Introduction
Renal fibrosis is the result of abnormal assembly of
the extracellular matrix, which ultimately leads to renal failure
progression, and advanced and end-stage renal disease (1). Renal fibrosis covers four overlapping
stages: The fibrogenic stage, activation, expansion and progression
(2). Transforming growth factor-β
(TGF-β) signaling is the important mediator in the
progression of renal fibrosis via inducing the extracellular matrix
to form renal scarring (3).
Mitogen-activated protein kinase (MAPK) serves a role in the
extracellular matrix in renal fibrosis as being downstream of
TGFβ (4). The underlying
molecular mechanisms (e.g. TGFβ and MAPK signaling)
are suggested as therapeutic targets of renal fibrosis (5).
MicroRNAs (miRNAs), characterized as noncoding RNAs
of 18–25 nucleotides, are revealed to take part in the regulation
of TGF-β-induced renal fibrosis (6). miR-21 is the TGFβ-induced
miRNA, which is upregulated in tubular epithelial cells and
promotes renal fibrosis (7).
However, miR-29 can be inhibited by TGF-β, thereby
facilitating the extensive accumulation of the extracellular matrix
in renal fibrosis (8). In
addition, miRNAs serve fundamental roles in tissue fibrosis by
either blocking the translation or inducing the degradation of
target mRNAs, therefore targeting specific miRNAs may lead to novel
therapeutic methods for treating renal fibrosis (9).
Unilateral ureteral obstruction (UUO) is a
representative model for researching renal fibrosis resulting from
non-inflammatory surgical insult (10). Previously, mice were treated with
UUO to identify the involved miRNAs in renal fibrosis (11). Using a different analysis process,
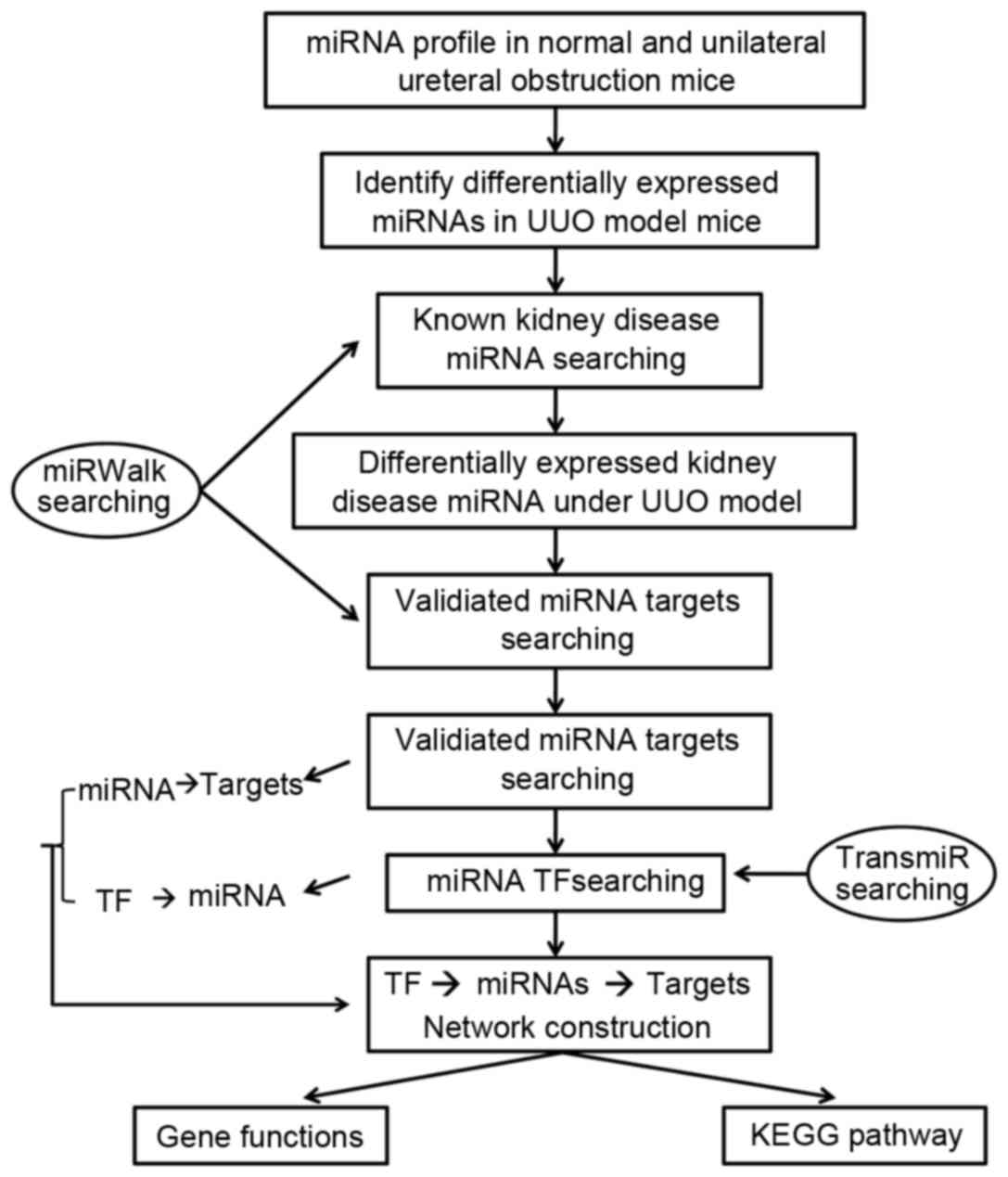
the current study aimed to identify candidate miRNAs specifically
associated with renal fibrosis based on microarray dataset
GSE42716, followed by screening of upstream transcription factors
and target genes of the candidate miRNAs. Additionally, enrichment
analysis of target genes was also performed in order to explain the
involvement of candidate miRNAs in renal fibrosis. The workflow of
the present study can be seen in Fig.
1.
Materials and methods
Microarray data of miRNA
Microarray dataset GSE42716 of miRNA extracted from
kidneys from four mice with UUO and four matched mice without UUO
were downloaded from the Gene Expression Omnibus database (12). These microarray expression
profiling data were previously investigated using GPL10834
Agilent-021828 Unrestricted Mouse miRNA Microarray (V2, Agilent
Technologies, Inc., Santa Clara, CA, USA). All animal studies have
been approved by Keio University Animal Ethics Committee
(Shinjuku-ku, Tokyo, Japan) and performed in accordance with the
ethical standards.
Data processing and screening of
differentially expressed miRNAs
The raw expression data on probe-level were
converted into recognizable miRNA expression data, followed by
median normalization and log2 transformation using Affy package in
R (13). The expression values of
probes corresponding to each miRNA were averaged to obtain the
expression level of the miRNA. Differentially expressed miRNAs in
mice with UUO compared with the normal mice were screened with the
criteria of |log2 fold-change (FC)|>1 and false
discovery rate (FDR) <0.05 using Limma package in R (14).
Screening of candidate miRNAs, their
target genes and transcription factors
The miRNAs related to renal disease were predicted
using miRWalk database (15).
Then, the candidate miRNAs specifically associated with renal
fibrosis were screened by taking the intersection of differentially
expressed miRNAs identified in the present study and the predicted
miRNAs using the miRWalk database. The persuasive target genes of
candidate miRNAs were subsequently predicted using the miRWalk
database, together with screening of upstream transcription factors
targeting candidate miRNAs using the TransmiR database (16).
Integrative regulatory network
construction
The interacting pairs among target genes were
identified using the Search Tool for Retrieval of Interacting Genes
database with the combined score >0.4 (17), followed by construction of
integrative regulatory network among transcription factors, miRNAs
and target genes using Cytoscape software (version 3.0.1;
http://www.cytoscape.org/) (18).
Enrichment analysis of target genes
from the integrative regulatory network
To reveal the potential biological functions or
pathways that may be changed by the differentially expressed
miRNAs, Gene Ontology (GO) term (19) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) enrichment analysis (20) were separately performed for the
target genes of candidate miRNAs using Database for Annotation,
Visualization and Integrated Discovery (DAVID) database (21) and KEGG Orthology Based Annotation
System (KOBAS) tool (22) with the
criterion of P<0.05.
Results
Differentially expressed miRNAs
With the criteria of |log2 FC| >1 and
FDR <0.05, 76 differentially expressed miRNAs were obtained,
including 15 downregulated and 61 upregulated miRNAs in kidneys
with UUO compared with the normal samples.
Screened candidate miRNAs, target
genes and their transcription factors
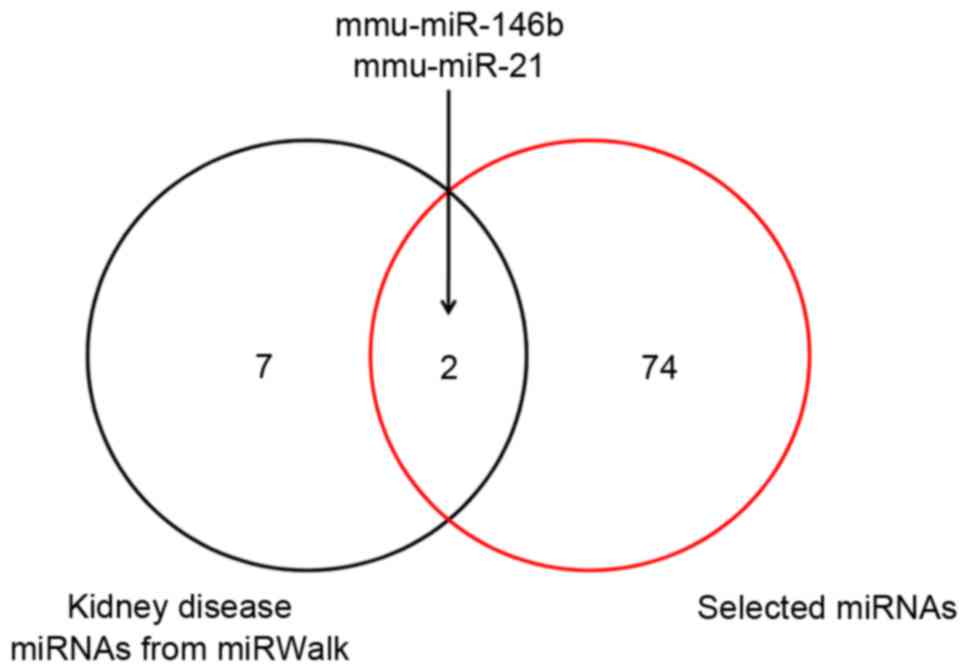
A total of nine miRNAs were predicted to be
associated with renal disease using the miRWalk database. By taking
the intersection of the nine predicted miRNAs and 76 differentially
expressed miRNAs analyzed by a Venn diagram, upregulated miR-146b
and miR-21 were identified as candidate miRNAs specifically
associated with the disease of renal fibrosis (Fig. 2). Then, 74 target genes of miR-146b
and 587 target genes of miR-21 were screened using the miRWalk
database, followed by identification of transcription factor
NFKB1 activating miR-146b and activating miR-21 using the
TransmiR database (Table I).
 | Table I.Upstream TFs of miR-146b and
miR-21. |
Table I.
Upstream TFs of miR-146b and
miR-21.
| TF | Entrezid | miR | Active | Pmid | Organism |
|---|
| NFKB1 |
4790 | miR-146b | Activation | 21981419 | Mouse |
| Gfi1 | 14581 | miR-21 | Repression | 19278956 | Mouse |
| AKT | 11651 | miR-21 | Activation | 20404348 | Mouse |
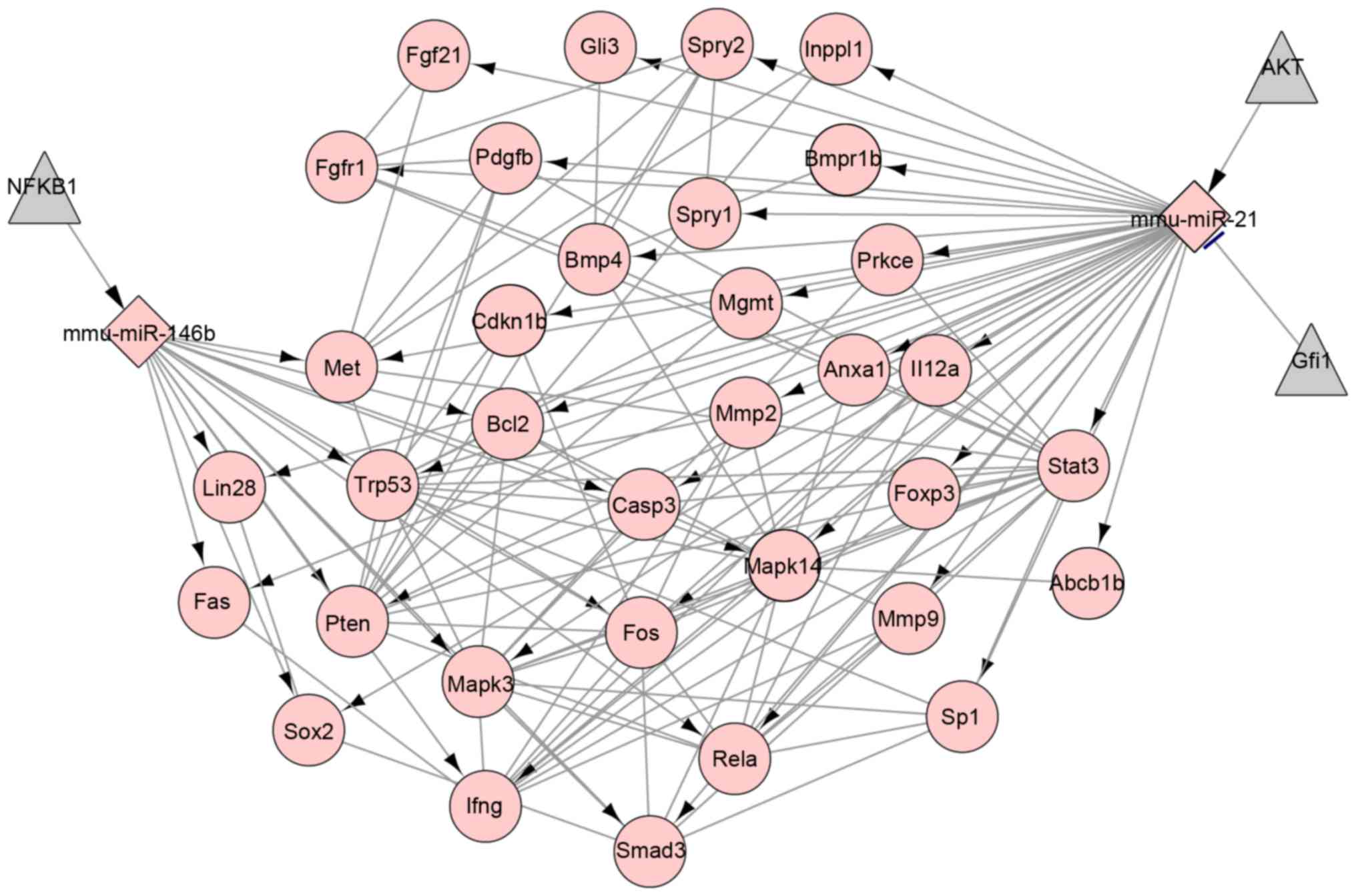
Integrative regulatory network
analysis
The interactions between target genes, regulatory
effect of miRNAs on the target genes and transcription factors on
miRNAs were presented in the integrative regulatory network which
included 39 nodes representing target genes, 140 edges representing
interactions between genes, two miRNAs and three transcription
factors (Fig. 3). Based on the
regulatory network, miR-21 had a regulatory effect on IFNG
expression and miR-146b may negatively regulate the expression of
BCL2, PTEN and IFNG.
GO functions and KEGG pathways
enrichment analysis
Enrichment analysis using DAVID and KOBAS databases
revealed the target genes of miR-146b and miR-21 were significantly
enriched in 14 GO terms including regulation of cell proliferation
(P=1.2E-17, such as BCL2, PTEN and IFNG), and two
KEGG pathways of the TGF-β signaling pathway (P=2.78E-03) and the
MAPK signaling pathway (P=1.11E-13; Table II).
 | Table II.The top 5 significantly enriched GO
terms and KEGG pathways of genes targeted by miR-146b and
miR-21. |
Table II.
The top 5 significantly enriched GO
terms and KEGG pathways of genes targeted by miR-146b and
miR-21.
| Term | P-value | Enriched genes |
|---|
| GO terms |
|
GO:0042127~regulation of cell
proliferation |
1.2×10−17 | TRP53, BMP4, FGFR1,
PDGFB, INPPL1, SOX2, ANX1, SMAD3, FGF21, FOXP3, PTEN, GLI3, SPRY2,
SPRY1, CASP3, CDKN1B, BCL2, IFNG, IL12A |
|
GO:0008285~negative regulation
of cell proliferation |
5.7×10−11 | BMP4, TRP53, SPRY2,
CASP3, SPRY1, CDKN1, INPP1, BCL2, FOXP3, PTEN, GLI3 |
|
GO:0010628~positive regulation
of gene expression |
7.5×10−9 | BMP4, TRP53, FOS,
SP1, RELA, MAPK14, IFNG, SOX, SMAD3, FOXP3, GLI3, STAT3 |
|
GO:0007167~enzyme linked
receptor protein signaling pathway |
8.8×10−9 | BMP4, TRP53, FGFR1,
PDGFB, MET, SMAD3, FGF21, BMPR1B, PTEN, STAT3 |
|
GO:0042325~regulation of
phosphorylation |
1.5×10−8 | BMP4, SPRY2, CASP3,
SPRY1, CDKN1B, PDGFB, BCL2, IFNG, MET, PRKCE |
| KEGG pathways |
|
mmu04350~TGF-β signaling
pathway |
2.78×10−3 | BMP4, SP1, IFNG,
MAPK3, SMAD3, BMPR1B |
|
mmu04010~MAPK signaling
pathway |
1.11×10−13 | TRP53, FOS, FGFR1,
CASP3, PDGFB, RELA, MAPK14, MAPK3, FGF21, FAS |
Discussion
miRNAs have been previously revealed to serve an
important role in initiating and promoting the pathologic processes
in the progression and development of renal fibrosis (23). By analyzing the microarray data of
miRNAs, the current study obtained 76 differentially expressed
miRNAs in mice with UUO compared with the control mice. Based on
the miRWalk database, nine miRNAs were predicted to be associated
with renal disease. By taking the interaction of the 76
differentially expressed miRNAs and 7 renal disease-related miRNAs,
miR-146b and miR-21 were identified to be candidate miRNAs.
Additionally, NFKB1 may activate miR-146b and AKT may
activate miR-21. Furthermore, target genes (e.g. BCL2,
PTEN and IFNG) of miR-146b were primarily involved in
the regulation of cell proliferation.
Highly expressed miR-21 is positively correlated
with kidney fibrosis (23). The
overexpression of miR-21 is induced by TGF-β signaling
(7), which regulates the
deposition and contraction of extracellular matrix in renal
fibrosis (6,8,24).
Similarly, in the present study, miR-21 was identified with
significant upregulation. Besides, based on the regulatory network,
miR-21 had a regulatory network on the expression of IFNG.
Previously, administration of IFNG was demonstrated to
reduce collagen III and IV expression and inhibit renal fibrosis in
rats with subtotal nephrectomy (25). Thus, the overexpressed miR-21 may
lead to inactive IFNG, thereby promoting renal fibrosis.
Additionally, AKT may activate miR-21 from the regulatory
network. In the mouse model of UUO, AKT2 expression
dramatically increases and AKT2 knockout inhibits
UUO-induced activation of epithelial-to-mesenchymal transition and
kidney fibrosis (26). It
therefore can be inferred that the overexpressed miR-21 in mice
with UUO may result from upregulated AKT2 and inversely
negatively regulate IFNG expression, which may make a
contribution to renal fibrosis.
In addition, miR-146b was also identified as
upregulated in mice with UUO, which had not been reported
previously. In the remodeling of atrial fibrosis, miR-146b is
overexpressed and the transfection of miR-146b into cardiac
fibroblasts markedly reduces expression of metallopeptidase
inhibitor 4 and reversely increases collagen content (27). In the present study, miR-146b may
negatively regulate the expression of BCL2, PTEN and
IFNG, which were enriched in the GO terms of regulation of
cell proliferation. Enhancive cell apoptosis and proliferation may
contribute to the progression of renal fibrosis and their
inhibition may help ameliorate renal fibrosis (28,29).
The expression of anti-apoptotic protein BCL2 gradually
reduced with time following UUO, thereby contributing to increased
cell apoptosis and renal fibrogenesis (30). In a previous study, loss of
PTEN expression, accompanied by increased TGF-β
signaling, led to failed differentiation and caused fibrosis in the
proximal tubules (31).
Additionally, IFNG exerts an inhibitory effect on renal
fibrosis in rats with subtotal nephrectomy (25). Thus, the overexpressed miR-146b in
mice with UUO may contribute to renal fibrosis via suppressing the
expression of BCL2, PTEN and IFNG. In
addition, transcription factor NFKB1 may activate miR-146b.
NFKB can be upregulated by the pro-fibrotic cytokines and
serve a role in the obstruction-induced cell apoptosis renal
fibrosis (32). Therefore,
miR-146b may be activated by NFKB1 and subsequently reduce
the expression of BCL2, PTEN and IFNG,
contributing to renal fibrosis. However, further research should be
conducted to validate these results.
In summary, based on the microarray data of miRNAs,
the present study identified two candidate miRNAs, miR-146b and
miR-21. The overexpression of miR-21 was in accordance with the
previous reports (7,23). However, the current study further
indicated that the overexpressed miR-21 in mice with UUO may be
activated by AKT2 and contribute to renal fibrosis via
negatively regulating IFNG expression. Notably, the study
also obtained another markedly expressed miRNA in mice with UUO,
namely miR-146b. Furthermore, miR-146b may be activated by
NFKB1 and subsequently reduce the expression of BCL2,
PTEN and IFNG in the progression of renal fibrosis.
These present findings suggested the potential usage of miR-146b
and miR-21 as diagnostic and therapeutic biomarkers for renal
fibrosis. However, further studies should be designed to confirm
these results.
Acknowledgements
The present study was supported by the Natural
Science Foundation of China (grant no. 81370810) and the Science
and Technology Development Plan Project of Jilin Province (grant
no. 20160101059JC).
References
|
1
|
Liu Y: Renal fibrosis: New insights into
the pathogenesis and therapeutics. Kidney Int. 69:213–217. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu Y: Cellular and molecular mechanisms
of renal fibrosis. Nat Rev Nephrol. 7:684–696. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chung AC and Lan HY: Molecular mechanisms
of TGF-β signaling in renal fibrosis. Current Pathobiology Reports.
1:291–299. 2013. View Article : Google Scholar
|
|
4
|
Stambe C, Atkins RC, Tesch GH, Masaki T,
Schreiner GF and Nikolic-Paterson DJ: The role of p38alpha
mitogen-activated protein kinase activation in renal fibrosis. J Am
Soc Nephrol. 15:370–379. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boor P, Ostendorf T and Floege J: Renal
fibrosis: Novel insights into mechanisms and therapeutic targets.
Nat Rev Nephrol. 6:643–656. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chung AC, Huang XR, Meng X and Lan HY:
miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc
Nephrol. 21:1317–1325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhong X, Chung AC, Chen HY, Meng XM and
Lan HY: Smad3-mediated upregulation of miR-21 promotes renal
fibrosis. J Am Soc Nephrol. 22:1668–1681. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qin W, Chung AC, Huang XR, Meng XM, Hui
DS, Yu CM, Sung JJ and Lan HY: TGF-β/Smad3 signaling promotes renal
fibrosis by inhibiting miR-29. J Am Soc Nephrol. 22:1462–1474.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiang X, Tsitsiou E, Herrick SE and
Lindsay MA: MicroRNAs and the regulation of fibrosis. FEBS J.
277:2015–2021. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ishidoya S, Morrissey J, Mccracken R,
Reyes A and Klahr S: Angiotensin II receptor antagonist ameliorates
renal tubulointerstitial fibrosis caused by unilateral ureteral
obstruction. Kidney Int. 47:1285–1294. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morizane R, Fujii S, Monkawa T, Hiratsuka
K, Yamaguchi S, Homma K and Itoh H: miR-34c attenuates
epithelial-mesenchymal transition and kidney fibrosis with ureteral
obstruction. Sci Rep. 4:45782014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barrett T and Edgar R: Gene expression
omnibus: Microarray data storage, submission, retrieval, and
analysis. Methods Enzymol. 411:352–369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smyth GK: Limma: Linear models for
microarray data, in Bioinformatics and computational biology
solutions using R and Bioconductor. Springer; pp. 397–420. 2005
|
|
15
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: A
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:(Database issue). D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9. 1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:(Database issue). D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: software for visualization and analysis of biological
networks, in Data Mining in Proteomics. Springer; pp. 291–303.
2011
|
|
19
|
Gene Ontology Consortium, . Blake JA,
Dolan M, Drabkin H, Hill DP, Li N, Sitnikov D, Bridges S, Burgess
S, Buza T, et al: Gene Ontology annotations and resources. Nucleic
Acids Res. 41:(Database issue). D530–D535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:(Database
issue). D109–D114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sherman BT, da Huang W, Tan Q, Guo Y, Bour
S, Liu D, Stephens R, Baseler MW, Lane HC and Lempicki RA: DAVID
Knowledgebase: A gene-centered database integrating heterogeneous
gene annotation resources to facilitate high-throughput gene
functional analysis. BMC Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:(Web Server issue). W316–W322. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zarjou A, Yang S, Abraham E, Agarwal A and
Liu G: Identification of a microRNA signature in renal fibrosis:
Role of miR-21. Am J Physiol Renal Physiol. 301:F793–F801. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lan HY: Diverse roles of TGF-β/Smads in
renal fibrosis and inflammation. Int J Biol Sci. 7:1056–1067. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oldroyd SD, Thomas GL, Gabbiani G and El
Nahas AM: Interferon-gamma inhibits experimental renal fibrosis.
Kidney Int. 56:2116–2127. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lan A, Zhang J, Xiao Z, Peng X, Qi Y and
Du J: Akt2 is involved in loss of epithelial cells and renal
fibrosis following unilateral ureteral obstruction. PLoS One.
9:e1054512014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Wang Y, Han J, Li Y, Xie C, Xie L,
Shi J, Zhang J, Yang B, Dong C and Xu M: Integrated analysis of
microRNA and mRNA expression profiles in the left atrium of
patients with non-valvular paroxysmal atrial fibrillation: Role of
miR-146b-5p in atrial fibrosis. Heart Rhythm. 12:1018–1026. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thomas GL, Yang B, Wagner BE, Savill J and
El Nahas AM: Cellular apoptosis and proliferation in experimental
renal fibrosis. Nephrol Dial Transplant. 13:2216–2226. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tan X, Li Y and Liu Y: Paricalcitol
attenuates renal interstitial fibrosis in obstructive nephropathy.
J Am Soc Nephrol. 17:3382–3393. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang G, Oldroyd SD, Huang LH, Yang B, Li
Y, Ye R and El Nahas AM: Role of apoptosis and Bcl-2/Bax in the
development of tubulointerstitial fibrosis during experimental
obstructive nephropathy. Exp Nephrol. 9:71–80. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lan R, Geng H, Polichnowski AJ, Singha PK,
Saikumar P, Mcewen DG, Griffin KA, Koesters R, Weinberg JM, Bidani
AK, et al: PTEN loss defines a TGF-β-induced tubule phenotype of
failed differentiation and JNK signaling during renal fibrosis. Am
J Physiol Renal Physiol. 302:F1210–F1223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Misseri R and Meldrum KK: Mediators of
fibrosis and apoptosis in obstructive uropathies. Curr Urol Rep.
6:140–145. 2005. View Article : Google Scholar : PubMed/NCBI
|