Introduction
Lung cancer is one of the leading causes of
cancer-associated mortality. Non-small cell lung cancer (NSCLC)
makes up ~80% of lung cancer cases (1). Multiple factors are involved in the
development of lung cancer, particularly environmental and genetic
factors. Although novel diagnostic and therapeutic strategies are
under development, the prognosis of lung cancer remains poor, with
the five-year overall survival rate <16% worldwide, due to early
distant metastasis and local invasion (2).
B7-H1, also known as programmed-death ligand
1 (PD-L1) was first identified in 1999 and belongs to the B7
superfamily of coinhibitory molecules. It is crucial as a negative
regulator for immune evasion of tumors, inhibiting T cell
activation and proliferation by engaging with the PD-1 receptor
(3,4). Previous studies have confirmed that
higher mRNA expression levels of B7-H1 are found in human
non-lymphoid tissues, compared with low or negligible protein
expression levels in these tissues, which has been observed in
activated lymphocytes, including T cells, B cells and macrophage
cells, previously (3,5). This observation indicated that
post-transcriptional regulation may be involved in regulating the
expression of B7-H1. Previous studies have reported that the
phosphatase and tensin homolog/phosphatidylinositol-3-kinase
pathway (6) and microRNA (miR)-513
(7) are important in the
post-transcriptional regulation of the cell surface expression of
B7-H1. Of note, Wang et al identified the functional
single nucleotide polymorphism (SNP) rs414815 within the miR-570
seed binding sequence in the 3′-untranslated region (3′-UTR) of
PD-L1 mRNA, which affected its expression in gastric cancer
(8).
MicroRNAs (miRNAs) are endogenous non-coding RNAs
consisting of 19–24 nucleotides, which bind to complementary sites
in the 3′-UTR of target messenger RNAs (mRNAs) to regulate diverse
biological functions, including cell differentiation,
proliferation, apoptosis and DNA repair (9–11).
Therefore, it has been suggested that miRNAs contribute to the
pathogenesis of various clinical diseases, including cancer. The
association between miRNAs and their target genes is complicated;
one miRNA has the ability to bind to various mRNAs simultaneously,
whereas the opposite is not observed (12,13).
Therefore, genetic variations, including those of miRNAs or the
3′-UTR of target mRNAs, can alter their interaction (14). There is increasing evidence
suggesting that SNPs in miRNA target genes, which represent genetic
variants of the miRNA-binding region, can modify miRNA binding,
affecting the risk and susceptibility of patients to cancer,
including gastric cancer (15),
ovarian carcinoma (16),
colorectal cancer (17,18) and skin carcinoma (19). However, no studies have focused on
SNPs in the 3-UTR of the B7-H1 gene or its association with
the risk of NSCLC.
Based on the aforementioned findings, three SNPs of
the miRNA-binding sites in the 3′-UTR of the B7-H1 gene,
including rs2297136, rs4143815 and rs4742098, were selected for
examination. The present case-control study was performed to
evaluate the relevance of these SNPs in terms of the risk of NSCLC
and its clinical characteristics.
Materials and methods
Study population
A total of 519 cases were enrolled, including 320
patients with NSCLC and 199 healthy controls, from The First
Affiliated Hospital of Soochow University (Suzhou, China) between
March 2010 and November 2014. The diagnosis of lung cancer was
histologically confirmed. For the patients with lung cancer,
several detailed clinical pathological characteristics, including
histological type, depth of tumor infiltration, distant metastasis,
lymph node metastasis and tumor-node-metastasis (TNM) stage, are
listed in Table I. None of the 199
age- and sex-matched healthy control individuals had any serious
respiratory disease or familial history of cancer. At recruitment,
written informed consent was obtained from participants. The
present study was approved by the Ethics Committee of the First
Affiliated Hospital of Soochow University.
 | Table I.Characteristics of participants in
the case-control study. |
Table I.
Characteristics of participants in
the case-control study.
| Characteristic | Variable | NSCLC n=320
(%) | Control n=199
(%) |
|---|
| Age, years | <65 | 165 (51.6) | 132 (66.3) |
|
| ≥65 | 155 (48.4) | 67
(33.7) |
| Sex | Male | 240 (75.0) | 121 (60.8) |
|
| Female | 80 (25.0) | 78
(39.2) |
| Pathological
type | Adenocarcinoma | 163 (50.9) | – |
|
| Squamous cell
carcinoma | 139 (43.4) | – |
|
| Other | 18 (5.7) | – |
| Depth of tumor
infiltration | T1 | 42 (13.1) | – |
|
| T2 | 70 (21.9) |
|
|
| T3 | 58 (18.1) |
|
|
| T4 | 150 (46.9) |
|
| Lymph node
metastasis | Negative | 53 (16.6) |
|
|
| Positive | 267 (83.4) |
|
| Distant
metastasis | Negative | 105 (32.8) |
|
|
| Positive | 215 (67.2) |
|
| TNM stage | I | 40 (12.5) |
|
|
| II | 20 (6.3) |
|
|
| III | 51 (15.9) |
|
|
| IV | 209 (65.3) |
|
Genomic DNA sample extraction
Total genomic DNA from venous blood samples of the
patients and controls were extracted at the time of recruitment
using a phenol/chloroform method. The isolated DNA was stored at
−20°C in TE buffer containing 10 mmol/l Tris-HCl and 1 mmol/l EDTA
(pH 8.0).
Cell lines and cell culture
The A549 lung adenocarcinoma cell line was selected
and obtained from the Shanghai Institutes of Biological Sciences
Cell Bank (Shanghai, China) for the present study. These cells were
cultured in Hyclone RPMI 1640 medium (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
L-glutamine and antibiotics (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified incubator with 5% CO2.
Cells in the logarithmic growth phase were used for
experiments.
miRNA SNP selection and genotyping
assays
Firstly, a search was performed of all potentially
functional SNPs in the 3′-UTR region of the B7-H1 gene from
the dbSNP Ensembl BUILD129, Ensembl, TSC1 and HGVbase1 databases
using Ensembl (www.ensembl.org). The miRNA target
prediction software programs miRanda 2010 (www.microrna.org) and TargetScan v5.1 (targetscan.org) were used to predict the possible
miRNA-binding sites in the 3′-UTR of the B7-H1 gene. Among
these, SNPs with a minor allele frequency (MAF) <0.05 were
excluded. The SNPs located in the putative miRNA binding sites were
regarded as candidate miRNA SNPs.
The genotypes of the specific primers were then
designed to amplify DNA segments of B7-H1 3′-UTR using
allele-specific oligonucleotide polymerase chain reaction (PCR) to
determine unknown genotypes. The 20 µl PCR reaction mixture
consisted of 10.55 µl H2O, 2 µl 10X buffer, 1.2 µl
MgCl2, 0.8 µl forward primer, 0.8 µl reverse primer, 0.4
µl dNTP, 0.25 µl Taq DNA polymerase and 1 µl genomic DNA model. The
PCR cycling procedure comprised of an initial predenaturation step
of 5 min at 94°C, followed by 35 cycles of denaturation at 94°C for
30 sec, annealing at 55°C for 30 sec, and extension at 70°C for 30
sec to form double-strand DNA. Elongation at 70°C for 7 min and
cooling at 4°C for 10 min were performed to render DNA product
stable.
3′-UTR luciferase reporter assays
To generate the luciferase reporter, two sequences
containing the potential binding sites for miR-324-5p, miR-296-5p
and miR-138, the 3′-UTRs of B7-H1 mRNA (rs2297136 and
rs4742098 SNPs) were synthesized (Genewiz, Inc., Suzhou, China),
corresponding either to the A haplotype or to the G haplotype. The
two sequences were cloned into the psiCHECK2 vector downstream of
the firefly luciferase gene, respectively.
For the luciferase reporter assays, the A549 cells
were plated at 5×104 cells per well in a 24-well plate
and were transfected 24 h later with Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Each co-transfection reaction contained
200 ng of psiCHECK2 constructs, 50 nM of chemically synthesized
miR-324-5p mimic, miR-296-5p mimic, miR-138 mimic, or negative
control miRNA, the latter serving as a normalizing control, and the
psiCHECK2-control vector as a blank control. The transfected A549
cells were maintained in RPMI 1640 medium with 10% FBS. Each
transfection was performed in triplicate. Following incubation for
48 h, the cells were collected and analyzed for luciferase activity
using the dual-luciferase reporter assay system (Promega
Corporation, Madison, WI, USA).
Statistical analysis
All statistical data were obtained using SPSS
software (version 11.5; SPSS, Inc., Chicago, IL, USA). The
Hardy-Weinberg equilibrium (HWE) was determined using the
goodness-of-fit χ2 test. Differences in genotype and
allelic frequencies of the involved SNPs between patients with lung
cancer and controls, and associations between SNPs and clinical
characteristics were calculated using χ2 tests. The
association between SNPs and lung cancer risk were estimated
according to the odds ratio (OR) and 95% confidence interval (95%
CI) using multivariate logistic regression. All statistical tests
were two-sided. P<0.05 was considered to indicate a
statistically significant difference.
Results
Basic characteristics of subjects
A total of 320 patients with NSCLC were enrolled in
the present study. The clinical and pathological variants are
listed in Table I. The control
group consisted of 199 individuals with no malignant respiratory
disease.
Polymorphic 3′-UTR of the B7-H1
gene
Three SNPs of the microRNA-binding sites in the
3′-UTR region of the B7-H1 gene, including rs2297136,
rs4143815 and rs4742098, were selected from the dbSNP BUILD129,
Ensembl, TSC1 and HGVbase15 databases. With the assistance of
miRanda and TargetScan v5.1 prediction software, several miRNAs
were identified with the potential ability to bind to the 3′-UTR
region of the B7-H1 gene. From these results, it was found
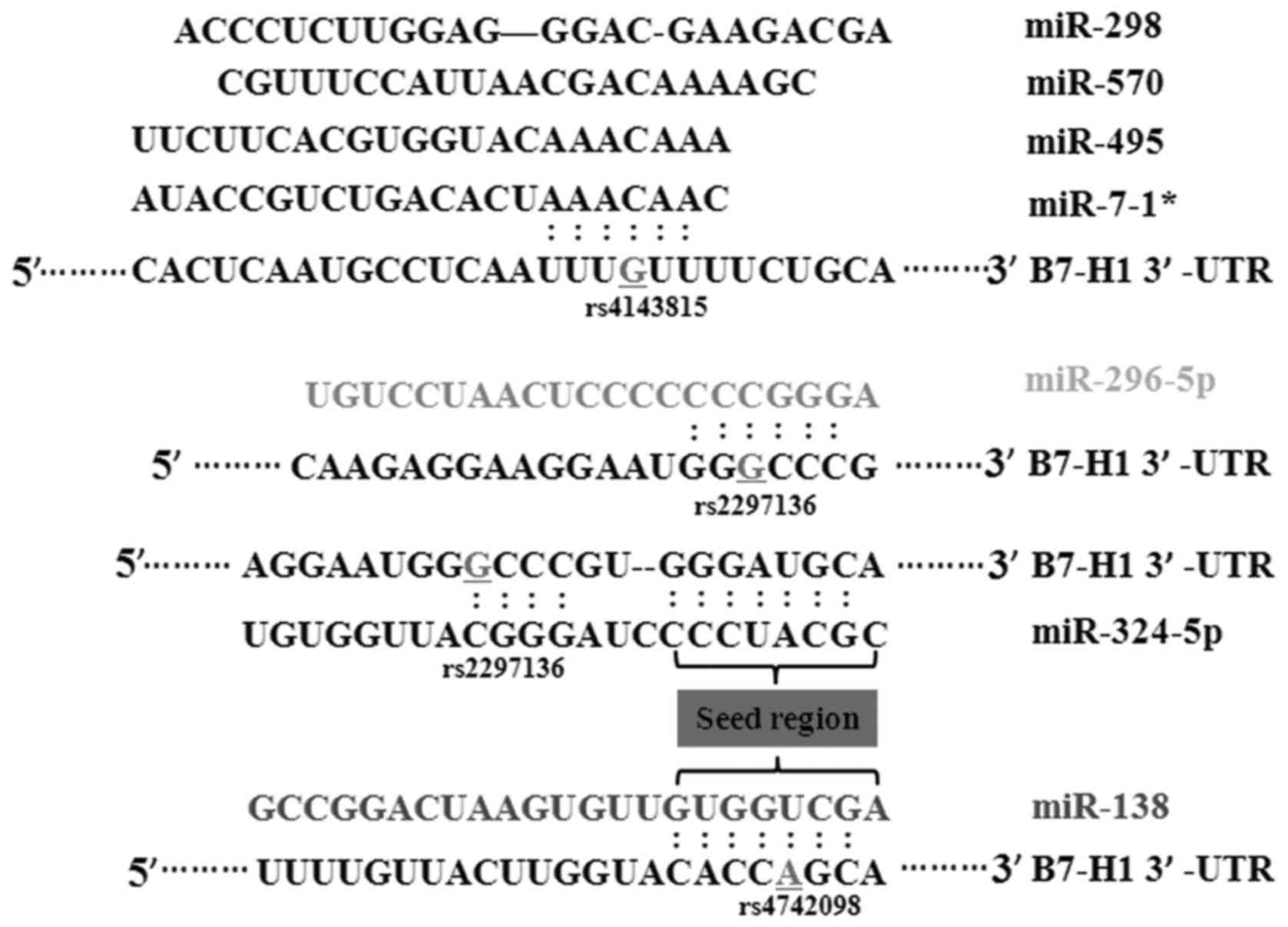
that miR-324-5p, miR-296-5p were able to bind to the rs2297136 SNP.
miR-570, miR-7-1*, miR-495 and miR-298 had certain binding sites
with the rs4143815 SNP. The rs4742098 SNP was located in the
binding sites of miR-138 (Fig. 1).
The detailed SNP IDs, wild-type and mutated gene transformations,
MAF and miRNAs are shown in Table
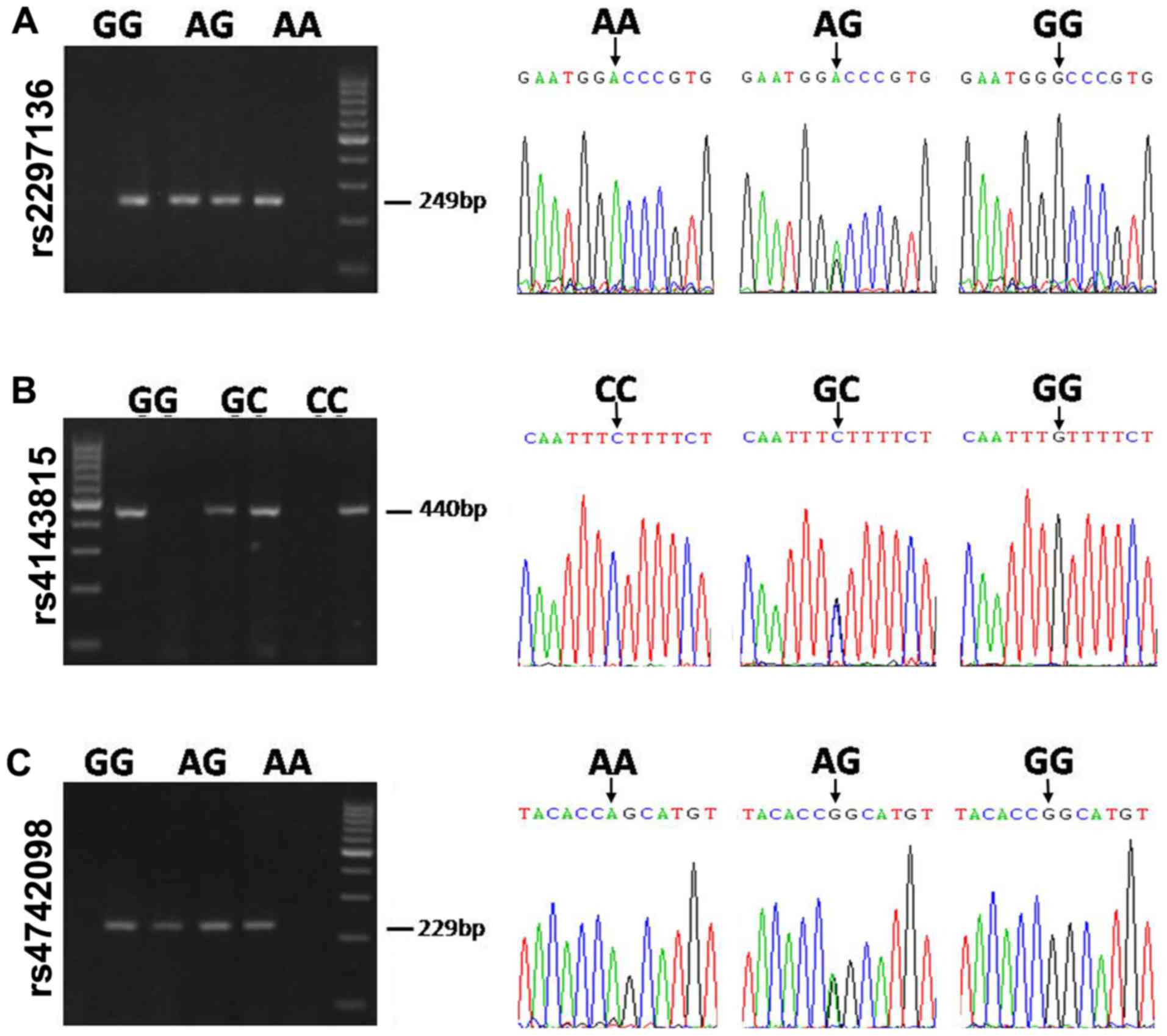
II. To confirm the findings, the three SNPs were genotyped
using a sequencing method. The typical genotyping results are
presented in Fig. 2.
 | Table II.SNPs located in the 3′-untranslated
region of B7-H1 and associated miRNAs. |
Table II.
SNPs located in the 3′-untranslated
region of B7-H1 and associated miRNAs.
| SNP | Nucleotide
change | MAFa | Position on
B7-H1 | miRNA |
|---|
| rs2297136 | G>A | G=0.3613 | 93 | miR-324-5p |
|
|
|
|
| miR-296-5p |
| rs4143815 | G>C | G=0.3320 | 395 | miR-570
miR-7-1* |
|
|
|
|
| miR-495
miR-298 |
| rs4742098 | A>G | G=0.3306 | 2,635 | miR-138 |
Association between SNPs in the B7-H1
gene and risk of NSCLC
All patients with NSCLC and healthy controls were
successfully genotyped for the three SNPs. The HWE was determined
using the goodness-of-fit χ2 test, and no deviations
were detected from the three SNPs, which had values of 0.133, 0.127
and 1.771. The overall genotype distributions and allele
frequencies of these three SNPs in all individuals are presented in
Table III.
 | Table III.Genotype distributions of SNPs in the
cases of NSCLC and controls, and risk estimate. |
Table III.
Genotype distributions of SNPs in the
cases of NSCLC and controls, and risk estimate.
| SNP | Genotype | NSCLC n=320
(%) | Control n=199
(%) |
P-valuea | HWE P-value | OR (95% CI) | P-value |
|---|
| rs2297136 | AA | 83 (25.9) | 84 (42.2) |
<0.001 | 0.133 | 1 (reference) |
|
|
| AG | 226 (70.6) | 100 (50.2) |
|
| 2.287
(1.558–3.358) | <0.001 |
|
| GG | 11 (3.5) | 15 (7.5) |
|
| 0.742
(0.322–1.711) | 0.532 |
|
| G allele
carrier | 237 (74.1) | 115 (57.8) |
|
| 2.086
(1.432–3.039) | <0.001 |
| rs4143815 | CC | 123 (38.4) | 79 (37.8) | 0.439 | 5.343 | 1 (reference) |
|
|
| GC | 145 (45.3) | 80 (41.5) |
|
| 1.076
(0.723–1.601) | 0.719 |
|
| GG | 52 (16.3) | 40 (20.7) |
|
| 1.296
(0.783–2.145) | 0.312 |
|
| G allele
carrier | 197 (61.6) | 120 (62.2) |
|
| 1.054
(0.734–1.515) | 0.782 |
| rs4742098 | AA | 67 (20.9) | 51 (25.6) | 0.007 | 1.771 | 1 (reference) |
|
|
| AG | 189 (59.1) | 90 (45.2) |
|
| 1.599
(1.027–2.488) | 0.037 |
|
| GG | 64 (20.0) | 58 (29.2) |
|
| 0.840
(0.505–1.390) | 0.502 |
|
| G allele
carrier | 253 (79.1) | 148 (74.4) |
|
| 1.301
(0.858–1.974) | 0.237 |
The results of the two-sided χ2 test
revealed that the genotype frequencies of the rs2297136 and
rs4742098 SNPs showed significant differences between cases and
controls (P<0.001 and P=0.007, respectively). In terms of
rs2297136 and rs4742098 the SNPs, individuals carrying the AG
genotype showed a significant association with risk of NSCLC
(OR=2.287; 95% CI=1.558–3.358; P<0.001), compared with those
carrying the AA genotype (OR=1.599; 95% CI=1.027–2.488; P=0.037),
respectively. As for the rs2297136 SNP alone, G allele carriers
(individuals with the AG or GG genotype) were also at a
significantly higher risk of NSCLC, compared with those carrying A
allele homozygotes (OR=2.086; 95% CI=1.432–3.039; P<0.001). No
significant differences in genotype distributions or allele
frequencies were found in the rs4143815 SNP between patients and
healthy controls (P=0.439).
Stratified analyses of the associations between each
SNP genotypes and the clinical characteristics of NSCLC were then
performed (Tables IV–VI). Significant associations were found
in rs2297136 genotypes with lymph node metastasis and distant
metastasis. Compared with the AA homozygote, individuals with the
GG homozygote were less likely to exhibit lymph node metastasis
(P=0.049) and more likely to exhibit distant metastases (P=0.035;
Table IV). For rs4742098, there
were associations between the variant genotypes and depth of tumor
infiltration (Table VI). However,
no statistically significant associations were found between the
rs4143815 genotypes and clinical characteristics (Table V).
 | Table IV.Association between the rs2297136
single nucleotide polymorphism and clinical characteristics. |
Table IV.
Association between the rs2297136
single nucleotide polymorphism and clinical characteristics.
|
| Genotype | AG | GG | G carrier |
|---|
|
|
|
|
|
|
|---|
| Characteristic | AA | AG | GG | G carrier | P-value | OR (95% CI) |
P-valuea | OR (95% CI) | P-value | OR (95% CI) |
|---|
| Sex |
|
Male/female | 63/20 | 171/55 | 6/5 | 177/60 | 0.965 | 1.013
(0.563–1.823) | 0.132 | 2.625
(0.723–9.527) | 0.825 | 0.937
(0.523–1.676) |
| Histological
type |
|
SCC/AC/diffuse | 40/40/3 | 95/118/13 | 4/5/2 | 99/123/15 | 0.534 |
| 0.124 |
| 0.460 |
|
| Depth of
infiltration |
|
|
|
|
|
|
|
| 0.177 | 1.452
(0.844–2.498) |
|
pT1+pT2/pT3+pT4 | 24/59 | 83/143 | 5/6 | 88/149 | 0.201 | 0.701
(0.406–1.210) | 0.264 | 0.488
(0.136–1.752) |
|
|
| TNM stage |
|
|
|
|
|
| 0.052 |
| 0.610 | 1.187
(0.614–2.293) |
|
I+II/III+IV | 14/69 | 41/185 | 5/6 | 46/191 | 0.795 | 0.916
(0.470–1.789) | 0.049 | 3.512
(0.936–13.175) |
|
|
| Lymph node
metastasis |
|
|
|
|
|
|
|
| 0.346 | 0.709
(0.346–1.453) |
|
Positive/negative | 72/11 | 188/38 | 7/4 | 195/42 | 0.448 | 1.323
(0.641–2.729) |
| 0.267
(0.067–1.066) |
|
|
| Distant
metastasis |
|
Positive/negative | 57/26 | 154/72 | 4/7 | 158/79 | 0.929 | 1.025
(0.596–1.762) | 0.035 | 3.83
(1.032–14.263) | 0.737 | 0.912
(0.533–1.560) |
 | Table VI.Association between the rs4742098
single nucleotide polymorphism and the clinical
characteristics. |
Table VI.
Association between the rs4742098
single nucleotide polymorphism and the clinical
characteristics.
|
| Genotype | AG | GG | G carrier |
|---|
|
|
|
|
|
|
|---|
| Characteristic | AA | AG | GG | G carrier |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
P-valuea | OR (95% CI) |
|---|
| Sex |
|
Male/female | 53/14 | 133/56 | 54/10 | 187/66 | 0.168 | 1.594
(0.818–3.104) | 0.436 | 0.701
(0.286–1.717) | 0.383 | 0.748
(0.390–1.437) |
| Histological
type |
|
SCC/AC/diffuse | 35/28/4 | 78/102/9 | 26/33/5 | 104/135/14 | 0.231 | 0.410 | 0.232 |
|
|
|
| Depth of tumor
infiltration |
|
pT1+pT2/pT3+pT4 | 30/37 | 52/137 | 14/50 | 66/187 | 0.009 | 2.136
(1.199–3.807) | 0.006 | 2.896
(1.349–6.214) | 0.003 | 2.297
(1.316–4.011) |
| TNM stage |
|
I+II/III+IV | 15/52 | 34/155 | 11/53 | 45/208 | 0.432 | 1.315
(0.664–2.606) | 0.456 | 1.390
(0.584–3.308) | 0.391 | 0.750
(0.388–1.449) |
| Lymph node
metastasis |
|
Positive/negative | 56/11 | 159/30 | 52/12 | 211/42 | 0.917 | 0.961
(1.451–2.044) | 0.726 | 1.175
(0.477–2.893) | 0.971 | 0.987
(0.477–2.040) |
| Distant
metastasis |
|
Positive/negative | 40/27 | 131/58 | 44/20 | 175/78 | 0.151 | 0.852
(0.481–1.508) | 0.280 | 0.673
(0.328–1.383) | 0.142 | 1.514
(0.868–2.642) |
 | Table V.Association between the rs4143815
single nucleotide polymorphism and clinical characteristics. |
Table V.
Association between the rs4143815
single nucleotide polymorphism and clinical characteristics.
|
| Genotype | AG | GG | G carrier |
|---|
|
|
|
|
|
|
|---|
| Characteristic | CC | GC | GG | G carrier | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) |
|---|
| Sex |
|
Male/female | 94/29 | 104/41 | 42/10 | 146/51 | 0.383 | 1.278
(0.736–2.218) | 0.528 | 0.772
(0.315–1.727) | 0.642 | 0.883
(0.523–1.492) |
| Histological
type | 50/64/9 | 66/74/5 | 23/25/4 | 89/99/9 | 0.319 |
| 0.891 |
| 0.491 |
|
|
SCC/AC/diffuse |
| Depth of tumor
infiltration |
|
pT1+pT2/pT3+pT4 | 39/84 | 49/96 | 24/28 | 73/124 | 0.717 | 0.910
(0.545–1.519) | 0.069 | 0.54 2
(0.279–1.053) | 0.329 | 1.268
(0.787–2.044) |
| TNM stage |
|
I+II/III+IV | 20/103 | 32/113 | 8/44 | 40/157 | 0.795 | 0.686
(0.369–1.274) | 0.885 | 1.068
(0.437–2.608) | 0.367 | 1.312
(0.726–2.371) |
| Lymph node
metastasis |
|
Positive/negative | 103/20 | 117/28 | 47/5 | 164/33 | 0.516 | 1.232
(0.655–2.319) | 0.251 | 0.548
(0.194–1.548) | 0.908 | 0.965
(0.526–1.772) |
| Distant
metastasis |
|
Positive/negative | 85/38 | 95/50 | 35/17 | 130/67 | 0.533 | 1.177
(0.705–1.967) | 0.815 | 1.086
(0.543–0.175) | 0.564 | 0.867
(0.535–1.406) |
Functional relevance of rs2297136 and
rs4742098 in the interaction between miRNA and the expression of
B7-H1
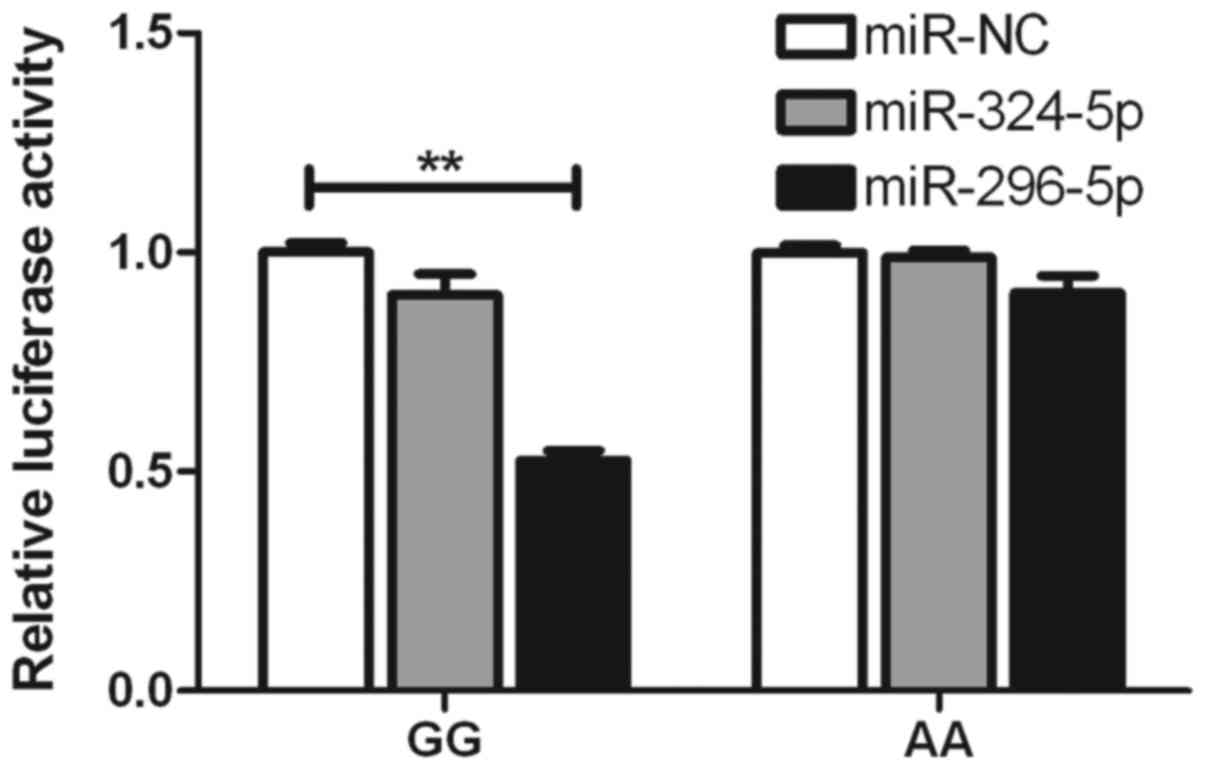
To determine whether the inhibitory roles of
miR-324-5p, miR-296-5p and miR-138 are affected by rs2297136 and
rs4742098, respectively, psiCHECK2 vectors were constructed,
including wild-type and mutated genotypes, which were then
co-transfected with these miRNAs into A549 cells. It was found that
the expression of the G-allele-specific psiCHECK2 construct was
significantly suppressed by miR-296-5p in terms of the rs2297136
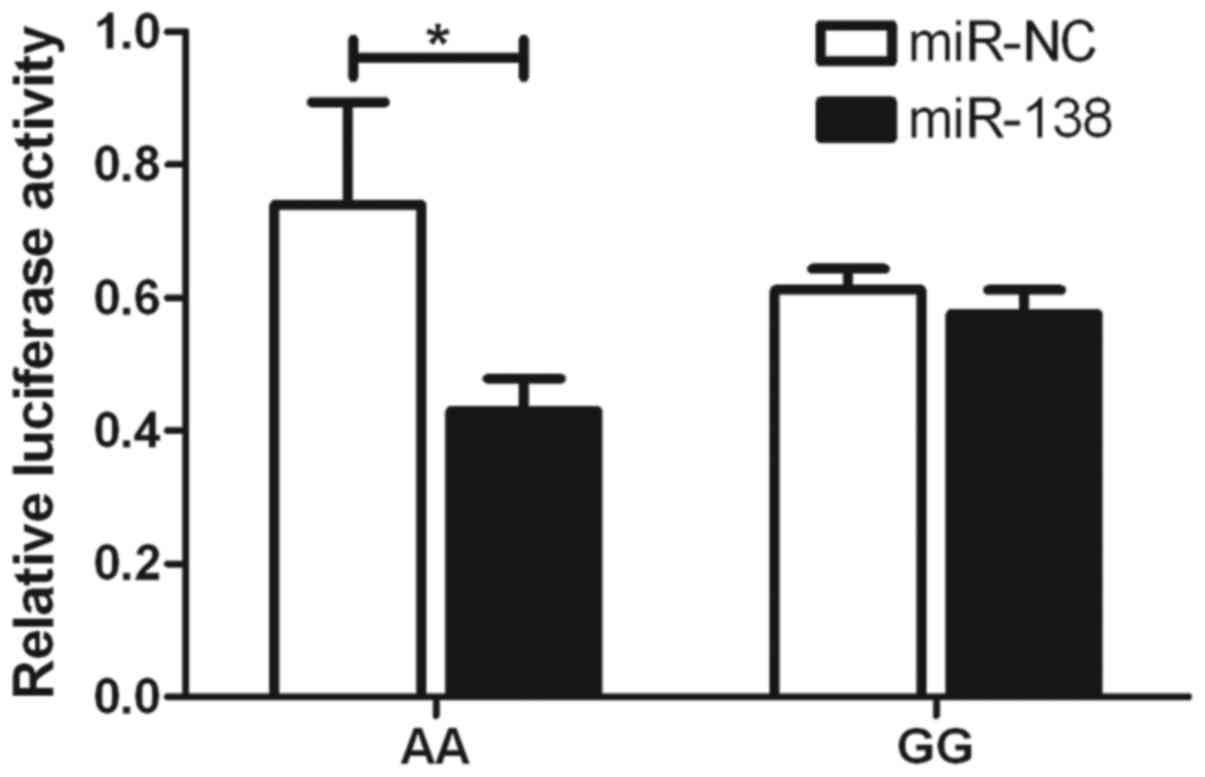
SNP (Fig. 3). With the rs4742098
SNP, the expression of A-allele-specific psiCHECK2 construct was
inhibited by miR-138 (Fig. 4).
These findings indicated that the two miRNAs inhibited the
expression of B7-H1 by binding to the two SNPs.
Discussion
To the best of our knowledge, genetic factors are
considered to contribute to the development of tumorigenesis in
lung cancer. However, the identification of prognostic markers for
the risk of lung cancer from the miR-SNP perspective is a novel
field in the progress of the cancer research (20). Based on this, the present study
aimed to investigate the potential association between three SNPs
of the miRNA-binding site in the 3′-UTR region of the B7-H1
gene (rs2297136, rs4143815 and rs4742098) and the risk of NSCLC
through a case-control approach. The results provided evidence that
rs2297136 and rs4742098 are functional SNPs, which contain genetic
variants associated with the increased risk of NSCLC. In addition,
the analysis revealed that these SNPs had significant associations
with clinicopathological features, including depth of tumor
infiltration, distant metastasis, lymph node metastasis and TNM
stage. Therefore, these two SNPs may be possible prognostic markers
for NSCLC.
B7-H1 exists as an inhibitory regulator of
cosignaling molecules involved in the pathway of activation of T
lymphoid cells. It can be expressed in diverse tissues, lymphoid
cells and nonlymphoid tissues, including smooth muscle cells,
epithelial cells and endothelial cells, in response to inflammatory
cytokines, including interferon (IFN)-γ, IFN-α and interleukin-1
(3,21,22).
It can also be induced in human solid tumors and is commonly
associated with poor prognosis, larger tumor size, distant
metastasis, deeper tumor infiltration, increased lymph node
metastasis and TNM stage, due to its ability to inhibit T-cell
proliferation and suppress its function to avoid immune
surveillance (23). Therefore, the
tumor-associated B7-H1 gene appears to have an important
role in tumor occurrence, promotion, invasion, angiogenesis and
metastasis (24,25).
miRNAs are involved in the regulation of a range of
biological function, including cell differentiation, proliferation,
apoptosis and DNA repair. Therefore, it has been suggested that it
contributes to the pathogenesis of various clinical diseases,
including cancer (26,27). Previous investigations have been
performed to examine the associations of SNPs in miRNAs target
genes, which locate in the miRNA-binding region and affect the
susceptibility of cancer. It has been reported that a genetic
mutation within the 3′-UTR region of the TNFAIP2 gene (rs8126)
contributes to the risk of esophageal squamous cell carcinoma
(28). In addition, the
miRNA-135a/b binding site polymorphism in the CD133 3′-UTR confers
decreased risk and favorable prognosis in lung cancer by reducing
the expression of CD133 (29). A
genetic variation in the miRNA target site of the KRT81 gene has
also been associated with survival rates in early-stage NSCLC
(30), and the reduced expression
of let-7 miRNAs in human lung cancer has been association with
shortened postoperative survival rates (31).
In the present study, genotyping of three SNPs
(rs2297136, rs4143815 and rs4742098) of the miRNA-binding sites in
the 3′-UTR region of the B7-H1 gene was performed in 320
patients with NSCLC and 199 healthy controls, to determine the
effect of these SNPs on the risk of NSCLC. Following genotyping,
significant differences were found between the cases and controls
in rs2297136 and rs4742098 SNPs (P<0.001 and P=0.007). For these
two SNPs, individuals carrying the AG genotype showed significant
associations with the risk of NSCLC, (OR=2.287; 95% CI=1.558–3.358;
P<0.001), as did those carrying the AA genotype (OR=1.599, 95%
CI=1.027–2.488, P=0.037). Furthermore, for the SNP rs2297136 alone,
individuals carrying genotype AA has a decreased risk of developing
NSCLC, compared with individuals carrying the G variant allele,
including AG and GG carriers (OR=2.086; 95% CI=1.432–3.039;
P<0.001). The present study then focused on clinical
characteristics, the GG homozygote of rs2297136 was significantly
more prevalent in patients with a reduced presence of lymph node
metastasis (P=0.049), and higher prevalence of distant metastasis
(P=0.035), compared with the AA homozygote. As for the rs4742098
SNP, all mutated alleles, including the AG, GG and G carriers,
showed significant association with deeper infiltration (P=0.009,
P=0.006 and P=0.003, respectively).
SNPs in the 3′UTR region of the B7-H1 gene
can affect the interaction of miRNAs and complementary sites, which
can result in the reduced expression of B7-H1 (32). In terms of the miRNAs of interest
in the present study, miR-324-5p, miR-296-5p and miR-138 were
selected to examine the association with the rs2297136 and
rs4742098 SNPs. It was found that the expression of the
G-allele-specific psiCHECK2 construct was significantly suppressed
by miR-296-5p in terms of the rs2297136 SNP. With the rs4742098
SNP, the expression of the A-allele-specific psiCHECK2 construct
was inhibited by miR-138. This may be a vital step in the mechanism
regulating the expression of B7-H1.
There were a number of limitations in the present
constructed case-control study. Firstly, the number of DNA samples
from all patients with NSCLC and healthy controls available for the
present study were low to a certain degree, therefore, the analyses
performed from these data may show minimal deviation from the
average. Secondly, all the samples collected were from the same
hospital, with the outcome that inherent selection bias may exist,
therefore a larger population-based study may be considered to
validate these results in the future.
In conclusion, the present study demonstrated that
the functional polymorphisms of the miRNA-binding sites in the
3′-UTR region of the B7-H1 gene were significantly
associated with higher risks of lung cancer, distant metastasis,
lymph node metastasis and deep infiltration (rs2297136 and
rs4742098). These results support the hypothesis that genetic
variants can interrupt the miRNA-mediated regulation of a target
gene, which may become a possible prognostic marker for the
prediction of NSCLC risk.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 31270940 to
Professor Jian-an Huang), the Jiangsu Provincial Special Program of
Medical Science (grant no. BL2012023) and the Clinical Key
Specialty Project of China and Clinical Medical Center of Suzhou
(grant no. Szzx201502).
References
|
1
|
Jemal A, Center MM, DeSantis C and Ward
EM: Global patterns of cancer incidence and mortality rates and
trends. Cancer Epidemiol Biomarkers Prev. 19:1893–1907. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong H, Strome SE, Salomao DR, Tamura H,
Hirano F, Flies DB, Roche PC, Lu J, Zhu G, Tamada K, et al:
Tumor-associated B7-H1 promotes T-cell apoptosis: A potential
mechanism of immune evasion. Nat Med. 8:793–800. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Greenwald RJ, Freeman GJ and Sharpe AH:
The B7 family revisited. Annu Rev Immunol. 23:515–548. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zou W and Chen L: Inhibitory B7-family
molecules in the tumour microenvironment. Nat Rev Immunol.
8:467–477. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parsa AT, Waldron JS, Panner A, Crane CA,
Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et
al: Loss of tumor suppressor PTEN function increases B7-H1
expression and immunoresistance in glioma. Nat Med. 13:84–88. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gong AY, Zhou R, Hu G, Li X, Splinter PL,
O'Hara SP, LaRusso NF, Soukup GA, Dong H and Chen XM: MicroRNA-513
regulates B7-H1 translation and is involved in IFN-gamma-induced
B7-H1 expression in cholangiocytes. J Immunol. 182:1325–1333. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang W, Li F, Mao Y, Zhou H, Sun J, Li R,
Liu C, Chen W, Hua D and Zhang X: A miR-570 binding site
polymorphism in the B7-H1 gene is associated with the risk of
gastric adenocarcinoma. Hum Genet. 132:641–648. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hoon DS, Ferris R, Tanaka R, Chong KK,
Alix-Panabières C and Pantel K: Molecular mechanisms of metastasis.
J Surg Oncol. 103:508–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Brodersen P and Voinnet O: Revisiting the
principles of microRNA target recognition and mode of action. Nat
Rev Mol Cell Biol. 10:141–148. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Makeyev EV and Maniatis T: Multilevel
regulation of gene expression by microRNAs. Science. 319:1789–1790.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song X, Zhong H, Zhou J, Hu X, Zhou Y, Ye
Y, Lu X, Wang J, Ying B and Wang L: Association between
polymorphisms of microRNA-binding sites in integrin genes and
gastric cancer in Chinese Han population. Tumour Biol.
36:2785–2792. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim M, Chen X, Chin LJ, Paranjape T, Speed
WC, Kidd KK, Zhao H, Weidhaas JB and Slack FJ: Extensive sequence
variation in the 3′ untranslated region of the KRAS gene in lung
and ovarian cancer cases. Cell Cycle. 13:1030–1040. 2014.Zhao Y, Du
Y, Zhao S and Guo Z: Single-nucleotide polymorphisms of microRNA
processing machinery genes and risk of colorectal cancer. Onco
Targets Ther 8: 421–425, 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Y, Du Y, Zhao S and Guo Z:
Single-nucleotide polymorphisms of microRNA processing machinery
genes and risk of colorectal cancer. Onco Targets Ther. 8:421–5.
2015.PubMed/NCBI
|
|
18
|
Ye P, Li Z, Jiang H and Liu T: SNPs in
microRNA-binding sites in the ITGB1 and ITGB3 3′-UTR increase
colorectal cancer risk. Cell Biochem Biophys. 70:601–607. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao Y, Zhang L, Ritprajak P, Tsushima F,
Youngnak-Piboonratanakit P, Kamimura Y, Hashiguchi M and Azuma M:
Immunoregulatory molecule B7-H1 (CD274) contributes to skin
carcinogenesis. Cancer Res. 71:4737–4741. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsushima F, Tanaka K, Otsuki N, Youngnak
P, Iwai H, Omura K and Azuma M: Predominant expression of B7-H1 and
its immunoregulatory roles in oral squamous cell carcinoma. Oral
Oncol. 42:268–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI,
Park YM, Oh S, Shin JG, Yao S, Chen L and Choi IH: Interferon
regulatory factor-1 is prerequisite to the constitutive expression
and IFN-gamma-induced upregulation of B7-H1 (CD274). FEBS Lett.
580:755–762. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Blank C, Gajewski TF and Mackensen A:
Interaction of PD-L1 on tumor cells with PD-1 on tumor-specific T
cells as a mechanism of immune evasion: Implications for tumor
immunotherapy. Cancer Immunol Immunother. 54:307–314. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Keir ME, Butte MJ, Freeman GJ and Sharpe
AH: PD-1 and its ligands in tolerance and immunity. Annu Rev
Immunol. 26:677–704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Youngnak-Piboonratanakit P, Tsushima F,
Otsuki N, Igarashi H, Machida U, Iwai H, Takahashi Y, Omura K,
Yokozeki H and Azuma M: The expression of B7-H1 on keratinocytes in
chronic inflammatory mucocutaneous disease and its regulatory role.
Immunol Lett. 94:215–222. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J, Yu H, Zhang Y, Zhang X, Zheng G,
Gao Y, Wang C and Zhou L: A functional TNFAIP2 3′-UTR rs8126
genetic polymorphism contributes to risk of esophageal squamous
cell carcinoma. PLoS One. 9:e1093182014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cheng M, Yang L, Yang R, Yang X, Deng J,
Yu B, Huang D, Zhang S, Wang H, Qiu F, et al: A microRNA-135a/b
binding polymorphism in CD133 confers decreased risk and favorable
prognosis of lung cancer in Chinese by reducing CD133 expression.
Carcinogenesis. 34:2292–2299. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee SY, Choi JE, Jeon HS, Hong MJ, Choi
YY, Kang HG, Yoo SS, Lee EB, Jeong JY, Lee WK, et al: A genetic
variation in microRNA target site of KRT81 gene is associated with
survival in early-stage non-small-cell lung cancer. Ann Oncol.
26:1142–1148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin
Y, Wang E, Wu M and Shen SH: Aberrant allele frequencies of the
SNPs located in microRNA target sites are potentially associated
with human cancers. Nucleic Acids Res. 35:4535–4541. 2007.
View Article : Google Scholar : PubMed/NCBI
|