppressor P53 (24).
P53 downregulates the expression of DNA methyltransferase 1 in NT2
cells, while overexpression of P53 restores the expression of
miR-199-3p/5p and miR-214 (25).
To determine how P53 regulates the expression of miR-199a-3p, the
effects of P53 on the expression of miR-199a-3p and SERPINE2 were
examined. Compared with control, overexpression of P53 in CRL-4025
cells decreased expression of SERPINE2 (Fig. 3A). By contrast, knockdown of P53
increased that of SERPINE2 compared with control (Fig. 3B). These results suggest that P53
is responsible for the regulation of miR-199a-3p and SERPINE2
expression. To determine whether P53 directly transcribes
miR-199a-3p, a putative P53-binding site upstream of the
transcriptional start site of miR-199a-3p was characterized: P53
stimulated the activity of the luciferase reporter containing the
putative P53-binding site but not the reporter with the mutated
binding site or without the putative P53-binding site (Fig. 3C). ChIP assays demonstrated the
miR-199a-3p promoter occupancy of P53 (Fig. 3D). Taken together, these data
strongly suggest that P53 activates miR-199a-3p transcription
through direct binding to its promoter.
Discussion
Since the discovery of miRNAs, many miRNAs have been
found to have different expression patterns in different diseases,
that are regulated by mechanisms as varied as the presence of
deletions, amplifications or mutations involving miRNA loci,
epigenetic silencing or the dysregulation of transcription factors
that target specific miRNAs (26,27).
There have been a number of miRNA-screening studies
in the plasma or serum of diabetes patients (28). Inducible transgenic overexpression
of miR-802 in mice causes impaired glucose tolerance and attenuates
insulin sensitivity, whereas reduction of miR-802 expression
improves glucose tolerance and insulin action (29). Another study suggested that a
defect in the NF-кB-miR-146a negative feedback loop may be involved
in the pathogenesis of DN through regulating TNF-α, interleukin 6
(IL-6), and interleukin 1β (IL-1β) in the sciatic nerve of diabetic
rats (30). The present study
examined miRNAs that are involved in DN occurrence from the
coagulation status angle. A previous study identified miR-1908,
miR-199a-5p and miR-199a-3p as endogenous promoters of metastatic
invasion, angiogenesis and colonization in melanoma by convergently
targeting apolipoprotein E and the heat shock factor DnaJ heat
shock protein family (Hsp40) member A4 (31). Furthermore, miR-199a-3p
dysregulation has been noted in liver cancer (32) and in leukemia (33). miR-199a-3p has been associated with
many kinds of cancer, but, to the best of our knowledge, its role
in diabetes has not previously been explored specifically. In the
present study, the expression of miR-199a-3p was upregulated in the
plasma of patients with diabetes and tissues of patients with DN
compared with volunteers. In addition, high expression was
significantly associated with high disease duration, HbA1C and
FIB.
Through manipulation of its expression, miR-199a-3p
was found to inhibit tPA in vitro through the regulation of
the expression of SERPINE2. SERPINE2 upregulates tPA, which is
associated with hypoxia-ischemia and excitotoxicity causes of DN
injuries and participates in the processes through proteolytic and
receptor-mediated pathways (34).
The present study observed downregulation of tPA protein expression
levels in endothelial cells and DN tissues, and tPA protein levels
were inversely associated with miR-199a-3p expression. Luciferase
activity assays demonstrated that miR-199a-3p bound to the 3′-UTR
of SERPINE2 mRNA. The effect of miR-199a-3p on SERPINE2 mRNA levels
was also investigated, and it was found that miR-199a-3p did not
modulate SERPINE2 mRNA expression levels, which indicated that
miR-199a-3p targets SERPINE2 through translation inhibition rather
than mRNA degradation. Furthermore, the miR-199a-3p-associated
inhibition of tPA expression could be offset by upregulating
SERPINE2 expression. Conversely, the increase in tPA expression
caused by downregulation of miR-199a-3p could be offset by
inhibition of SERPINE2.
miRNA studies have demonstrated that P53 expression
affects the expression of many miRNAs, however, the mechanisms
through which miRNAs are regulated remain poorly understood
(35). P53 accumulation may be
responsible for impaired wound healing in patients with diabetes
(36). The present study
demonstrated a close association between miR-199a-3p expression and
P53: Overexpression of P53 enhanced miR-199a-3p expression and
downregulated SERPINE2 expression. miR-199a-3p was, therefore,
identified as a downstream target of P53. These results
demonstrated that P53 regulated miR-199a-3p expression through
interacting with the promoter section as a transcriptional factor.
The fact that P53 regulated miR-199a-3p expression suggests that
miR-199a-3p may serve a role in DN.
In summary, the present study suggests that
miR-199a-3p, induced by P53, functions as a pro-coagulating factor
in DN by targeting SERPINE2, and subsequently suppresses expression
of downstream tPA. miR-199a-3p, as a fascinating molecule involved
in the pathogenesis and progression of DN, may serve as both a
biomarker in early diagnosis of DN and a directing factor in
treatment of DN.
References
|
1
|
Shi TJ, Zhang MD, Zeberg H, Nilsson J,
Grünler J, Liu SX, Xiang Q, Persson J, Fried KJ, Catrina SB, et al:
Coenzyme Q10 prevents peripheral neuropathy and attenuates neuron
loss in the db-/db- mouse, a type 2 diabetes model. Proc Natl Acad
Sci USA. 110:690–695. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haw JS, Tantry S, Vellanki P and Pasquel
FJ: National strategies to decrease the burden of diabetes and its
complications. Curr Diab Rep. 15:652015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palmer BF and Clegg DJ: Electrolyte and
acid-base disturbances in patients with diabetes mellitus. N Engl J
Med. 373:548–559. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bouton MC, Boulaftali Y, Richard B, Arocas
V, Michel JB and Jandrot-Perrus M: Emerging role of
serpinE2/protease nexin-1 in hemostasis and vascular biology.
Blood. 119:2452–2457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valiente M, Obenauf AC, Jin X, Chen Q,
Zhang XH, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E and Massagué
J: Serpins promote cancer cell survival and vascular co-option in
brain metastasis. Cell. 156:1002–1016. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Santoro A, Conde J, Scotece M, Abella V,
Lois A, Lopez V, Pino J, Gomez R, Gomez-Reino JJ and Gualillo O:
SERPINE2 inhibits IL-1α-induced MMP-13 expression in human
chondrocytes: Involvement of ERK/NF-κB/AP-1 pathways. PLoS One.
10:e01359792015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alsaweed M, Lai CT, Hartmann PE, Geddes DT
and Kakulas F: Human milk miRNAs primarily originate from the
mammary gland resulting in unique miRNA profiles of fractionated
milk. Sci Rep. 6:206802016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yuan Y, Kang R, Yu Y, Liu J, Zhang Y, Shen
C, Wang J, Wu P, Shen C and Wang Z: Crosstalk between miRNAs and
their regulated genes network in stroke. Sci Rep. 6:204292016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka
H, Boland CR and Goel A: Circulating microRNA-203 predicts
prognosis and metastasis in human colorectal cancer. Gut.
66:654–665. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang
EL, Wu ZB, Huang ZY and Chen XP: MiR-216b is involved in
pathogenesis and progression of hepatocellular carcinoma through
HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis.
6:e16702015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu FY, Deng YL, Li Y, Zeng D, Zhou ZZ,
Tian DA and Liu M: Down-regulated KLF17 expression is associated
with tumor invasion and poor prognosis in hepatocellular carcinoma.
Med Oncol. 30:4252013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang X, Li W, Ma L, Gao J, Liu J, Ping F
and Nie M: Association study of the miRNA-binding site
polymorphisms of CDKN2A/B genes with gestational diabetes mellitus
susceptibility. Acta Diabetol. 52:951–958. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen Y, Wang X and Shao X: A combination
of human embryonic stem cell-derived pancreatic endoderm transplant
with LDHA-repressing miRNA can attenuate high-fat diet induced type
II diabetes in mice. J Diabetes Res. 2015:7969122015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song C, Ji Y, Zou G and Wan C: Tetrandrine
down-regulates expression of miRNA-155 to inhibit signal-induced
NF-κB activation in a rat model of diabetes mellitus. Int J Clin
Exp Med. 8:4024–4030. 2015.PubMed/NCBI
|
|
16
|
Chattopadhyay M, Zhou Z, Hao S, Mata M and
Fink DJ: Reduction of voltage gated sodium channel protein in DRG
by vector mediated miRNA reduces pain in rats with painful diabetic
neuropathy. Mol Pain. 8:172012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-(Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nam JW, Rissland OS, Koppstein D,
Abreu-Goodger C, Jan CH, Agarwal V, Yildirim MA, Rodriguez A and
Bartel DP: Global analyses of the effect of different cellular
contexts on microRNA targeting. Mol Cell. 53:1031–1043. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:(Database Issue). D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:(Database Issue). D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: Tools for microRNA genomics. Nucleic Acids
Res. 36:(Database Issue). D154–D158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:(Database issue).
D140–D144. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Griffiths-Jones S: The microRNA registry:
Nucleic Acids Res. 32:(Database issue). D109–D111. 2004.
|
|
24
|
Tornovsky-Babeay S, Dadon D, Ziv O,
Tzipilevich E, Kadosh T, Haroush R Schyr-Ben, Hija A,
Stolovich-Rain M, Furth-Lavi J, Granot Z, et al: Type 2 diabetes
and congenital hyperinsulinism cause DNA double-strand breaks and
p53 activity in beta cells. Cell Metab. 19:109–121. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen BF, Suen YK, Gu S, Li L and Chan WY:
A miR-199a/miR-214 self-regulatory network via PSMD10, TP53 and
DNMT1 in testicular germ cell tumor. Sci Rep. 4:64132014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seviour EG, Sehgal V, Lu Y, Luo Z, Moss T,
Zhang F, Hill SM, Liu W, Maiti SN, Cooper L, et al: Functional
proteomics identifies miRNAs to target a p27/Myc/phospho-Rb
signature in breast and ovarian cancer. Oncogene. 35:8012016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thomas H: Diabetes: Enterovirus
dysregulates islet miRNAs. Nat Rev Endocrinol. 12:22016. View Article : Google Scholar
|
|
28
|
Jansen F, Wang H, Przybilla D, Franklin
BS, Dolf A, Pfeifer P, Schmitz T, Flender A, Endl E, Nickenig G and
Werner N: Vascular endothelial microparticles-incorporated
microRNAs are altered in patients with diabetes mellitus.
Cardiovasc Diabetol. 15:492016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kornfeld JW, Baitzel C, Könner AC,
Nicholls HT, Vogt MC, Herrmanns K, Scheja L, Haumaitre C, Wolf AM
and Knippschild U: Obesity-induced overexpression of miR-802
impairs glucose metabolism through silencing of Hnf1b. Nature.
494:111–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yousefzadeh N, Alipour MR and Soufi FG:
Deregulation of NF-кB-miR-146a negative feedback loop may be
involved in the pathogenesis of diabetic neuropathy. J Physiol
Biochem. 71:51–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pencheva N, Tran H, Buss C, Huh D,
Drobnjak M, Busam K and Tavazoie SF: Convergent multi-miRNA
targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis
and angiogenesis. Cell. 151:1068–1082. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee CG, Kim YW, Kim EH, Meng Z, Huang W,
Hwang SJ and Kim SG: Farnesoid X receptor protects hepatocytes from
injury by repressing miR-199a-3p, which increases levels of LKB1.
Gastroenterology. 142:1206–17.e7. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Alemdehy MF, Haanstra JR, de Looper HW,
van Strien PM, Verhagen-Oldenampsen J, Caljouw Y, Sanders MA,
Hoogenboezem R, de Ru AH, Janssen GM, et al: ICL-induced miR139-3p
and miR199a-3p have opposite roles in hematopoietic cell expansion
and leukemic transformation. Blood. 125:3937–3948. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Omouendze PL, Henry VJ, Porte B, Dupré N,
Carmeliet P, Gonzalez BJ, Marret S and Leroux P: Hypoxia-ischemia
or excitotoxin-induced tissue plasminogen activator- dependent
gelatinase activation in mice neonate brain microvessels. PLoS One.
8:e712632013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Garibaldi F, Falcone E, Trisciuoglio D,
Colombo T, Lisek K, Walerych D, Del Sal G, Paci P, Bossi G, Piaggio
G and Gurtner A: Mutant p53 inhibits miRNA biogenesis by
interfering with the microprocessor complex. Oncogene.
35:3760–3770. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Morimoto Y, Bando YK, Shigeta T, Monji A
and Murohara T: Atorvastatin prevents ischemic limb loss in type 2
diabetes: Role of p53. J Atheroscler Thromb. 18:200–208. 2011.
View Article : Google Scholar : PubMed/NCBI
|