Introduction
Colorectal carcinoma (CRC) is one of the highest
incident malignant tumors in the world. The five-year survival rate
is less than 10%, and 50–60% of the CRC patients eventually
progress to metastatic colorectal carcinoma (mCRC) (1,2).
Although FOLFIRI, FLOFLOX, and other chemotherapy alone or in
combination with anti-VGFR monoclonal antibody (e.g., Bevacizumab)
can improve the prognosis of patients with mCRC, who may produce
drug resistance, application of epidermal growth factor receptor
(EGFR) monoclonal antibody (e.g., Panitumumab) at the moment is
still effective (3,4). As a member of ErbB transmembrane
tyrosine kinase receptor family, EGFR activates MEK/ERK, PI3K/AKT,
and STAT signaling pathways to induce cell proliferation,
dedifferentiation and blocking of apoptosis (5). Anti-EGFR monoclonal antibodies,
mainly include Cetuximab and Panitumumab, have been used in the
clinical therapy of mCRC (6).
KRAS is a downstream EGFR-signaling pathway-core component
of MAPK pathway. Previous studies have shown that its KRAS
mutation leads to failure of Erbitux, Panini, or other monoclonal
antibody therapy due to ineffective inhibition of EGFR signaling
pathway using those monoclonal antibodies, indicating that mutant
KRAS is critical for EGFR monoclonal and targeted antibody
therapy (7). The European
Medicines Agency requires clinicians to detect KRAS mutation
in the patients before administrating monoclonal antitumor drug
(8).
A variety of detection methods have been available
for the screening of mutant KRAS, including sequencing,
single-stranded confirmation polymorphism (SSCP), AS-PCR, TaqMan
probe PCR, beads, emulsion, amplification, and magnetics (BEAMing),
LigAmp assay, and clamping-based PCR (9,10).
Clamping-based PCR is the most sensitive method for the detection
of low abundance mutations by selectively adding the wild-type
amplified nucleic acid into the reaction system to block the
amplification of wild-type gene (11,12).
The related techniques have been successfully applied to detect the
trace amount of gene mutation in some tumors (12,13).
However, the application of clamping-based PCR also has a problem:
Susceptible to the interference of DNA polymerase in the reaction
system, which may result in base mismatch in PCR with high cycle
number (14–16). To solve this problem, the present
study modified the existing clamping-based PCR by adding internal
competitive amplified fragments to enhance the inhibition of
wild-type KRAS via locked nucleic acid (LNA) probe and
established a method for detecting the trace amount of mutant
KRAS in colorectal neoplasms. The method was applied to
detect mutant KRAS from the colorectal biopsies of 50
patients with suspected colorectal cancer, followed by DNA
sequencing and pathological analysis to validate the test results.
The present study provided a reference for effectively predicting
the therapeutic outcomes of colorectal cancer patients through our
methods on mutant KRAS detection.
Materials and methods
Patients and DNA extraction from
colorectal biopsies
The present study recruited Han Chinese patients
from the outpatient and inpatient clinics of the Southwest Hospital
(Chongqing City, China). Our study protocol was approved by the
Ethics Committee of the Southwest Hospital. All patients or their
guardians signed the informed consents before participating in the
present study.
Fresh colorectal biopsies obtained during
colonoscopy from the patients with suspected colorectal cancer were
washed in PCR and subsequently placed in new Eppendorf tubes,
followed by DNA extraction using Tissue DNA kit (Catalogue no.
536-050, Gene Tech Biotechnology Co., Ltd., Shanghai, China) with
reference to the manufacturer's instructions and DNA quantification
using Nanodrop to obtain mean value of total DNA of each sample
from the triplicated measurements.
Amplification of mutant KRAS using
WIRE-PCR
As shown in Table
I, the PCR system in the present study contained 500 nM primer
set (SW-329/330) and 100 nM florescent probe (SW-1294) for the
detection of internal reference gene; 500 nM primer set
(SW-1595/1596) and 250 nM fluorescent probe (SW-1438) for the
detection of KRAS; and 500 nM LNA probe (SW-144). The PCR
conditions were 50°C for 2 min and 60 cycles of 95°C for 2 min,
95°C for 15 sec, and 60°C for 1 min. The 2Χ SuperMix-UDG was used
as the enzyme for the PCR system. To determine the sensitivity of
WIRE-PCR system, a concentration gradient of mutant template was
prepared by mixing increasing concentrations of plasmid
(105, 104, 103, 102,
101, 100, copies/µl) from the previously
constructed containing mutated single nucleotides
(c34G>C;G12R.1R) with human WT-gDNA in total 50 ng. The reaction
conditions were 50°C for 2 min and 60 cycles of 95°C for 2 min,
95°C for 15 sec, and 60°C for 1 min in total 20 µl.
 | Table I.Sequences of οligonucleotides used in
the present study. |
Table I.
Sequences of οligonucleotides used in
the present study.
| Oligo ID | Oligo sequences
(5′-3′) |
|---|
| SW-329 |
CAGTCTCCTCCAAACAGAAAGTCA |
| SW-330 |
GTCCATCTTGGATAAGGTCAGGA |
| SW-1294 | (Texas Red)
CGGTTTGGACTTCATTCCTGGGCTCC (BHQ2) |
| SW-1595 |
TTTATTATAAGGCCTGCTGAAAATGAC |
| SW-1596 |
CGTCAAGGCACTCTTGCCTAC |
| SW-1438 | (VIC)
ACTACCACAAGTTTATATTC (MGB) |
| SW-144 | TACGCCACCAGCT |
Results
Establishment and optimization of
WIRE-PCR
Given the importance of the annealing temperature in
PCR system, we optimized the best anneal temperature for the primer
sets used in the present study. For example, anneal temperatures
for the amplification of KRAS gene using SW-1595/1596
primers were set as 60–68°C to amplify the PCR products at eight
gradient temperatures. The results of agarose gel electrophoresis
separating the PCR products of different annealing temperatures,
and the results showed the optimal temperature 60°C, with good
amplification efficacy and the corresponding annealing, which was
suggested to be the optimal annealing temperature for the reaction
system.
The mean CT values ± standard deviation in the
internal reference gene, LEPTIN-involved amplification
reaction using 500 and 250 nM KRAS primer set were 25.6±0.23
and 26.52±0.36, respectively. The small CT value of the reaction
system using 500 nM KRAS primer set showed good
reproducibility, and thus we used 500 nM KRAS primer set to
optimize the KRAS amplification. Subsequent test using
different concentrations of fluorescent probe of the internal
reference gene (i.e., 50, 100, and 200 nM) showed that the
wild-type blocking (WTB) probe in our WIRE-PCR using different
concentrations of fluorescent probe of the internal reference gene
could effectively block the amplification of wild-type KRAS.
With reference to the results of LEPTIN and KRAS
amplifications, the mean CT values of LEPTIN amplification
group and KRAS amplification group were relatively large
when using 200 nM fluorescent probe of the internal reference gene
(Table II). Therefore, 50 nM and
100 nM fluorescence probe for the internal reference gene were
considered to be the optimal concentrations in the system.
Application of 100 nM fluorescent probe of the internal reference
gene better enhanced the fluorescence intensity of the KRAS
amplification than the other two concentrations of the probe. In
addition, the fluorescence signal was stable. Therefore, a final
concentration of 100 nM fluorescent probe of the internal reference
gene was selected to optimize the PCR reaction, which contained 500
nM primer set (SW-329/330) and 100 nM florescent probe (SW-1294)
for the detection of internal reference gene, LEPTIN; and
500 nM primer set (SW-1595/1596) and 250 nM fluorescent probe
(SW-1438) for the detection of KRAS.
 | Table II.Comparison of the mean CT values of
the LEPTIN and KRAS amplification groups. |
Table II.
Comparison of the mean CT values of
the LEPTIN and KRAS amplification groups.
| Mean CT value ±
(SD) | Fluorescent probe
50 nM | Fluorescent probe
100 nM | Fluorescent probe
200 nM |
|---|
| LEPTIN (with
WTB) | 24.4±0.24 | 24.33±0.68 | 26.15±0.15 |
| LEPTIN
(without WTB) | 23.75±0.46 | 23.81±0.76 | 25.62±0.27 |
| KRAS (with
WTB) | NA | NA | NA |
| KRAS
(without WTB) | 25.55±0.23 | 25.68±0.55 | 26.22±0.21 |
Blocking effect of different
concentrations of wild-type template in the reaction system
The concentrations of most DNA samples extracted
from the clinical biopsies ranged from 50 to 150 ng. Evaluation if
the WTB concentration used in the reaction system could effectively
block the amplification of different amounts of DNA templates was
necessary at the early experimental stage. Different concentrations
of DNA template (i.e., 50, 100, 150, and 200 ng/µl) used in the
KRAS amplification group imparted an effective blocking
effect on 50–200 ng of the wild-type template under the WTB
reaction. Non-specific amplification of KRAS was found in
the system using 200 ng/µl template and near 40 cycles (with the
mean CT value of 36.78±0.61), indicating that application of 500 nM
WTB probe did not completely block the amplification of wild-type
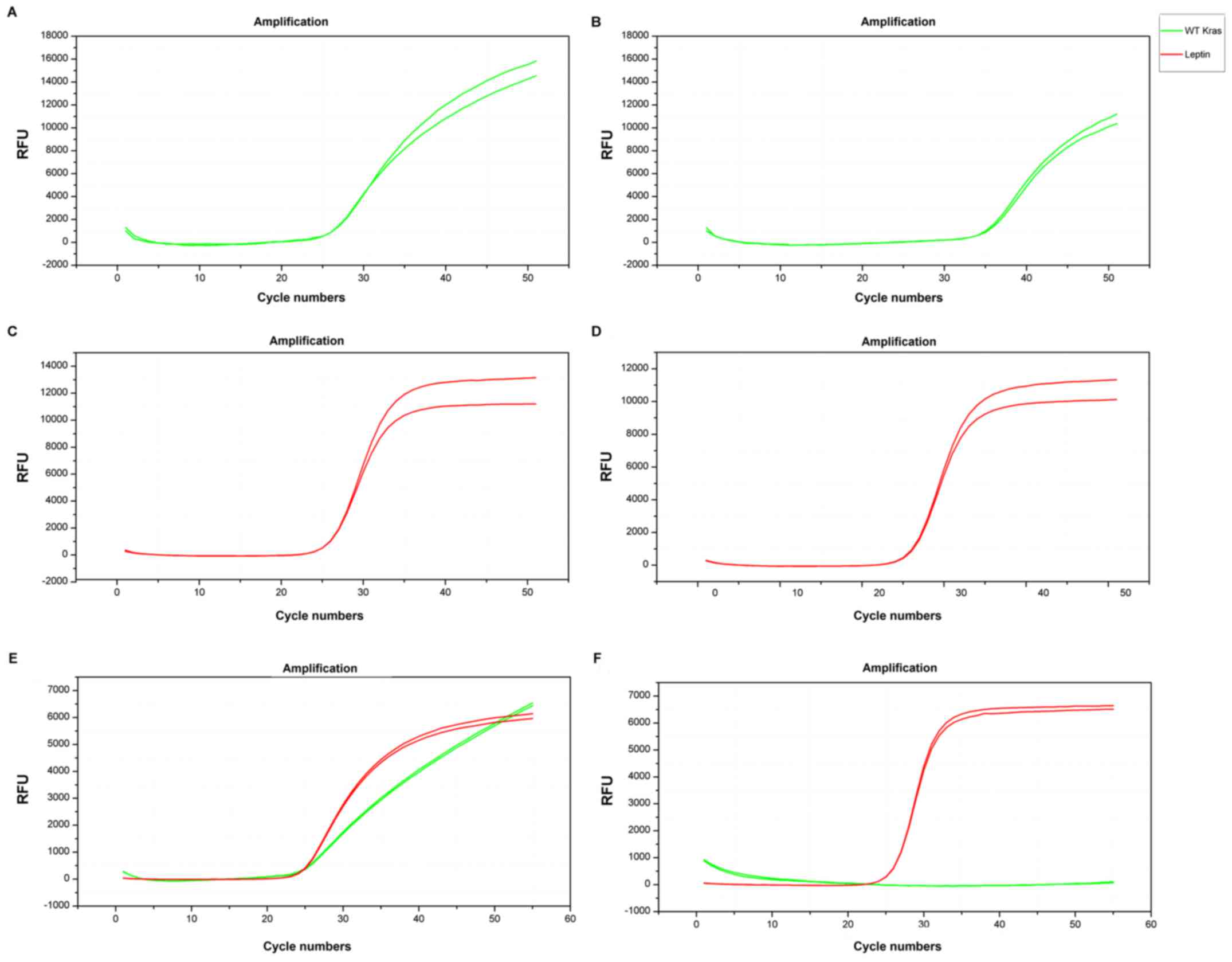
gene when using 200 ng wild-type template (Table III and Fig. 1A and B). No significant difference
of the mean CT values of LEPTIN amplification was found
before and after adding WTB (25.39±0.21 and 25.37±0.04,
respectively P>0.05), suggesting that WTB had no significant
effect on LEPTIN amplification (Fig. 1C and D). The results with and
without WTB when added LEPTIN showed the internal amplified
fragment were amplified together with the target gene, which were
used to reduce base mismatch due to high number of cycles in PCR
and enhanced the specificity (Fig. 1E
and F).
 | Table III.Blocking effects of different
concentrations of wild-type template in the constructed wire PCR
system. |
Table III.
Blocking effects of different
concentrations of wild-type template in the constructed wire PCR
system.
| Mean CT value ±
(SD) | Template 50
ng/µl | Template 100
ng/µl | Template 150
ng/µl | Template 200
ng/µl |
|---|
| LEPTIN (with
WTB) | 24.46±0.63 | 23.75±0.20 | 24.21±0.25 | 23.54±0.2 |
| LEPTIN
(without WTB) | 24.07±0.08 | 23.41±0.11 | 24.86±0.15 | 23.21±0.13 |
| KRAS (with
WTB) | NA | NA | NA | 36.78±0.61 |
| KRAS
(without WTB) | 24.16±0.90 | 23.49±0.60 | 22.75±0.11 | 22.64±0.35 |
The sensitivity capabilities of the
WIRE-PCR system
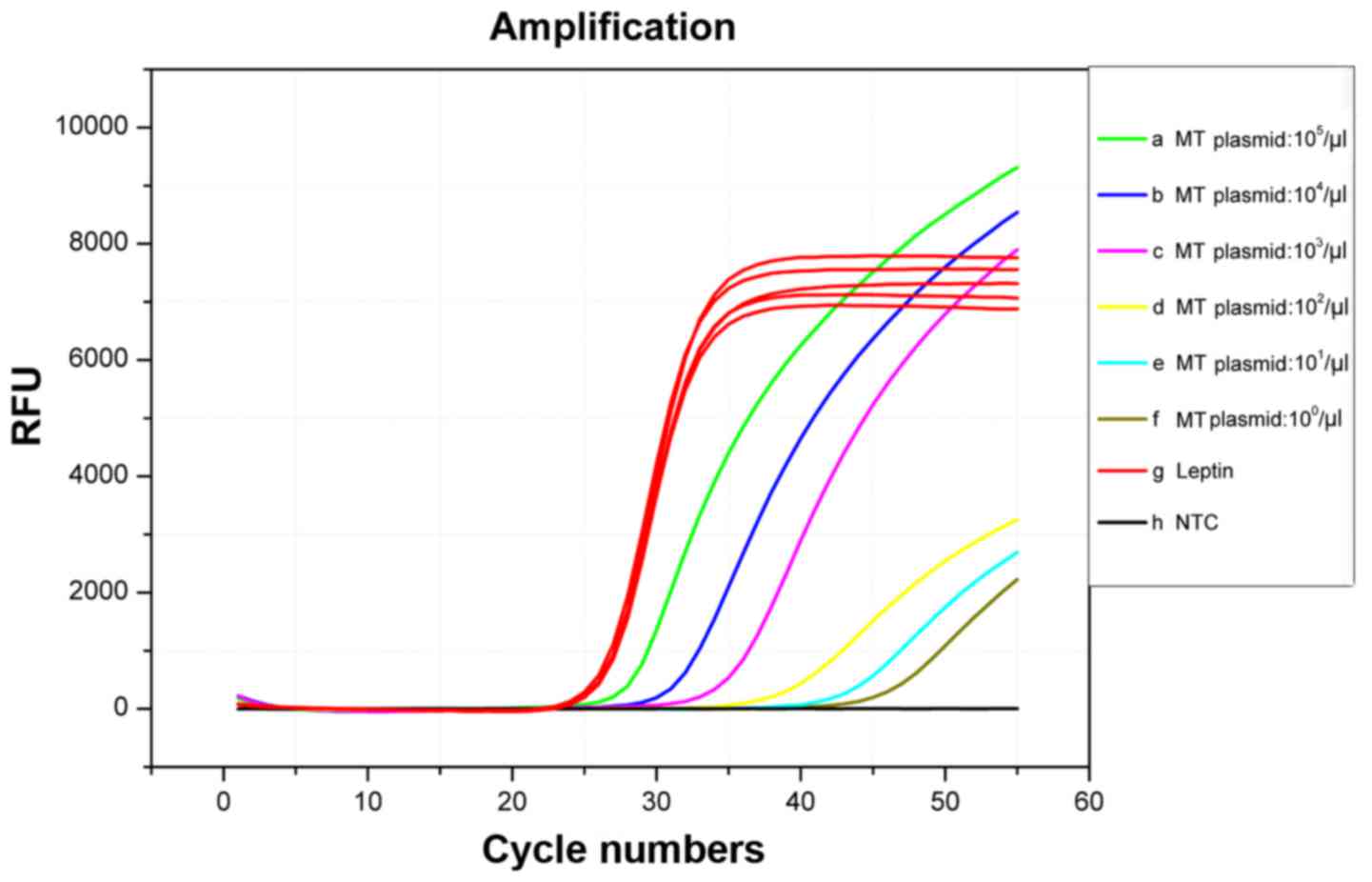
According to the blocking efficiency curves in
Fig. 2, we used the concentration
gradient (105, 104, 103,
102, 101, 100, copies/µl) plasmid
(c34G>C;G12R.1R) mixed with WT-gDNA in total 50 ng as template
to assessment the sensitivity. The result shown in Fig. 4 indicated that in the WIRE-PCR
system, different concentration result in different Cq values even
at the level of single base pair of Plasmid. However, a previous
study showed that traditional PCR always lead to consistent Cq
values despite the differences in the mutated template
concentrations associated with the KRAS MT-alleles because
of both WT-alleles and MT-alleles could be equivalently amplified
(17). We used the Cq values
associated with traditional PCR to indicate the total quantity of
input DNA in previous research. When WIRE-PCR utilized, the Cq
values increased with the quantity of KRAS mutation
template, indicating that the amplification of KRAS
WT-alleles was efficiently inhibited. And we added internal
competitive amplified gene, not only can it amplified together with
the target gene, but can used to reduce base mismatch due to high
number of cycles in PCRs and quantify the total amount of DNA. The
result showed the high blocking efficiency and indicated that the
WIRE-PCR promotes the detection of KRAS MT-alleles with high
sensitivity even at single base pair level (Fig. 2).
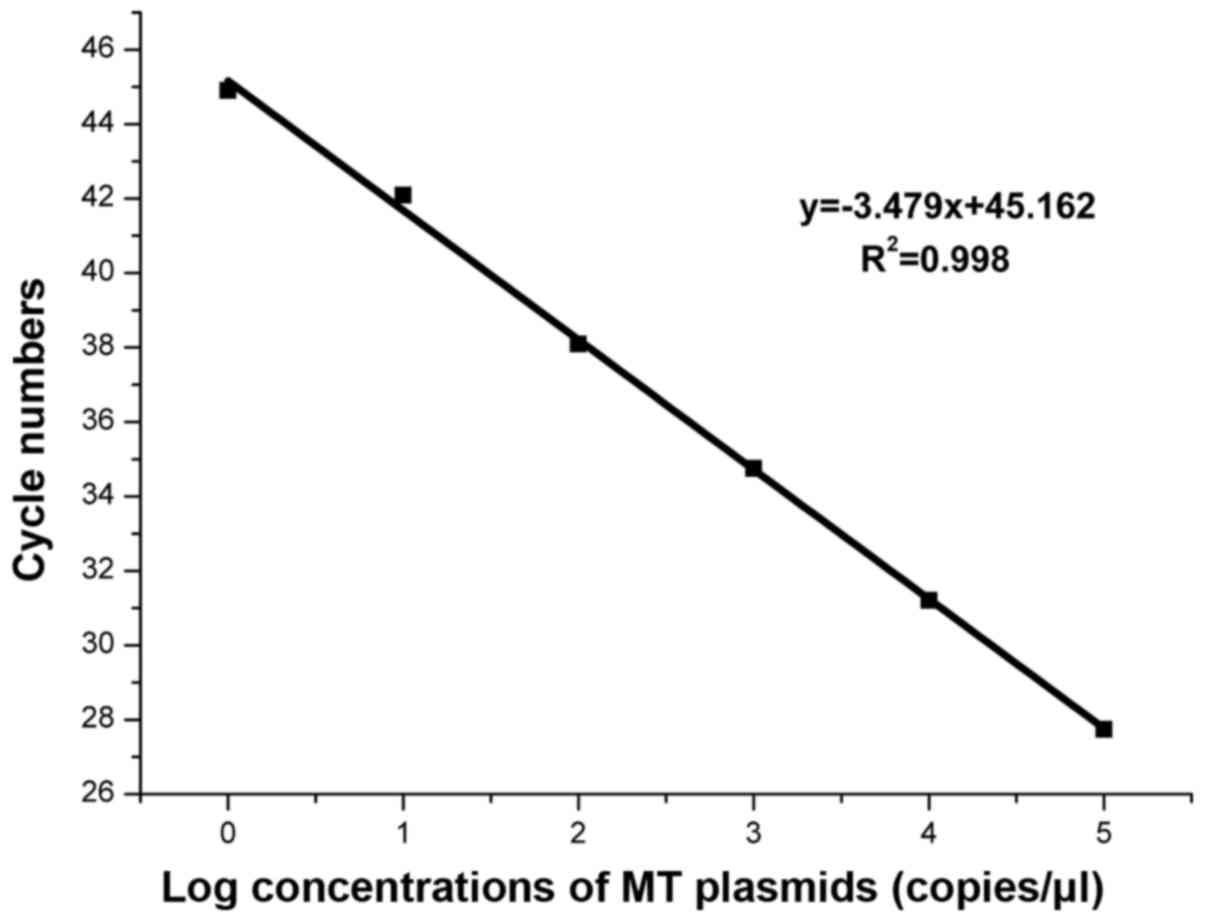
Based on the amplification curves in Fig. 2, we concluded that the WIRE-PCR was
capable of detecting a single copy of KRAS MT-allele in high
presence of WT-alleles with an amplification efficiency of 93.8%
and R2=0.998 (Fig. 3).
The linear association suggest the amount of MT-alleles in a given
sample could be quantified by the real-time PCR standard curve.
The race amount of mutant KRAS in the
clinical biopsies
In the present study, WIRE-PCR was used to detect
the trace amount of mutant KRAS in 50 colorectal biopsies
collected during colonoscopy and find 18 positive cases, indicating
that approximately 36% of the colorectal biopsies from the 50
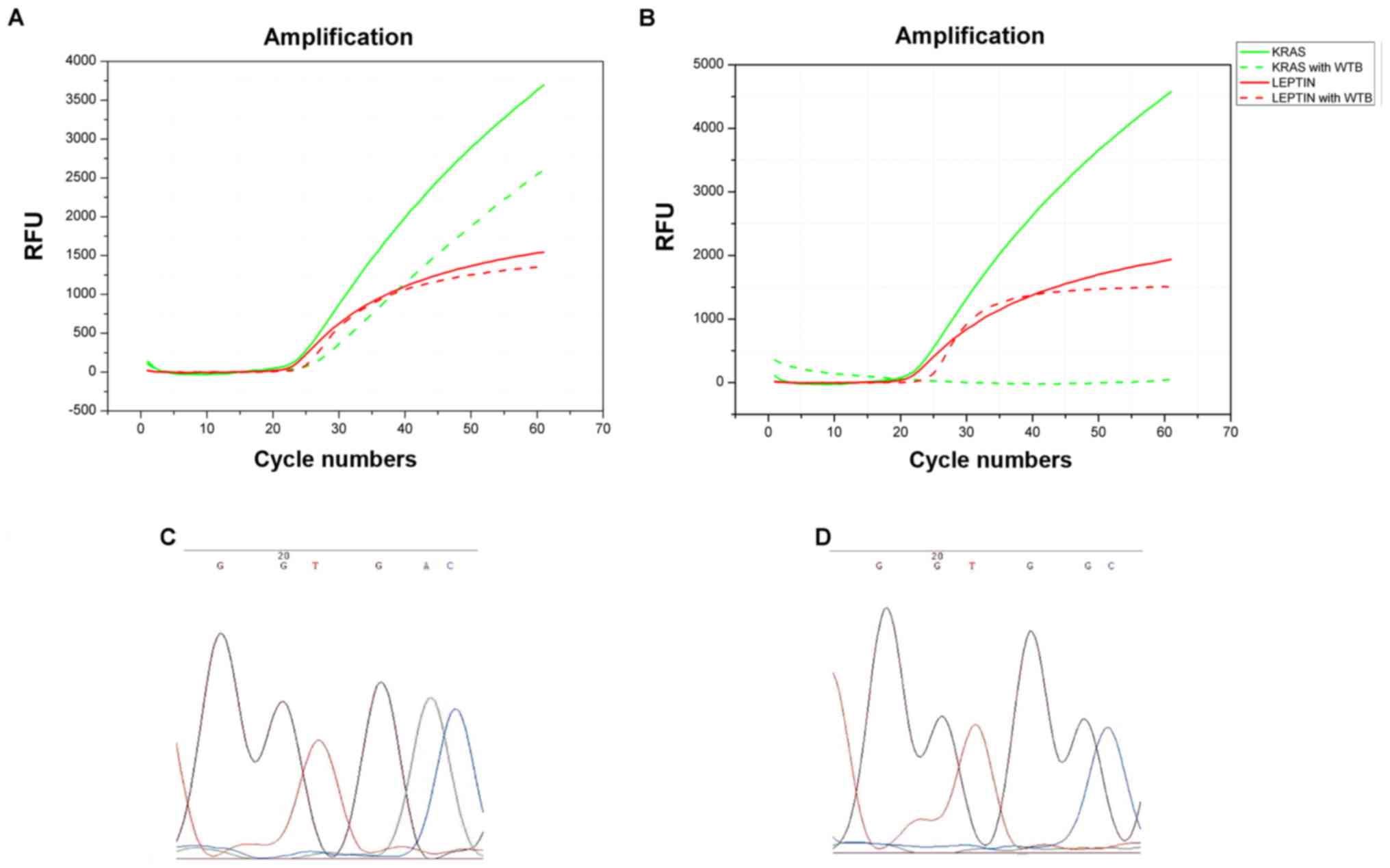
patients had trace amount of mutant KRAS (18). Fig.
4A shows a colorectal biopsy that harbored a mutant KRAS
detected by our early constructed system. The green dashed line
shows continued amplification of mutant KRAS after adding
LNA, indicating that gene mutation in KRAS occurred.
Fig. 4B shows no specific gene
amplification after adding LNA, suggesting no mutant KRAS is
contained in that particular sample. Moreover, the red lines in
Fig. 4A indicated that the
amplification of internal competitive fragment was not affected
before and after the addition of LNA, indicating a good
reproducibility of our experimental results. To further confirm the
mutation in 18 colorectal biopsies, we sent the PCR products of
these 18 cases with positive KRAS mutation for sequencing a
biotechnology company in Shanghai, China. The results showed that
18 specimens contained KRAS gene mutation. As shown in
Fig. 4C and D, the green peak
demonstrates the G to A mutation, which was consistent with the
immunohistochemical analysis of the corresponding specimens under
20x magnification of light microscopy. Calculation of the changes
of CT values of KRAS amplification group before and after
the addition of LNA as well as the CT values of the internal
competitive amplified fragment, LEPTIN, helped assess the
KRAS mutation rate in each positive sample. Among the 18
biopsies with positive mutations, the KRAS mutation rate
ranged from 18.6 to 64.2%. In the present study, the constructed
WIRE-PCR effectively detected the trace amount of mutant
KRAS in the clinical biopsies.
Discussion
Different proportions of KRAS mutation have
been discovered in a variety of human malignancies, such as
malignant melanoma, lung cancer, colorectal cancer, and thyroid
cancer. The KRAS mutation rate in the patients with rectal
cancer is approximately 40% that included the point mutations in
codons 12, 13, 15, 18, 61, 117, and 146 of exon 2, in which, the
point mutations in codons 12 and 13 of exon 2 were common,
accounting for approximately 40% (19,20).
Mutant KRAS has become one of the markers affecting the
prognosis of the colorectal cancer patients, and detection of
mutant KRAS is particularly important (21,22).
In a large number of detection assays for gene mutation,
clamp-based PCR has given us good inspiration. Application of
peptide nucleic acid (PNA) or LNA forms LNA/DNA chimeras, which
closely bind with wild-type template and prevent the amplification
of the wild-type template based on the high affinity binding
between LNA/PNA and DNA (23). In
clinical practice, LNA has been used as a substitute for PNA.
According to the high affinity binding between LNA and DNA, LNA
probe has been used to inhibit the wild-type PCR amplification
(16). The principle is that the
design of upstream and downstream primers is outside the LNA/DNA
chimeras. In this reaction system, the polymerase lacking 5′ to 3′
end exonuclease activity ensured that LNA/DNA chimera probes were
not hydrolyzed in the reaction system and LNA/DNA chimera probes
were used for the inhibition of wild-type gene (24). To target the design of
complementary oligonucleotide of sense strand KRAS, the
present study added WTB probe into the reaction system. The added
WTB probe sequence was partially overlapped with the wild-type
template. KRAS and WTB probes competitively bound to the
wild-type template in the same reaction system. Binding efficiency
between LNA of the WTB probe and template was higher, occupying the
base complementary binding region of primers and templates, thereby
interfering the binding between primers and wild-type templates and
inhibiting the amplification of the wild-type gene (11). In the 20 µl PCR system, the
addition of polymerase and bases is often superfluous and necessary
for common PCR amplification reactions, However, non-targeted
mutant gene amplification occurred between LNA/DNA chimeras and
template binding region in the WTB-involved PCR leads to false
positive results. With increasing number of PCR cycles, the
products of non-targeted mutant gene amplification are continuously
increased, which seriously affects the reading in the detection of
trace amount of the single-point mutant gene.
In our previous study, we found that the KRAS of the
wild-type samples also amplified under the WTB reaction when PCR
system near high cycles. Under the thermodynamic driving force of
DNA polymerase, the single base terminal mismatch between primers
and template could easily trigger the non-specific amplification of
an input DNA having opposite genotype (e.g., WT genotype) (25,26).
Moreover, weak-destabilization effects of terminal mismatches could
further promote non-specific amplification (27). Although stringent reaction
conditions can be used to dramatically reduce or eliminate
non-specific amplification, boptimization is time-consuming, and
sometimes unsuccessful. Internal reference gene added as internal
competitive amplified fragments in the reaction system consumed any
excess DNA polymerase and free base fragments, thereby enhancing
the blocking effect of WTB probe on wild-type KRAS template
and improving the detection efficiency for trace amount of
KRAS gene. In addition, since the internal competitive
amplified fragment had no complementary binding site with WTB
probe, WTB probe exclusively bound to the complementary template
region but not the internal competitive amplified fragment, which
did not affect the amplification of the internal competitive
amplified fragment. Addition of the primer sets of internal
reference gene, LEPTIN, massively produce the internal
competitive amplified fragments, which helped consuming any excess
DNA polymerase and free base fragment and reducing the likelihood
of false mismatch, thereby increasing the blocking effect of LNA
probe on the wild-type gene. Because this was the same reaction
system amplifying the same template, this method was able to
quantify the total amount of DNA template and reduce contamination
by simplifying the steps of the reaction. The present study
analyzed the CT values and fluorescence intensity obtained from the
amplification to optimize the WIRE-PCR system by adjusting the
final concentrations of primers of the internal reference gene and
its probe, the primer for KRAS gene and its probe, and the
LNA in the reaction system as follows: 500 nM of the primer sets
and 100 nM of the fluorescent probe of the internal reference gene;
500 nM of the primer sets and 250 nM of the fluorescent probe of
the KRAS gene; and 500 nM of the LNA probe. Subsequent
evaluation of the blocking effect on the common DNA quantity in the
clinical sample under the optimized reaction system showed that
when the wild-type templates ranged from 50 ng to 150 ng, the WTB
probe effectively blocked the amplification of the wild-type
template in the reaction system. Subsequent detection of the trace
amount of mutant KRAS in the 50 colorectal biopsies of the
patients with suspected colorectal cancer showed that 36% of the
specimens had mutant KRAS, with the mutation rate ranging
from 18.6 to 64.2%.
In summary, the constructed internal competitive
amplified fragment improved the detection of trace amount of mutant
KRAS by WTB in a real-time fluorescence-based quantitative
detection assay. Series optimization in primer concentrations,
fluorescent probe concentrations, and LNA concentration effectively
block the wild-type DNA templates of the specimens used in the PCR
system, which in turn, effectively enriched the mutant gene. The
resulted showed the sensitivity is as low as single base pair level
and completely inhabited WT-alleles of KRAS. Among the 50
colorectal biopsies collected during colonoscopy, the mutation rate
of detected trace amount of mutant KRAS was 36%. The wire
PCR in the present study was highly sensitive and specific, easily
operated and inexpensive method compared to the direct sequencing
approach. It could be extensively used to monitor gene mutation in
clinical practice and provide references for tumor monitoring and
individualized drug therapies.
Acknowledgements
This work was supported in part by grants from the
National 863 Program of China (no. 2013AA020204), and the
Scientific Foundation of Chongqing (no. CSTC2014YYKFA110029;
CSTC2015JCSF0105;CSTC2015ZDCY-ZTZX0065;
CSTC2015SHMS-ZTZXX0001).
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Venook AP: Epidermal growth factor
receptor-targeted treatment for advanced colorectal carcinoma.
Cancer. 103:2435–2446. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wadlow RC, Hezel AF, Abrams TA,
Blaszkowsky LS, Fuchs CS, Kulke MH, Kwak EL, Meyerhardt JA, Ryan
DP, Szymonifka J, et al: Panitumumab in patients with KRAS
wild-type colorectal cancer after progression on cetuximab.
Oncologist. 17:142012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Karapetis CS, Khambata-Ford S, Jonker DJ,
O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD,
Robitaille S, et al: K-ras mutations and benefit from cetuximab in
advanced colorectal cancer. N Engl J Med. 359:1757–1765. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amado RG, Wolf M, Peeters M, Van Cutsem E,
Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, et
al: Wild-type KRAS is required for panitumumab efficacy in patients
with metastatic colorectal cancer. J Clin Oncol. 26:1626–1634.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kranenburg O: The KRAS oncogene: Past,
present, and future. Biochim Biophys Acta. 1756:81–82.
2005.PubMed/NCBI
|
|
8
|
De Roock W, Jonker DJ, Di Nicolantonio F,
Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M,
Piessevaux H, et al: Association of KRAS p.G13D mutation with
outcome in patients with chemotherapy-refractory metastatic
colorectal cancer treated with cetuximab. JAMA. 304:1812–1820.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anderson SM: Laboratory methods for KRAS
mutation analysis. Expert Rev Mol Diagn. 11:635–642. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lenz HJ: Testing for RAS mutations in
patients with metastatic colorectal cancer. Clin Adv Hematol Oncol.
12:48–49. 2014.PubMed/NCBI
|
|
11
|
Oldenburg RP, Liu MS and Kolodney MS:
Selective amplification of rare mutations using locked nucleic acid
oligonucleotides that competitively inhibit primer binding to
wild-type DNA. J Invest Dermatol. 128:398–402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang Q, Wang GY, Huang JF, Zhang B and Fu
WL: High sensitive mutation analysis on KRAS gene using LNA/DNA
chimeras as PCR amplification blockers of wild-type alleles. Mol
Cell Probes. 24:376–380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dominguez PL and Kolodney MS: Wild-type
blocking polymerase chain reaction for detection of single
nucleotide minority mutations from clinical specimens. Oncogene.
24:6830–6834. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang JF, Zeng DZ, Duan GJ, Shi Y, Deng
GH, Xia H, Xu HQ, Zhao N, Fu WL and Huang Q: Single-tubed wild-type
blocking quantitative pcr detection assay for the sensitive
detection of codon 12 and 13 KRAS mutations. PLoS One.
10:e1456982015. View Article : Google Scholar
|
|
15
|
Di Giusto DA and King GC: Strong
positional preference in the interaction of LNA oligonucleotides
with DNA polymerase and proofreading exonuclease activities:
Implications for genotyping assays. Nucleic Acids Res. 32:e322004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
You Y, Moreira BG, Behlke MA and Owczarzy
R: Design of LNA probes that improve mismatch discrimination.
Nucleic Acids Res. 34:e602006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Custodio A and Feliu J: Prognostic and
predictive biomarkers for epidermal growth factor receptor-targeted
therapy in colorectal cancer: Beyond KRAS mutations. Crit Rev Oncol
Hematol. 85:45–81. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kimura T, Okamoto K, Miyamoto H, Kimura M,
Kitamura S, Takenaka H, Muguruma N, Okahisa T, Aoyagi E, Kajimoto
M, et al: Clinical benefit of high-sensitivity KRAS mutation
testing in metastatic colorectal cancer treated with anti-EGFR
antibody therapy. Oncology. 82:298–304. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Smit VT, Boot AJ, Smits AM, Fleuren GJ,
Cornelisse CJ and Bos JL: KRAS codon 12 mutations occur very
frequently in pancreatic adenocarcinomas. Nucleic Acids Res.
16:7773–7782. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eberhard DA, Johnson BE, Amler LC, Goddard
AD, Heldens SL, Herbst RS, Ince WL, Jänne PA, Januario T, Johnson
DH, et al: Mutations in the epidermal growth factor receptor and in
KRAS are predictive and prognostic indicators in patients with
non-small-cell lung cancer treated with chemotherapy alone and in
combination with erlotinib. J Clin Oncol. 23:5900–5909. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lievre A, Bachet JB, Boige V, Cayre A, Le
Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al: KRAS
mutations as an independent prognostic factor in patients with
advanced colorectal cancer treated with cetuximab. J Clin Oncol.
26:374–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Itonaga M, Matsuzaki I, Warigaya K, Tamura
T, Shimizu Y, Fujimoto M, Kojima F, Ichinose M and Murata S: Novel
methodology for rapid detection of KRAS mutation using PNA-LNA
mediated loop-mediated isothermal amplification. PLoS One.
11:e1516542016. View Article : Google Scholar
|
|
24
|
Dono M, Massucco C, Chiara S, Sonaglio C,
Mora M, Truini A, Cerruti G, Zoppoli G, Ballestrero A, Truini M, et
al: Low percentage of KRAS mutations revealed by locked nucleic
acid polymerase chain reaction: Implications for treatment of
metastatic colorectal cancer. Mol Med. 18:1519–1526.
2013.PubMed/NCBI
|
|
25
|
Chen D, Yang Z, Xia H, Huang JF, Zhang Y,
Jiang TN, Wang GY, Chuai ZR, Fu WL and Huang Q: Enhanced
specificity of TPMT*2 genotyping using unidirectional wild-type and
mutant allele-specific scorpion primers in a single tube. PLoS One.
9:e918242014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yuryev A: PCR primer design using
statistical modeling. Methods Mol Biol. 402:93–104. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wangkumhang P, Chaichoompu K, Ngamphiw C,
Ruangrit U, Chanprasert J, Assawamakin A and Tongsima S: WASP: A
Web-based Allele-Specific PCR assay designing tool for detecting
SNPs and mutations. BMC Genomics. 8:2752007. View Article : Google Scholar : PubMed/NCBI
|