Introduction
Osteosarcoma, the most common primary bone cancer,
occurs mainly in adolescents and young adults, which is
characterized by high malignant and metastatic potentials. It
primarily occurs in actively growing long bone metaphysis (1). Despite an intensive search for new
therapeutic strategies, survival rates have not improved over the
past two decades (2). Because the
metastatic process comprises a series of steps all of which require
the participation of specific molecules. Therefore, it is crucial
to identify novel molecules and novel alternative therapeutic
strategies to improve the clinical outcome of patients with
osteosarcoma.
Homeobox genes (HOX) encode a large family of
transcriptional factors, which are essential for embryonic
development and tumorigenesis (3,4). In
addition, they are frequently deregulated in cancer where they
variably influence tumor cell proliferation, apoptosis, stem cell
renewal, differentiation, motility and angiogenesis (5–7). The
authors previously reported that HOXB7 overexpression confers
tamoxifen-resistance through upregulation of EGFR signaling in
breast cancer (8). Increased
expression of HOXB7 has also been described in oral squamous cell
carcinoma, where it induces cell proliferation and has been
indicated to be associated with poor prognosis (9). Moreover, in colorectal cancer, the
protein encoded by HOXB7 was considered as a prognostic factor and
mediator of tumor development and progression (10). Besides, the present study
demonstrates that decreasing the HOXB7 expression level by small
interfering (si)RNA could significantly increases cell cycle arrest
and apoptosis in pancreatic ductal adenocarcinomas (11). In addition, previous studies have
demonstrated that overexpression of HOXB7 is closely associated
with the clinical progression and poor prognosis of patients with
lung adenocarcinoma, esophageal squamous cell cancer (12,13)
and gastric cancer (14). However,
the role of HOXB7 in osteosarcoma has not been reported.
In the current study, the authors demonstrated that
the expression of HOXB7 was increased in osteosarcoma tissues and
cell lines compared with paired adjacent non-tumor bone tissues and
osteoblastic cells. Following this, knockdown of HOXB7 expression
was presented to inhibit cell viability, proliferation and
migration, and suppress epithelial-mesenchymal transition (EMT), in
an attempt to elucidate the potential influence of HOXB7 in the
development of osteosarcoma.
Materials and methods
Tissue samples and cell lines
The 32 paired osteosarcoma specimens and adjacent
non-tumor tissues used in the present study were obtained from
surgically excised samples from Affiliated Hospital of Nantong
University (Nantong, China). All research involving human tissue
samples was approved by the Ethics Review Committee of Affiliated
Hospital of Nantong University (Nantong, China) and written
informed consent was obtained from all participating patients.
Saos-2, MG-63, HOS and U2OS osteosarcoma cell lines,
and immortalized human fetal osteoblastic cell line hFOB 1.19, were
obtained from the American Type Culture Collection (Manassas, VA,
USA). Cells were cultured RPMI-1640 medium supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific Inc., Waltham,
MA, USA) and maintained at 37°C under 5% CO2.
Transfection of siRNA
For siRNA silencing of HOXB7, RNA interference was
performed by using synthetic siRNA duplexes. HOXB7 siRNA and
scrambled siRNA (NC-siRNA) were purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China). The targeting sequences were as
follows: siRNA1, 5′-GGAGCCTTCCCAGAACAAA-3′; siRNA2,
5′-CCCTTTGAGCAGAACCTCT-3′; siRNA3, 5′-GCCTCACGGAAAGACAGAT-3′. In
the present study, siRNA3 was used as it can effectively reduce
endogenous HOXB7 expression. The target sequence for scrambled
siRNA was 5′-GCAGATAGGTAGGCGTTAT-3′. Si-HOXB7 or scramble siRNA
were transfected into MG63 cells at 400 pmol respectively using
Lipofectamine RNAiMAX transfection reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocols.
Following 24 h of transfection, cells were harvested for cell
proliferation, migration and colony formation assays.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues lysate using a
TRIzol kit (Invitrogen; Thermo Fisher Scientific, Inc.), and cDNA
was subsequently synthesized from total RNA using an Omniscript RT
kit (Qiagen, Inc., Valencia, CA) following the supplier's
protocols. RT-qPCR was conducted on the Mastercycler Ep Realplex
(Eppendorf, Hamburg, Germany). The reactions used the following
cycling conditions: Incubation at 96°C for 2 min, 40 cycles at 96°C
for 15 sec and 60°C for 1 min. The Cq value was defined as the
cycle number at which the fluorescence intensity reached a certain
threshold where amplification of each target gene was within the
linear region of the reaction amplification curves. GAPDH gene
served as an internal control. Relative expression level for each
target gene was normalized by the Ct value of GAPDH using a
2−ΔΔCq relative quantification method (15). The sequences of the primers for
HOXB7 as follows: HOXB7 forward, 5′-ACACGCTCTGCCTCACG-3′; HOXB7
reverse, 5′-GCTTCAGCCCTGTCTTGG-3′.
Western blot analysis
Equal amounts of protein (20 µg) were separated by
10% SDS-PAGE and transferred onto polyvinylidene difluoride
membranes. The membranes were blocked with 5% non-fat milk in TBS.
Following blocking, the target proteins were probed with the
following: Anti-HOXB7 antibody (cat. no. ab196007; 1:500; Abcam,
Cambridge, MA, USA), anti-E-cadherin antibody (cat. no. ab1416;
1:1,000; Abcam), anti-vimentin antibody (cat. no. ab92547; 1:1,000;
Abcam), anti-N-cadherin antibody (cat. no. ab18203; 1:1,000,
Abcam), anti-Snail antibody (cat. no. ab180714; 1:1,000; Abcam),
anti-Slug antibody (cat. no. ab27568; 1:1,000; Abcam) and
anti-Twist antibody (cat. no. ab49254; 1:1,000; Abcam) overnight at
4°C. Then, the blots were washed and incubated with a horseradish
peroxidase-conjugated secondary antibody (cat. no. ab6789 or cat.
no. ab6721; 1:5,000; Abcam). The membrane was incubated for 1 h at
room temperature and washed with PBS three times, each time for 10
min. An enhanced chemiluminescence kit (cat. no. 32106; Thermo
Fisher Scientific, Inc.) was used to visualize the membrane. The
densities of protein bands were analyzed using PDQuest software
version 7.2.0 (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
expression of proteins was normalized to β-actin.
Cell viability assay
Cell viability was analyzed using an MTT assay.
Cells transfected with HOXB7 siRNA, scrambled siRNA (NC) or no
transfection (untreated) were seeded into 96-well plates
(5×103 cells/well) and incubated for 1, 2, 3, 4 and 5
days, respectively. Following incubation with 25 µl MTT (5 mg/ml)
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C for 4 h,
the supernatants were removed, and 150 µl dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) was added to each well. The absorbance
value of each well was measured at 490 nm. Experiments were
repeated at least three times.
Colony formation assay
Cells (5×104 cells/well) were separately
plated in a 24-well plate. At 24 h, the cells were collected and
seeded (1,000–1,500/well) in a fresh six-well plate for 14 days.
Surviving colonies (>50 cells per colony) were counted following
fixing with methanol/acetone and stained with 5% Gentian Violet
(ICM Pharma, Singapore), and then rinsed three times with PBS to
remove excess dye, photographed and counted. The experiment was
carried out in triplicate.
Transwell assay
The migration ability of cells was measured in
Transwell chambers with 8.0 mm pore polycarbonate membrane insert
(Corning Incorporated, Corning, NY, USA) according to the
manufacturer's protocols. Cells (5×104/ml) suspended in
Dulbecco's' modified Eagle's medium (DMEM) were added to the upper
chamber, and the plate was incubated with 5% CO2 for 12
h at 37°C. The lower chamber of the plate was filled with 500 µl
DMEM containing 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells on the upper surface of the filters were
removed using cotton swabs. The migrated cells to the lower surface
of the filters were washed, fixed, stained with Giemsa and counted
under a microscope. Experiments were repeated at least three
times.
Statistical analysis
All data were analyzed using SPSS software version
17.0 (SPSS, Inc., Chicago, IL, USA) and presented as the mean ±
standard error of the mean. Statistical analysis was determined
using Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
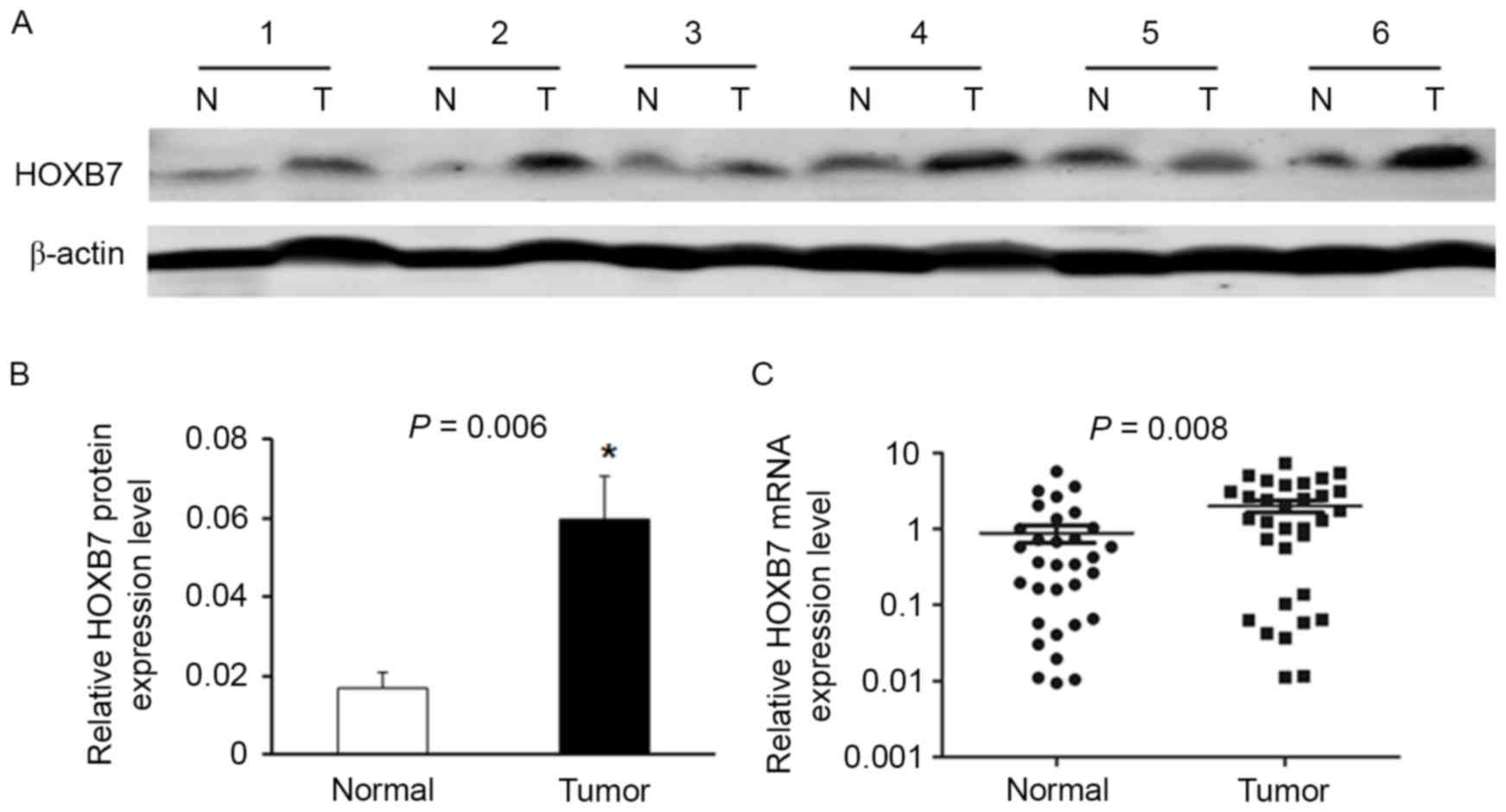
Expression of HOXB7 was upregulated in
osteosarcoma
Western blotting indicated that the expression of
HOXB7 in osteosarcoma tissues was significantly upregulated
compared with corresponding adjacent non-tumor tissues. A total of
6 representative pairs of the western blotting results are
presented in Fig. 1A. In addition,
the protein level of HOXB7 was higher in osteosarcoma tissues than
in their adjacent non-tumor tissues in the 24 randomly selected
pairs (Fig. 1B; P=0.006).
Moreover, to measure HOXB7 mRNA level in osteosarcoma tissues,
RT-qPCR was performed in the 32 tumor tissues and corresponding
non-tumor samples. The results demonstrated that HOXB7 expression
was significantly increased in tumor samples when compared with
that in their matched non-tumor tissues (Fig. 1C; P=0.008).
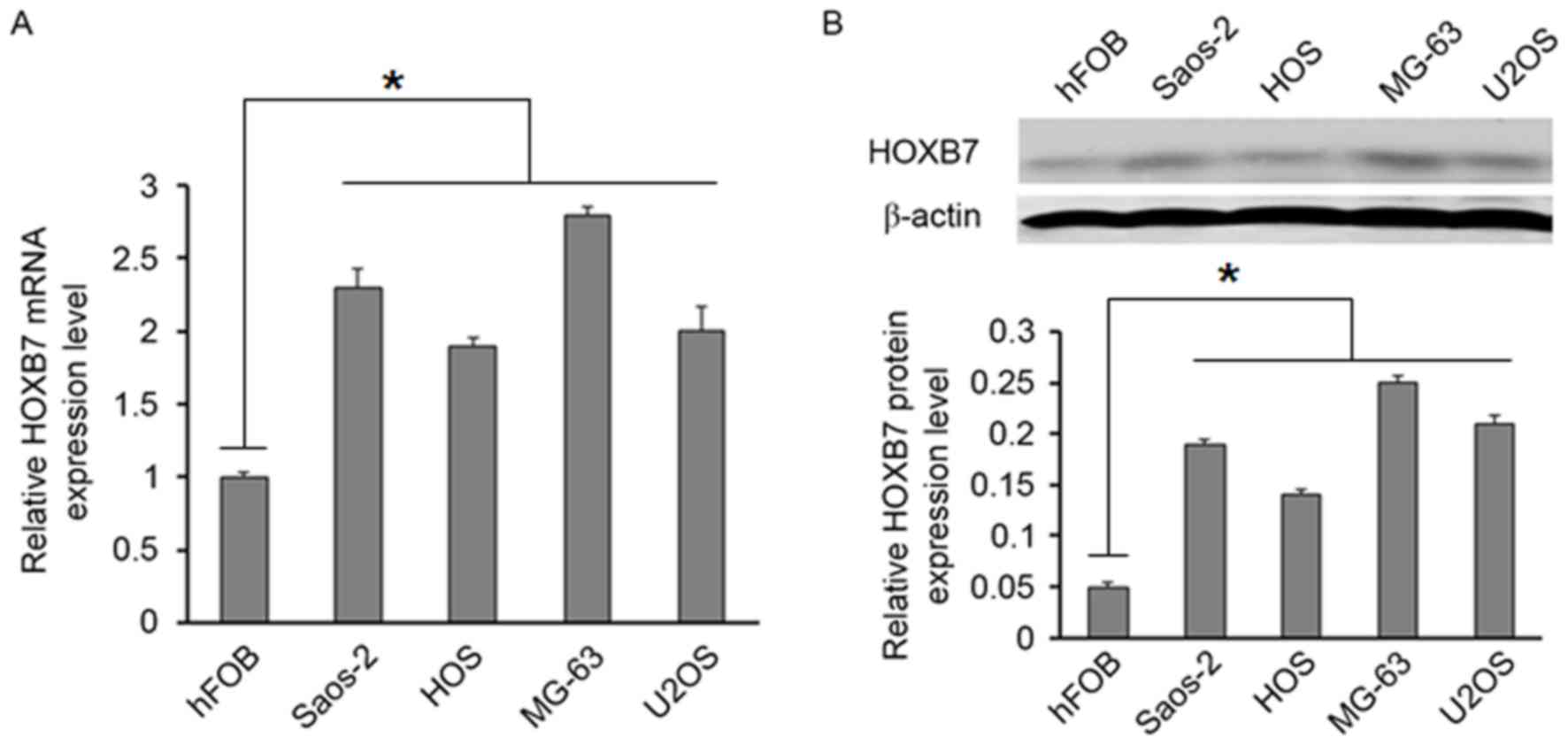
Furthermore, the expression of HOXB7 was
investigated in four osteosarcoma cell lines (MG63, U2OS, Saos-2
and HOS). RT-qPCR and western blot analysis indicated that the
HOXB7 level was higher in four osteosarcoma cell lines than in the
hFOB 1.19 cell line (Fig. 2A and
B).
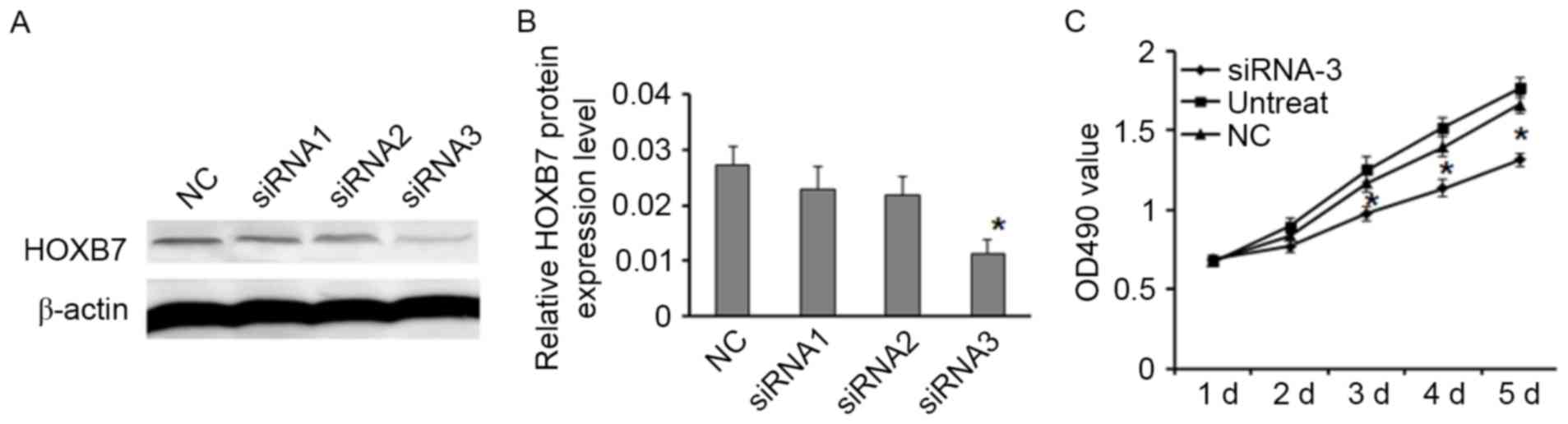
Downregulation of HOXB7 inhibited cell
viability, proliferation and migration
Western blotting indicated that HOXB7 siRNA-3
decreased the level of HOXB7 expression more effectively than
control and other siRNAs (Fig. 3A and
B). To obtain a further insight into the role of HOXB7 in the
tumorigenesis of osteosarcoma, the effect of HOXB7 knockdown on
cell viability, proliferation and migration was examined by MTT,
Transwell and colony formation assays. The results of MTT assay
indicated that downregulation of HOXB7 suppressed MG63 cells
proliferation (Fig. 3C;
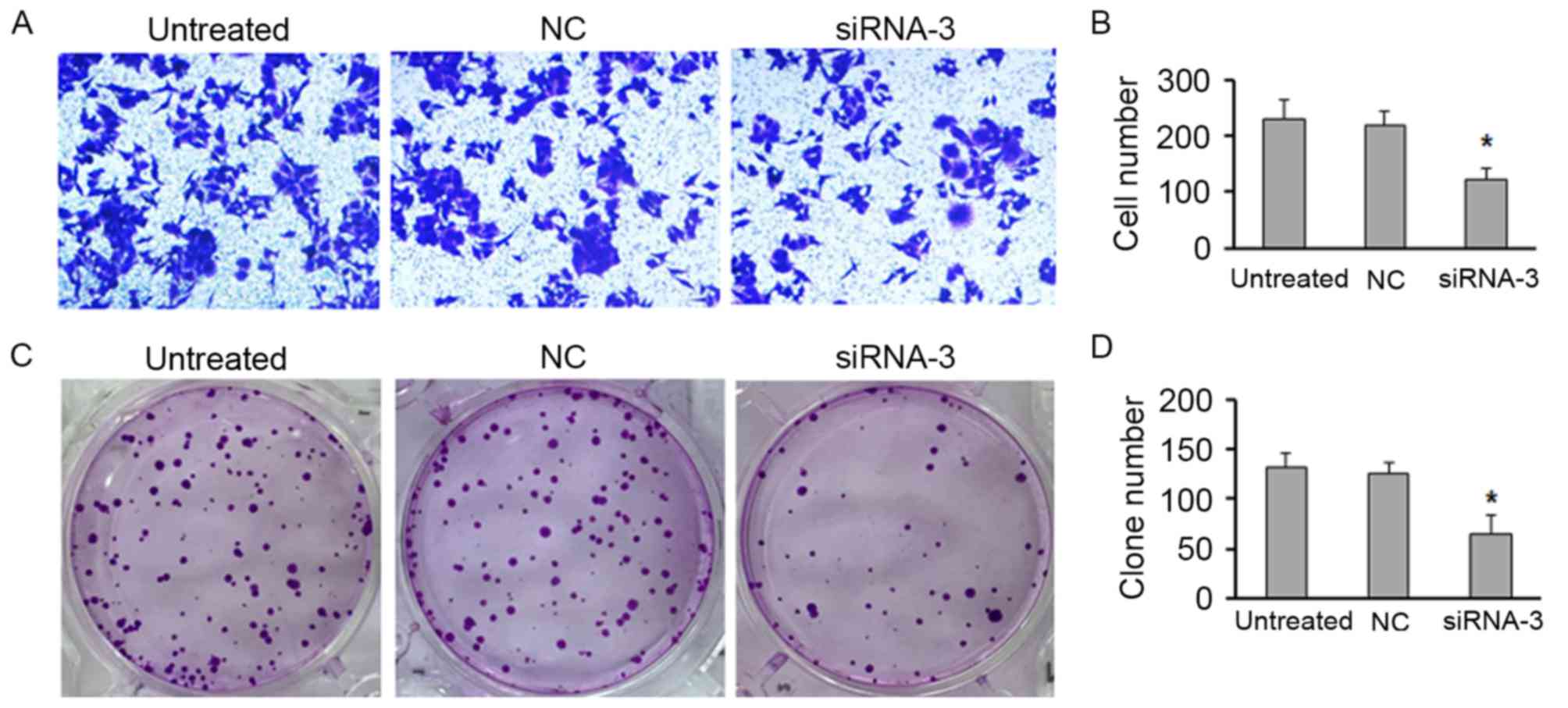
P<0.05). Moreover, the Transwell assay demonstrated that
knockdown of HOXB7 expression significantly inhibited the migratory
capacity of MG63 cells compared with that of control cells
(Fig. 4A and B; P<0.05).
Furthermore, colony formation assay displayed that downregulation
of HOXB7 significantly decreased the proliferation, leading to the
more less numbers of colonies compared with the control (Fig. 4C and D; P<0.05).
Overexpression of HOXB7 induced
EMT
EMT has previously been linked to tumor progression
by which the epithelial cells acquire mesenchymal properties and
show reduced intercellular adhesion and increased motility
(16). As measured by western
blotting, downregulation of HOXB7 expression induced the protein
expression of E-cadherin and suppressed the protein expression of
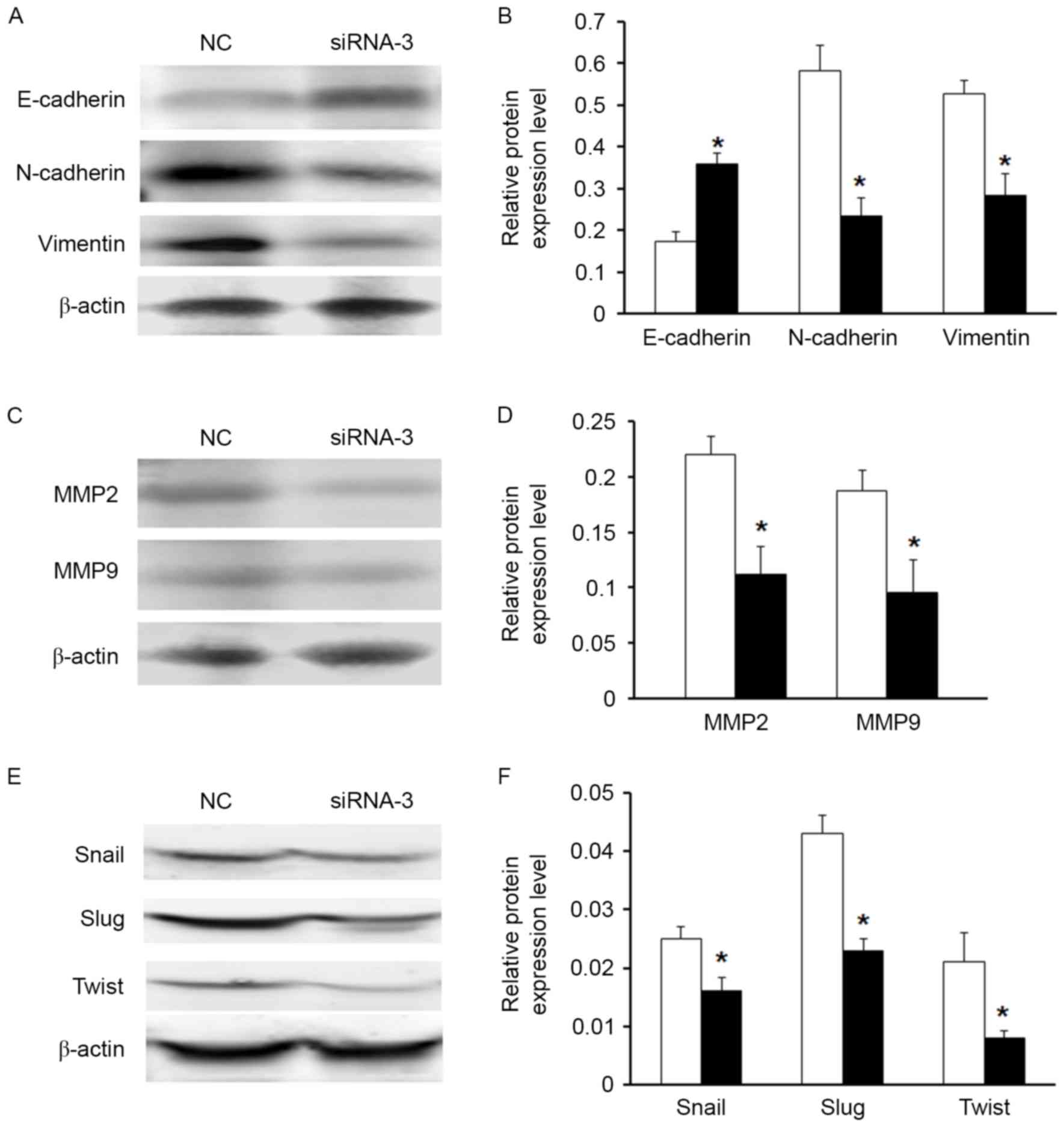
vimentin and N-cadherin in the MG63 cell line (Fig. 5A and B). Moreover, downregulation
of HOXB7 expression significantly inhibited MMP2 and MMP7 protein
levels in MG63 cells (Fig. 5C and
D). It was further investigated whether other EMT markers were
regulated following knockdown of HOXB7 expression by western blot
analysis. The results demonstrated that cells with knockdown
expression of HOXB7 expressed loss of mesenchymal markers (Snail,
Slug and Twist), which is consistent with vimentin and N-cadherin
expression (Fig. 5E and F).
Discussion
Transcriptional factors encoded by HOX genes
regulate cell cycle, proliferation, apoptosis and cell mobility
(7), and their abnormal expression
is often associated with diseases, thus attracting increasing
attention in cancer research (6,17).
In particular, aberrant expression of HOXB7 has been presented in
different tumor types, including breast cancer (18), ovarian cancer (19), oral cancer (20), colorectal cancer (10), lung cancer (12), melanoma (21) and pancreatic cancer (11). To the best of the authors'
knowledge, the present study is the first to demonstrate the
potential role of HOXB7 in osteosarcoma. HOXB7 was identified as
being generally overexpressed in osteosarcoma tissues and cell
lines. Therefore, theoretically, HOXB7 may serve an oncogenic role
in osteosarcoma pathogenesis.
Furthermore, previous studies indicated that an
enforced expression of HOXB7 in hematopoietic progenitors
stimulates self-renewal, sustaining proliferation and
differentiation (22). In the
current study, the current results indicated that ectopic
expression of HOXB7 promoted osteosarcoma cells viability,
proliferation and migration. In addition, overexpression of HOXB7
induces EMT. So, the present results was corroborated by previous
findings where HOXB7 is overexpressed in a number of cancers and
encompasses many oncogenic functions, which has been demonstrated
to promote cell migration and invasion, and induce EMT and
angiogenesis (21). Taken
together, previous efforts were used to elucidate that HOXB7
promotes tumor progression in a cell-autonomous and
non-cell-autonomous manner through activation of the transforming
growth factor-β signaling pathway (23). Modulation of the tumor
proliferation effect through inhibiting PI3K/AKT or
mitogen-associated protein kinase activation mediated by HOXB7
overexpression may be used as a potential target for cancer
prevention and therapy (10).
Finally, they collectively provide compelling circumstantial
evidence that HOXB7 functions dominantly to facilitate tumor
progression in many solid tumor types, including osteosarcoma.
In summary, the present study demonstrated that
HOXB7 was increased in osteosarcoma tissues and cell lines.
Overexpression of HOXB7 promoted the cell proliferation and
migration. Moreover, knockdown of HOXB7 expression resulted in the
increase of epithelial markers E-cadherin, and decrease of
mesenchymal marker vimentin. To the best of our knowledge, the
present study is the first to demonstrate that the HOXB7 regulates
the proliferation and migration of osteosarcoma cells.
References
|
1
|
Damron TA, Ward WG and Stewart A:
Osteosarcoma, chondrosarcoma, and Ewing's sarcoma: National cancer
data base report. Clin Orthop Relat Res. 459:40–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel HJ and Pressey JG: Current concepts
on the surgical and medical management of osteosarcoma. Expert Rev
Anticancer Ther. 8:1257–1269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Corsetti MT, Levi G, Lancia F, Sanseverino
L, Ferrini S, Boncinelli E and Corte G: Nucleolar localisation of
three hox homeoproteins. J Cell Sci. 108:187–193. 1995.PubMed/NCBI
|
|
4
|
Inamori K, Takeshita K, Chiba S, Yazaki Y
and Hirai H: Identification of homeobox genes expressed in human
T-lymphocytes. Biochem Biophys Res Commun. 196:203–208. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Samuel S and Naora H: Homeobox gene
expression in cancer: Insights from developmental regulation and
deregulation. Eur J Cancer. 41:2428–2437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abate-Shen C: Deregulated homeobox gene
expression in cancer: Cause or consequence? Nat Rev Cancer.
2:777–785. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shah N and Sukumar S: The Hox genes and
their roles in oncogenesis. Nat Rev Cancer. 10:361–371. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin K, Kong X, Shah T, Penet MF, Wildes F,
Sgroi DC, Ma XJ, Huang Y, Kallioniemi A, Landberg G, et al: The
HOXB7 protein renders breast cancer cells resistant to tamoxifen
through activation of the EGFR pathway. Proc Natl Acad Sci USA.
109:2736–2741. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bitu CC, Carrera M, Lopes MA, Kowalski LP,
Soares FA and Coletta RD: HOXB7 expression is a prognostic factor
for oral squamous cell carcinoma. Histopathology. 60:662–665. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liao WT, Jiang D, Yuan J, Cui YM, Shi XW,
Chen CM, Bian XW, Deng YJ and Ding YQ: HOXB7 as a prognostic factor
and mediator of colorectal cancer progression. Clin Cancer Res.
17:3569–3578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chile T, Fortes MA, Corrêa-Giannella ML,
Brentani HP, Maria DA, Puga RD, de Paula Vde J, Kubrusly MS, Novak
EM, Bacchella T and Giorgi RR: HOXB7 mRNA is overexpressed in
pancreatic ductal adenocarcinomas and its knockdown induces cell
cycle arrest and apoptosis. Bmc Cancer. 13:4512013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan W, Zhang X, Xu Y, Li S, Hu Y and Wu
S: Role of HOXB7 in regulation of progression and metastasis of
human lung adenocarcinoma. Mol Carcinog. 53:49–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie X, Zhang SS, Wen J, Yang H, Luo KJ,
Yang F, Hu Y and Fu JH: Prognostic value of HOXB7 mRNA expression
in human oesophageal squamous cell cancer. Biomarkers. 18:297–303.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tu W, Zhu X, Han Y, Wen Y, Qiu G and Zhou
C: Overexpression of HOXB7 is associated with a poor prognosis in
patients with gastric cancer. Oncol Lett. 10:2967–2973.
2015.PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grier DG, Thompson A, Kwasniewska A,
Mcgonigle GJ, Halliday HL and Lappin TR: The pathophysiology of HOX
genes and their role in cancer. J Pathol. 205:154–171. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu X, Chen H, Parker B, Rubin E, Zhu T,
Lee JS, Argani P and Sukumar S: HOXB7, a homeodomain protein, is
overexpressed in breast cancer and confers epithelial-mesenchymal
transition. Cancer Res. 66:9527–9534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Naora H, Yang YQ, Montz FJ, Seidman JD,
Kurman RJ and Roden RB: A serologically identified tumor antigen
encoded by a homeobox gene promotes growth of ovarian epithelial
cells. Proc Natl Acad Sci USA. 98:4060–4065. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
De Souza Setubal Destro MF, Bitu CC,
Zecchin KG, Graner E, Lopes MA, Kowalski LP and Coletta RD:
Overexpression of HOXB7 homeobox gene in oral cancer induces
cellular proliferation and is associated with poor prognosis. Int J
Oncol. 36:141–149. 2010.PubMed/NCBI
|
|
21
|
Caré A, Silvani A, Meccia E, Mattia G,
Stoppacciaro A, Parmiani G, Peschle C and Colombo MP: HOXB7
constitutively activates basic fibroblast growth factor in
melanomas. Mol Cell Biol. 16:4842–4851. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Carè A, Valtieri M, Mattia G, Meccia E,
Masella B, Luchetti L, Felicetti F, Colombo MP and Peschle C:
Enforced expression of HOXB7 promotes hematopoietic stem cell
proliferation and myeloid-restricted progenitor differentiation.
Oncogene. 18:1993–2001. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu S, Jin K, Hui Y, Fu J, Jie C, Feng S,
Reisman D, Wang Q, Fan D, Sukumar S and Chen H: HOXB7 promotes
malignant progression by activating the TGFβ signaling pathway.
Cancer Res. 75:709–719. 2015. View Article : Google Scholar : PubMed/NCBI
|