Introduction
Chondrocyte hypertrophic differentiation is a
physiological stage in endochondral ossification which starts from
the condensation of mesenchymal stem cells (MSCs) (1). Then MSCs which are located within the
centre of condensation differentiate to chondrocytes (2). After proliferation, chondrocytes
present hypertrophic morphology and secrete metal matrix proteinase
to facilitate angiopoiesis and the formation of bone collar
(3). However, the pathogenesis of
osteoarthritis (OA) is that hyaline cartilages situated in the
surface of osteoarticular manifest unbalanced homeostasis and
chondrocytes differentiate into hypertrophy (4,5).
Additionally, utilizing tissue engineering to repair cartilage
defects, the regenerated cartilages are fibrocartilages and the
chondrocytes from MSCs express hypertrophic marker protein
(6,7). Thus it can be seen that inhibition of
pathological chondrocyte hypertrophy is a very important
proposition.
Xanthotoxin (XAT) is mainly isolated in plants such
as Umbelliferae and Rutaceae which are used for the treatment of
vitiligo, psoriasis and other skin diseases (8,9).
Besides, XAT can also induce apoptosis of tumor cells (10,11).
It was demonstrated that XAT had anticonvulsant pharmacological
effects in the maximal electroshock-induced seizure test (12). However, the effect of XAT on
chondrocyte differentiation remains unclear. Therefore, the purpose
of this study is to investigate the effect of XAT on chondrocyte
hypertrophy of MSCs.
Here, we set out to explore whether XAT could
inhibit hypertrophic differentiation of MSCs. Furthermore, we also
screened the upstream transcription factors of Runx2, a marker of
chondrocyte hypertrophy, to find the molecular mechanism involved
in the effect of XAT on chondrocyte hypertrophy.
Materials and methods
Reagents and cell culture
XAT (catalog no. M3501) was purchased from Sigma
(St. Louis, MO, USA) which was dissolved in DMSO. Besides, the
experimental concentration of DMSO is 0.1% and the control groups
were given a DMSO vehicle treatment. C3H10T1/2 mesenchymal stem
cells were purchased from the American Type Culture Collection
(ATCC; Manassas, VA, USA). The cells were cultured in DMEM-F12
containing 10% fetal bovine serum (FBS) (both from Hyclone, Logan,
UT, USA) and 1% penicillin-streptomycin solution (Beyotime,
Shanghai, China) at 37°C with 5% CO2. The culture medium
was replaced every 2–3 d until the cells reached 90% confluence.
Cells were passaged by 0.25% trypsin (Hyclone) for 2 min at room
temperature. The second generation cells were used for the
following experiments.
In vitro cell proliferation assay
C3H10T1/2 cells were seeded (2×103/well)
into 96-well plates (Corning, Corning, NY, USA) in triplicates and
allowed to adhere overnight. On the following day, the medium was
replaced by fresh medium with different concentrations of XAT (10,
50, 100, 150, and 250 nM). Cultures were incubated for 24 and 72 h.
Then, 10 µl Cell Counting Kit-8 (CCK-8; Dojindo Laboratories,
Japan) reagents were added to each well and incubated for an
additional 4 h. The absorbance was read at the wavelength of 450 nm
in an automated plate reader. Wells containing the CCK-8 reagents
without cells were used as the blank control. Cell proliferation
was assessed by the absorbance values according to the
manufacturer's protocol.
Apoptosis assay
MSCs were seeded (5×105/well) into 6-well
plates. XAT (50, 100, 150, 200, 250 nM) induced apoptosis of MSCs
was detected by flow cytometry using Annexin V-FITC Apoptosis
Detection kit (KGA; KeyGEN Biotechnology) according to the
manufacturer's instructions. The apoptosis rate was assayed by
using FACSCalibur flow cytometry (Becton Dickinson and
Beckman-Coulter, San Jose, CA, USA) at 488 nm.
Chondrogenic differentiation assay and
hypertrophic differentiation assay
C3H10T1/2 cells were trypsinized by 0.25% trypsin
and modulated at a density of 105 cells/ml. Two
millilitres of the suspension was placed into the center of each
well on a 6-well plate (Corning). After incubation for 24 h at 37°C
and 5% CO2, the medium was replaced by 2 ml chondrogenic
differentiation medium (Hyclone). The chondrogenic differentiation
medium composed of dexamethasone (100 nmol/l), ascorbate (50
µg/ml), ITS supplement, proline (40 µg/ml) and TGF-β3 (10 ng/ml)
was replaced every 3 days.
C3H10T1/2 cells were treated by chondrogenic
differentiation medium as previously described for 14 days. Then
the cells were inducted by hypertrophic differentiation medium
composed of dexamethasone (1 nmol/l), ascorbate (50 µg/ml), ITS
supplement, proline (40 µg/ml) and triiodothyronine (T3, 100 ng/ml)
for another 14 days. Each medium was replaced every 3 days.
Quantitative RT-PCR analysis
Total RNA was isolated using TRIzol reagent (Life
Technologies, Carlsbad, NY, USA). Single-stranded cDNA was prepared
from 1 µg of total RNA using reverse transcriptase with oligo-dT
primer according the manufacturer's instructions (Takara, Liaoning,
Dalian, China). Two microlitres of each cDNA was subjected to PCR
amplification using specific primers with detailed information in
Table I. The cycling conditions
were set as 95°C for 30 sec, 40 cycles of 95°C for 5 sec, and 60°C
for 30 sec (13). The relative
mRNA level was calculated by the normalization to that of
GAPDH.
 | Table I.Primer sequences for qPCR. |
Table I.
Primer sequences for qPCR.
| Genes | Forward (5′-3′) | Reverse (5′-3′) | Tm (°C) |
|---|
| HDAC4 |
CACTCCTCTACGGCACAAATCC |
CCAACACCACCACAAGGAAGC | 60 |
| Sox9 |
CCCAGCGAACGCACATCA |
TGGTCAGCGTAGTCGTATT | 61 |
| Col2a1 |
TGGTGGAGCAGCAAGAGC |
TGGACAGTAGACGGAGGAAA | 60 |
| GAPDH |
GTTGTCTCCTGCGACTTCA |
GGTGGTCCAGGGTTTCTTA | 62 |
| Col10a1 |
CTTTCTGGGATGCCGCTTGT |
GGGTCGTAATGCTGCTGCCTA | 61 |
| Runx2 |
CCAACTTCCTGTGCTCCGTG |
ATAACAGCGGAGGCATTTCG | 63 |
| Mmp13 |
TTGATGCCATTACCAGTCTCCG |
CACGGGATGGATGTTCATATGC | 61 |
Western blot analysis
The cells were extracted with lysis buffer
containing 50 mM Tris (pH 7.6), 150 mM NaCl, 1% Triton X-100, 1%
deoxycholate, 0.1% SDS, 1 mM PMSF and 0.2% Aprotinin (Beyotime).
After we measured the protein concentration, the equal protein
samples were mixed with 5X sample buffer (Beyotime) and boiled. The
proteins (30 µg) were resolved by 10% SDS-PAGE gel and transferred
on PVDF membrane (Millipore, Hong Kong, China) by using the
semi-dry transfer method. After blocking in 10% nonfat dried milk
in TBST for 2 h, the blots were incubated with primary antibodies
including HDAC4 (rabbit polyclonal 1:500, ab12172), p38 (rabbit
polyclonal 1:500, ab27986), Runx2 (rabbit polyclonal 1:500,
ab23981), Col10a1 (rabbit polyclonal 1:500, ab58632) (all from
Abcam), p-p38 (rabbit polyclonal 1:500, bs-50486R), Mmp13 (rabbit
polyclonal 1:500, bs-0575R) (both from Bioss) and GAPDH (rabbit
polyclonal 1:1,000, ab37168; Abcam) at 4°C overnight. After washing
by TBST, the blots were incubated with a horseradish
peroxidase-conjugated secondary antibody (diluted 1:2,000, sc-2374;
Santa Cruz Biotechnology) at room temperature for 1 h. Blots
against GAPDH served as loading control.
Glycosaminoglycan (GAG) synthesis
analysis by alcian blue staining
To demonstrate the deposition of cartilage matrix
proteoglycans, representative cultures were collected at day 28,
Then sulfated cartilage glycosaminoglycans (GAGs) were measured by
alcian blue (ScienCell) staining. The cells were fixed in 4%
paraformaldehyde for 15 min and incubated in 3% acetic aicd for 3
min. Then the cells were stained with alcian blue for 30 min. The
mean density was normalized to total cell number.
Immunohistochemistry
The cells were fixed in 4% paraformaldehyde for 15
min. This was followed by washing in phosphate-buffered saline
(PBS) and treated with H2O2 (ZSGB-BIO,
Peking, China) for 10 min to inactivate endogenous peroxidase.
After treatment with normal goat serum (ZSGB-BIO) at room
temperature for 15 min, cells were incubated with primary
antibodies including Runx2 (rabbit polyclonal 1:150, ab23981) and
Col10a1 (rabbit polyclonal 1:150, ab58632) (both from Abcam) at 4°C
overnight. After washing, the cells were incubated with
biotinylated goat anti-rabbit (ZSGB-BIO) secondary antibodies for
30 min, followed by washing and incubation with horseradish
peroxidase (HRP) (ZSGB-BIO) for 15 min. The area of the
immunocomplex was visualized by chromogen 3,3′-diaminobenzidine
(DAB) for 5 min. The cells were investigated under the Olympus
microscope. Image-Pro Plus 6.0 software was used to analyze the IOD
and area to calculate mean density of images.
Statistics
All data are representative of at least three
experiments of similar results performed in triplicate unless
otherwise indicated. Data are expressed as mean ± SEM. One-way
ANOVA followed by Student-Newman-Keuls post hoc tests was used to
determine the significance of difference between results, with
P<0.05, P<0.01 and P<0.001 being regarded as
significant.
Results
Toxicity evaluation of XAT on
MSCs
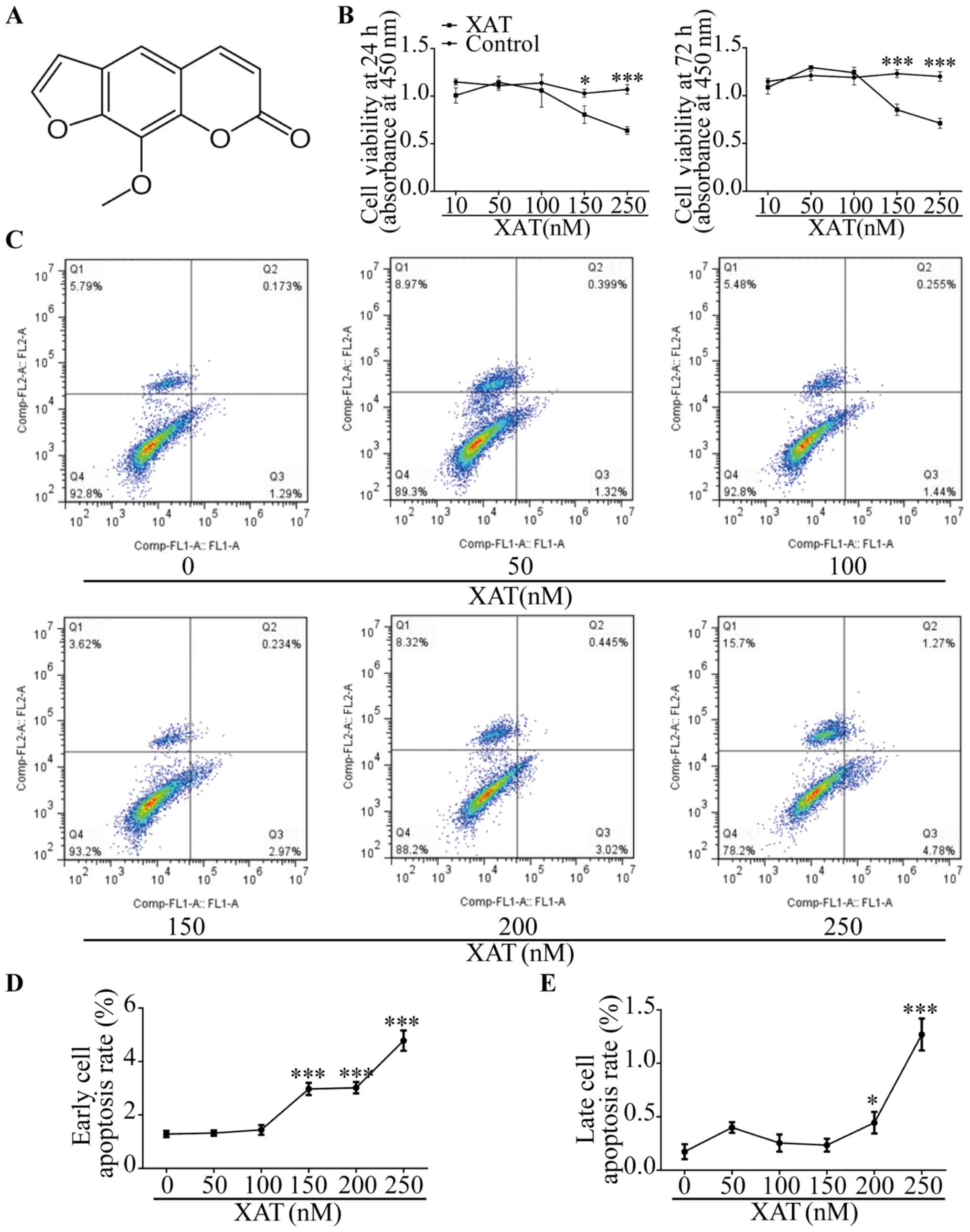
The chemical formula of XAT is shown (Fig. 1A) (14). MSCs were treated with basic medium
for 24 h and 72 h with different concentrations of XAT followed by
CCK-8 assays (Fig. 1B). The
results showed that concentration of XAT higher than 100 nM
inhibited proliferation of MSCs (P<0.05). After culturing MSCs
with basic medium for 72 h, we also investigated effect of XAT on
cell apoptosis by FCM (Fig. 1C).
In the plots, X axis represented Annexin V-FITC and Y axis
represented Propidium iodide. The results revealed that XAT
concentration above 100 nM affected early cell apoptosis and above
150 nM promoted late cell apoptosis (P<0.05, Fig. 1D and E). Therefore, the safe
concentration of XAT used in following experiments was 100 nM.
XAT inhibited expression of
chondrocyte hypertrophic marker genes
MSCs were treated with chondrogenic differentiation
medium containing TGF-β3 (10 ng/ml) for 14 days. RT-PCR was
performed to evaluate the expression of chondrogenic genes. The
results showed that expressions of Sox9 and Col2a1 were increased
(P<0.01) on day 14 compared with day 0 (Fig. 2A). Then the cells were induced by
hypertrophic medium containing T3 (5 ng/ml) for another 14 days.
Results of RT-PCR revealed that hypertrophic markers including
Runx2 and Col10a1 were increased (P<0.01) on day 28 compared
with day 14 (Fig. 2B). According
to this in vitro culture system, we further research the
effect of XAT on hypertrophic differentiation of MSCs. After
treated with chondrogenic differentiation medium for 14 days
without XAT, the cells were induced by hypertrophic medium with
DMSO or with XAT (100 nM) for another 7 and 14 days. From the
alcian blue stain, we found that the groups treated with XAT had
higher (P<0.05) intensity than control groups. In addition, the
intensity at the concentration of 100 nM was higher (P<0.05)
than 50 nM (Fig. 2C and D).
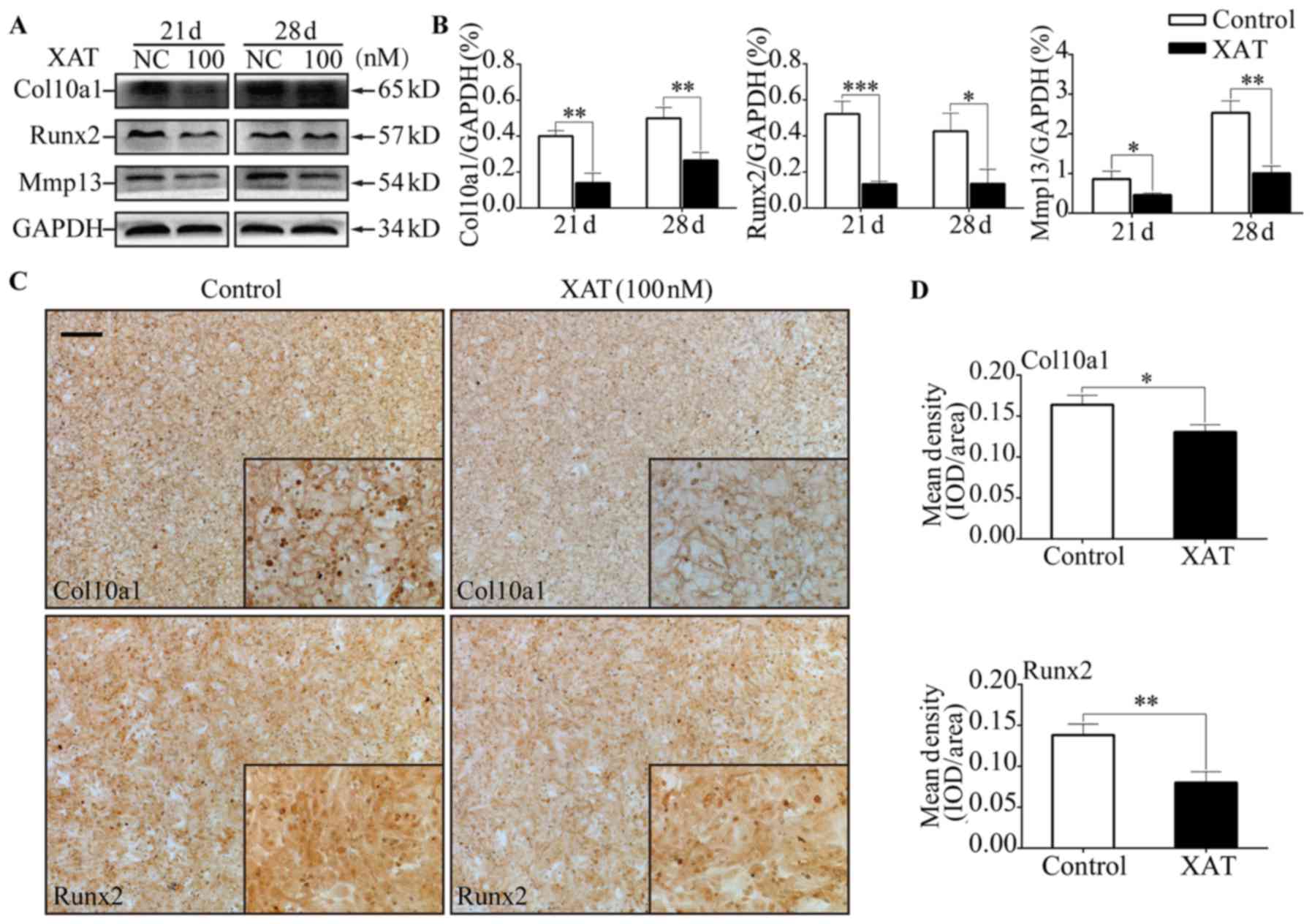
Besides, after treatment with XAT, expressions of Col10a1, Runx2
and Mmp13 on mRNA level were decreased (P<0.05) on day 17, 21
and 28 (Fig. 2E). Results of
western blotting revealed that hypertrophic markers including
Col10a1, Runx2 and Mmp13 were decreased (P<0.05) in cells
treated with XAT on day 21 and 28 (Fig. 3A and B). We also performed
immunohistochemistry to test the effect of XAT on expression of
hypertrophic marker genes. The results showed that the mean density
of Col10a1 and Runx2 in groups treated with XAT on day 28 (Fig. 3C and D) were lower (P<0.05) than
those in groups without XAT. Taken together, these results
demonstrated that chondrocyte hypertrophic differentiation of MSCs
was inhibited by XAT at the concentration of 100 nM.
XAT suppressed phosphorylation of p38
and promoted HDAC4 expression during chondrocyte hypertrophic
differentiation of MSCs
To explore the molecular mechanisms involved in the
effect of XAT on of MSCs, qPCR was performed to test the key
transcriptional factors related to chondrocyte hypertrophy.
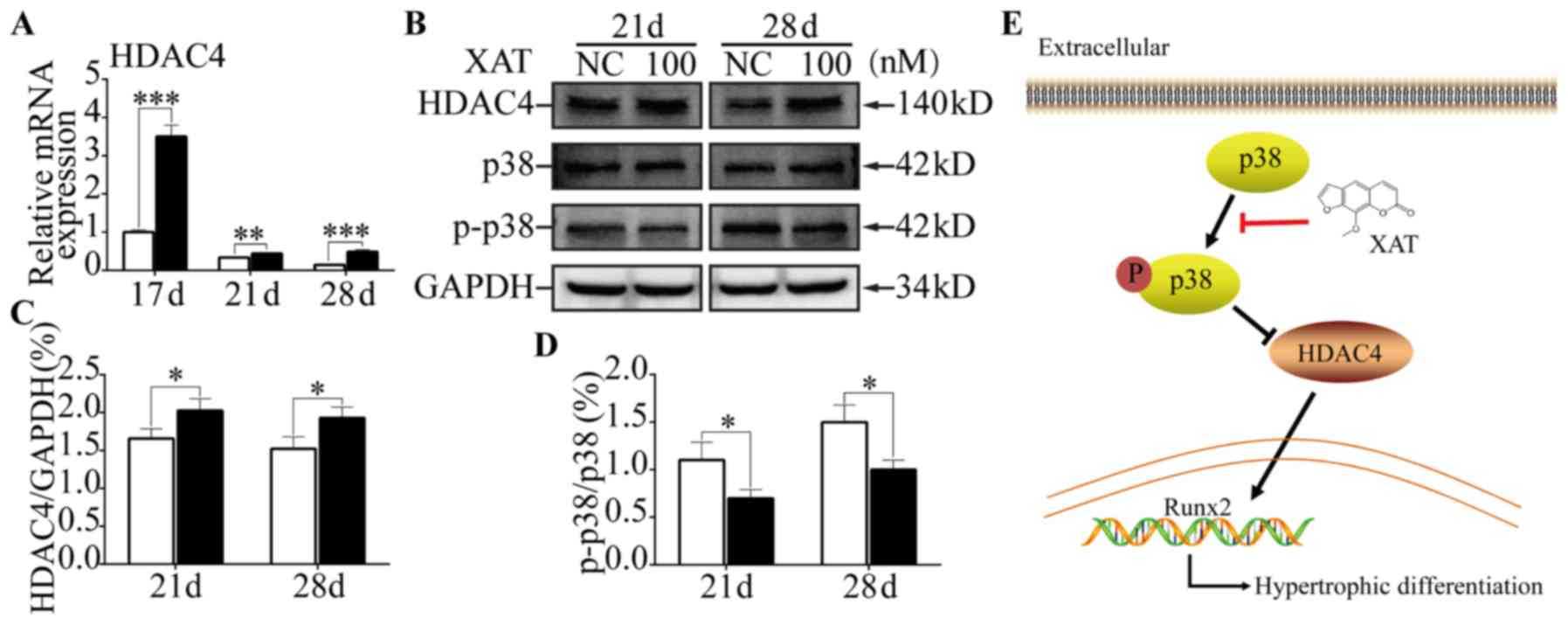
Compared with control groups, we found expression of HDAC4 on mRNA
level was increased (P<0.01) with XAT on day 21 and 28 (Fig. 4A). From the results of western
blotting (Fig. 4B), HDAC4
expression was increased (P<0.05) on day 21 and 28 in the
presence of XAT (Fig. 4C). To
further illustrate the mechanism of XAT upregulating expression of
HDAC4, we next performed western blotting to test the
phosphorylation of p38-MAPK (Fig.
4B). The results revealed that, following XAT treatment,
phosphorylation proportion of p38 was lower (P<0.05) than
control groups on day 21 and 28 (Fig.
4D). Taken together, these results indicated that XAT
suppressed phosphorylation of p38 and promoted HDAC4 expression
during hypertrophic differentiation of MSCs.
Discussion
XAT also called Methoxsalen, has various functions,
such as anti-inflammatory, analgesic and anti-oxidation (15–17).
It is well established that chondrocyte hypertrophy is closely
related to inflammation in osteoarticular (18). Based on the characteristics of XAT,
we hypothesized that XAT might suppress chondrocyte hypertrophic
differentiation of MSCs. A study has found XAT is a new class of
anti-glioma drug via evaluating the effect of XAT in MDA-MB-231,
CT-26 and SCC-3 cell lines (19).
Besides, our group also confirmed that XAT prevents bone loss by
inhibiting RANKL-induced osteoclastogenesis (20).
This is the only study to our knowledge to research
the role of XAT on chondrocytes. In this study, we provide evidence
that XAT could decrease glycosaminoglycan degradation and suppress
expressions of chondrocyte hypertrophic genes including Runx2,
Mmp13 and Col10a1. Then we draw a conclusion that XAT suppresses
chondrocyte hypertrophic differentiation of MSCs. The transcription
factor Runx2 is crucial for regulating chondrocyte hypertrophy
(21,22). Overexpression of Runx2 in all
chondrocytes resulted in premature chondrocyte hypertrophy
(23). We screened several
upstream genes of Runx2 to explore the mechanism such as Bapx1,
Hoxa2 and HDAC4 through RT-PCR. A recent study showed that Sox9
directly promoted Bapx1 gene expression to repress Runx2 in
chondrocytes (24). Besides, Hoxa2
restricted the chondrogenic domain and inhibited bone formation
through suppressed Runx2 expression (25). Studies have found HDAC4, a member
of the histone deacetylases, interacts with RUNX2 and decreases
RUNX2 binding to DNA. Deletion of HDAC4 greatly promoted
chondrocyte hypertrophy while forced expression of HDAC4 in
chondrocytes suppressed hypertrophy (2,26).
It has been confirmed that HDAC4, which was expressed in
prehypertrophic chondrocytes, regulated chondrocyte hypertrophy by
interacting with and inhibiting the activity of Runx2 (27). Besides, a study revealed that HDAC4
inhibited Runx2 promoter activity in a dosage-dependent manner
(28). Therefore, Bapx1, Hoxa2 and
HDAC4 might prevent hypertrophy of chondrocytes through reducing
RUNX2 activity. Following XAT treatment, Bapx1 and Hoxa2 expression
on mRNA level was not changed (data not shown) while HDAC4
expression was increased on day 21 and 28. The protein level was
consistent with RT-PCR. Jun N-terminal kinase (JNK) and p38
mitogen-activated protein kinase (MAPK) signaling is associated
with cancers and innate immunity. Inhibition of the p38-MAPK
significantly reduced the proliferation and invasion of pancreatic
cancer cells under high-glucose conditions (29). Besides, it has been confirmed that
neospora caninum increased p38 phosphorylation to downregulate the
host's innate immune responses (30). A recent study showed that p38
promotes HDAC4 degradation by increasing caspase-mediated cleavage
(31). Our results showed XAT
could increase phosphorylation of p38 in MSCs. So we figured out
that XAT inhibited chondrocyte hypertrophy through down-regulating
p38-MAPK signaling pathway, then reducing degradation of HDAC4
(Fig. 4E).
In OA, chondrocytes exhibit hypertrophic morphology
and the homeostasis of chondrocyte is broken down. Hhpertrophic
chondrocytes express crucial transcription factor Runx2 and secrete
Mmp13, which leads to degradation of extracellular matrix (32). Then the function of articular
cartilage is severely impaired. Moreover, regenerated cartilage
which is constructed by tissue engineering always presents
fibrocartilage but hyaline cartilage. Chondrocyte hypertrophic
differentiation is the root to cause above phenomenon (33). Hence, inhibition of chondrocyte
hypertrophy is the key point to treat arthritis and maintain the
function of regenerated cartilage.
XAT, from the common spices, could suppress
expressions of chondrocyte hypertrophic genes by inactivating
p38-MAPK/HDAC4 signaling pathway. The above data indicate that XAT
can be used as a potential drug for the treatment of arthritis and
also for maintaining the homeostasis of regenerated cartilage in
construction of tissue engineered cartilage.
Acknowledgements
This work was funded by grants from the Nature
Science Foundation of China (no. 81571893 and 81271980), the
National High-Tech R&D Program of China (863 Program,
2015AA020315) and the Key Project of Logistics Research Plan of PLA
(BWS13C014).
References
|
1
|
Hinton RJ, Jing Y, Jing J and Feng JQ:
Roles of chondrocytes in endochondral bone formation and fracture
repair. J Dent Res. 96:23–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Long F and Ornitz DM: Development of the
endochondral skeleton. Cold Spring Harb Perspect Biol.
5:a0083342013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancedda R, Castagnola P, Cancedda FD,
Dozin B and Quarto R: Developmental control of chondrogenesis and
osteogenesis. Int J Dev Biol. 44:707–714. 2000.PubMed/NCBI
|
|
4
|
Glyn-Jones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bianchi A, Guibert M, Cailotto F, Gasser
A, Presle N, Mainard D, Netter P, Kempf H, Jouzeau JY and Reboul P:
Fibroblast growth factor 23 drives MMP13 expression in human
osteoarthritic chondrocytes in a Klotho-independent manner.
Osteoarthritis Cartilage. 24:1961–1969. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang T, Lai JH and Yang F: Effects of
hydrogel stiffness and extracellular compositions on modulating
cartilage regeneration by mixed populations of stem cells and
chondrocytes in vivo. Tissue Eng Part A. 22:1348–1356. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Di Luca A, Szlazak K, Lorenzo-Moldero I,
Ghebes CA, Lepedda A, Swieszkowski W, Van Blitterswijk C and Moroni
L: Influencing chondrogenic differentiation of human mesenchymal
stromal cells in scaffolds displaying a structural gradient in pore
size. Acta Biomater. 36:210–219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Garg BJ, Garg NK, Beg S, Singh B and
Katare OP: Nanosized ethosomes-based hydrogel formulations of
methoxsalen for enhanced topical delivery against vitiligo:
Formulation optimization, in vitro evaluation and preclinical
assessment. J Drug Target. 24:233–246. 2016. View Article : Google Scholar
|
|
9
|
Archier E, Devaux S, Castela E, Gallini A,
Aubin F, Le Maître M, Aractingi S, Bachelez H, Cribier B, Joly P,
et al: Carcinogenic risks of psoralen UV-A therapy and narrowband
UV-B therapy in chronic plaque psoriasis: A systematic literature
review. J Eur Acad Dermatol Venereol. 26:(Suppl 3). S22–S31. 2012.
View Article : Google Scholar
|
|
10
|
Yang H, Xiong J, Luo W, Yang J and Xi T:
8-Methoxypsoralen induces intrinsic apoptosis in HepG2 cells:
Involvement of reactive oxygen species generation and ERK1/2
pathway inhibition. Cell Physiol Biochem. 37:361–374. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Panno ML, Giordano F, Palma MG, Bartella
V, Rago V, Maggiolini M, Sisci D, Lanzino M, De Amicis F and Andò
S: Evidence that bergapten, independently of its photoactivation,
enhances p53 gene expression and induces apoptosis in human breast
cancer cells. Curr Cancer Drug Targets. 9:469–481. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zagaja M, Andres-Mach M, Patrzylas P,
Pyrka D, Szpringer M, Florek-Łuszczki M, Żółkowska D,
Skalicka-Woźniak K and Łuszczki JJ: Influence of xanthotoxin
(8-methoxypsoralen) on the anticonvulsant activity of various novel
antiepileptic drugs against maximal electroshock-induced seizures
in mice. Fitoterapia. 115:86–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian W, Cai J, Xu Y, Luo X, Zhang J, Zhang
Z, Zhang Q, Wang X, Hu L and Lin G: Determination of xanthotoxin
using a liquid chromatography-mass spectrometry and its application
to pharmacokinetics and tissue distribution model in rat. Int J
Clin Exp Med. 8:15164–15172. 2015.PubMed/NCBI
|
|
15
|
Wolnicka-Glubisz A, Sarna T, Klosner G,
Knobler R and Trautinger F: UVA activated 8-MOP and chlorpromazine
inhibit release of TNF-alpha by post-transcriptional regulation.
Photochem Photobiol Sci. 3:334–336. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen YF, Tsai HY and Wu TS:
Anti-inflammatory and analgesic activities from roots of
Angelica pubescens. Planta Med. 61:2–8. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ng TB, Liu F and Wang ZT: Antioxidative
activity of natural products from plants. Life Sci. 66:709–723.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Caron MMJ, Emans PJ, Surtel DA, van der
Kraan PM, van Rhijn LW and Welting TJ: BAPX-1/NKX-3.2 acts as a
chondrocyte hypertrophy molecular switch in osteoarthritis.
Arthritis Rheumatol. 67:2944–2956. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
de Oliveira DM, Lima RM Ferreira,
Clarencio J, Velozo Eda S, de Amorim IA, da Andrade Mota TH, Costa
SL, Silva FP and El-Bachá Rdos S: The classical photoactivated drug
8-methoxypsoralen and related compounds are effective without UV
light irradiation against glioma cells. Neurochem Int. 99:33–41.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dou C, Chen Y, Ding N, Li N, Jiang H, Zhao
C, Kang F, Cao Z, Quan H, Luo F, et al: Xanthotoxin prevents bone
loss in ovariectomized mice through the inhibition of RANKL-induced
osteoclastogenesis. Osteoporosis Int. 27:2335–2344. 2016.
View Article : Google Scholar
|
|
21
|
Liu CF, Samsa WE, Zhou G and Lefebvre V:
Transcriptional control of chondrocyte specification and
differentiation. Semin Cell Dev Biol. 62:34–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo F, Han X, Wu Z, Cheng Z, Hu Q, Zhao Y,
Wang Y and Liu C: ATF6a, a Runx2-activable transcription factor, is
a new regulator of chondrocyte hypertrophy. J Cell Sci.
129:717–728. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takeda S, Bonnamy JP, Owen MJ, Ducy P and
Karsenty G: Continuous expression of Cbfa1 in nonhypertrophic
chondrocytes uncovers its ability to induce hypertrophic
chondrocyte differentiation and partially rescues Cbfa1-deficient
mice. Genes Dev. 15:467–481. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamashita S, Andoh M, Ueno-Kudoh H, Sato
T, Miyaki S and Asahara H: Sox9 directly promotes Bapx1 gene
expression to repress Runx2 in chondrocytes. Exp Cell Res.
315:2231–2240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanzler B, Kuschert SJ, Liu YH and Mallo
M: Hoxa-2 restricts the chondrogenic domain and inhibits bone
formation during development of the branchial area. Development.
125:2587–2597. 1998.PubMed/NCBI
|
|
26
|
Chen W, Sheng P, Huang Z, Meng F, Kang Y,
Huang G and Zhang Z, Liao W and Zhang Z: MicroRNA-381 regulates
chondrocyte hypertrophy by inhibiting histone deacetylase 4
expression. Int J Mol Sci. 17:E13772016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Vega RB, Matsuda K, Oh J, Barbosa AC, Yang
X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et
al: Histone deacetylase 4 controls chondrocyte hypertrophy during
skeletogenesis. Cell. 119:555–566. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li P, Wei X, Guan Y, Chen Q, Zhao T, Sun C
and Wei L: MicroRNA-1 regulates chondrocyte phenotype by repressing
histone deacetylase 4 during growth plate development. FASEB J.
28:3930–3941. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L, Bai YY, Yang Y, Hu F, Wang Y, Yu
Z, Cheng Z and Zhou J: Diabetes mellitus stimulates pancreatic
cancer growth and epithelial-mesenchymal transition-mediated
metastasis via a p38 MAPK pathway. Oncotarget. 7:38539–38550. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mota CM, Oliveira AC, Davoli-Ferreira M,
Silva MV, Santiago FM, Nadipuram SM, Vashisht AA, Wohlschlegel JA,
Bradley PJ, Silva JS, et al: Neospora caninum activates p38 MAPK as
an evasion mechanism against innate immunity. Front Microbiol.
7:14562016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou JM, Li PC, Chen Q, Wei X, Zhao T,
Wang Z and Wei L: Mitogen-activated protein kinase p38 induces
HDAC4 degradation in hypertrophic chondrocytes. Biochim Biophys
Acta. 1853:370–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kung LH, Zaki S, Ravi V, Rowley L, Smith
MM, Bell KM, Bateman JF and Little CB: Utility of circulating serum
miRNAs as biomarkers of early cartilage degeneration in animal
models of post-traumatic osteoarthritis and inflammatory arthritis.
Osteoarthritis Cartilage. 25:426–434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Doran PM: Cartilage tissue engineering:
What have we learned in practice? Methods Mol Biol. 1340:3–21.
2015. View Article : Google Scholar : PubMed/NCBI
|