Introduction
AT motif binding factor 1 (ATBF1) was originally
identified as an inhibitory transcription regulator of the
α-fetoprotein (AFP) gene (1–4).
ATBF1 competes with hepatocyte nuclear factor 1 (HNF1) to bind to
AT-rich elements in the enhancer and promoter regions of the
AFP gene. There are two isoforms of ATBF1, ATBF1-A and
ATBF1-B, which are produced by alternative splicing (4). ATBF1-A is a 404-kDa protein
containing four homeodomains, 23 zinc finger motifs and a number of
segments thought to be involved in transcriptional regulation.
ATBF1-B is a 306-kDa protein that possesses the same four
homeodomains, however, it contains five fewer zinc finger motifs
due to the absence of 920 amino acid residues at the N-terminus.
ATBF1-B binds to AT-rich enhancer elements in the region flanking
the promoter of the AFP gene and downregulates promoter
activity. ATBF1 negatively regulates cancer cell growth (5), and a number of genetic alterations to
ATBF1 have been reported in several cancers (6). ATBF1 is currently recognized as a
novel tumour suppressor (7).
Due to the role of ATBF1 in transcriptional
regulation, it is required to translocate from the cytoplasm to the
nucleus. In a previous study investigating the subcellular
localization of ATBF1 in gastric cancer, ATBF1 was demonstrated to
bind to the AT motif in the promoter region of the mucin 5AC gene
and negatively regulate its transcription (8). In addition, ATBF1 was observed to
translocate to the nucleus by forming a complex with runt domain
transcription factor 3 (RUNX3), in response to transforming growth
factor (TGF)-β signal transduction (9). Previous studies have demonstrated
that the subcellular localization of ATBF1 may be a potential
prognostic maker for skin cancer and head and neck cancer (10,11).
However, information regarding the post-transcriptional
modifications of the ATBF1 protein and their association with the
nuclear translocation of ATBF1 remain to be elucidated.
In order to investigate the subcellular localization
of ATBF1 and it post-transcriptional modifications in detail, four
different polyclonal antibodies raised against four individual
epitopes of the ATBF1 protein were generated. These were used for
the immunohistochemical analysis of different types of colon cancer
tissue samples, in order to determine the subcellular localization
and post-transcriptional modifications of ATBF1 in colon cancer
cells.
Materials and methods
Polyclonal antibodies
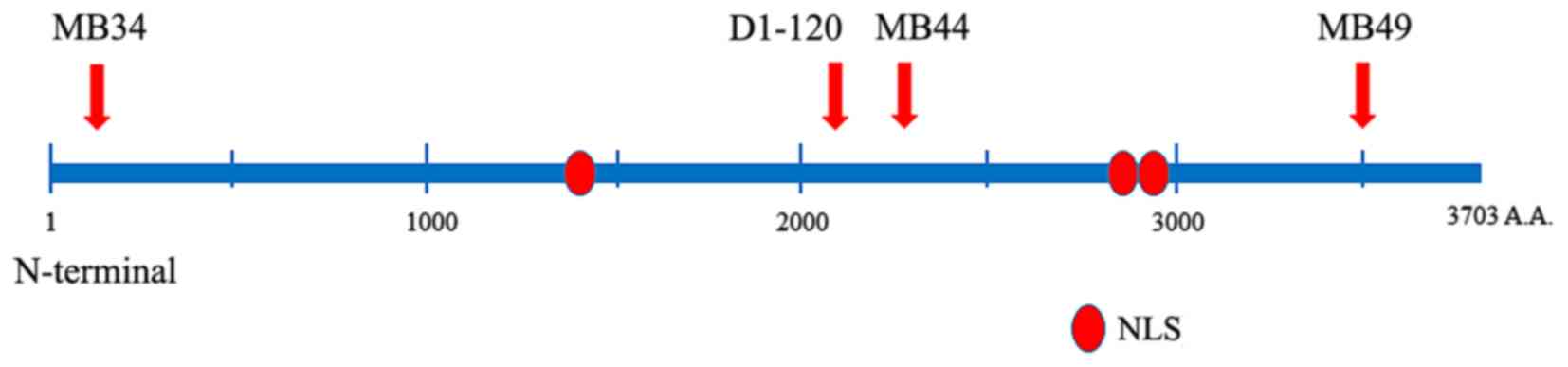
As shown in Fig. 1,
the following 4 anti-ATBF1-A rabbit polyclonal antibodies were
produced against independent epitopes: MB34, which recognizes the
N-terminal region of ATBF1 (amino acids, 238-255); D1-120, which
recognizes a middle region of ATBF1 (amino acids, 2114–2147); MB44,
which recognizes a middle region of ATBF1 (amino acids, 2229–2245);
and MB49, which recognizes the C-terminal region (amino acids,
3410–3426). The antibodies were produced as described previously
(12). The specificity of all the
antibodies used for the purposes of this study was confirmed by
western blot analysis in a previous study (12), using whole cell protein and
fractionated protein lysates from the nuclear and cytoplasm.
Tissue samples
Immunohistochemical analysis was performed on 191
human colon samples obtained from endoscopic polypectomy, mucosal
resection, submucosal dissection or surgical procedures from 111
patients admitted to Nagoya City University Hospital (Nagoya,
Japan) from November 2006 to December 2010. The histological
profiles of these samples are presented in Table I. The present study was performed
in accordance with the Declaration of Helsinki and was approved by
the Ethics Committee of Nagoya City University Graduate School of
Medical Sciences (Nagoya, Japan; reference no. 00-00-1312). Written
informed consent was provided by all patients prior to surgical
procedures, endoscopic examinations or surgeries, and included an
opt-out system.
 | Table I.Histological profiles of the samples
examined. |
Table I.
Histological profiles of the samples
examined.
| Colon sample | No. of samples |
|---|
| Normal colon
mucosa | 80 |
| Hyperplastic
polyp | 4 |
| Serrated adenoma | 11 |
| Tubular adenoma | 13 |
| Tubulovillous
adenoma | 17 |
| Tubular
adenocarcinoma (tub 1) |
|
| m | 11 |
| sm | 22 |
| mp | 1 |
| ss | 4 |
| Tubular
adenocarcinoma (tub 2) |
|
| m | 2 |
| sm | 4 |
| mp | 3 |
| ss | 10 |
| Poorly differentiated
adenocarcinoma |
|
| mp | 1 |
| ss | 2 |
| Mucinous
adenocarcinoma |
|
| sm | 1 |
| ss | 5 |
| Total | 191 |
Immunohistochemistry
Samples were fixed in 10% formalin for
immunohistochemical examination using the four aforementioned
polyclonal antibodies raised against ATBF1-A. All the following
incubations were performed at room temperature. Consecutive tissue
sections (4-µm in thickness) were deparaffinised in xylene and
hydrated using a graded ethanol series. Antigens were retrieved in
a pressure cooker with citrate buffer (0.01 M, pH 6.0) at 110°C for
4 min, sections were subsequently cooled down to room temperature.
The sections were incubated with methanol containing 0.3%
H2O2 and 1.0% sodium azide for 5 min to block
endogenous peroxidase activity. Antibodies were diluted with
Ready-to-Use Dako Antibody Diluent (S0809; Dako; Agilent
Technologies, Inc., Santa Clara, CA, USA) containing carrier
protein to reduce background binding as follows: MB34, 1:1,500;
D1-120, 1:3,000 (PD010; Medical & Biological Laboratories Co.,
Ltd., Nagoya, Japan); MB44, 1:1,200; MB49, 1:500. The sections were
incubated with the primary antibodies for 60 min, washed in PBS and
covered with undiluted EnVision+ System HRP Labelled Polymer
(K4003; Dako; Agilent Technologies, Inc.) for 60 min. The immune
complexes were visualized by incubating the samples with 0.01%
H2O2 and 0.05% 3,3′-diaminobenzidine
tetrachloride (DAB) for 4 min. Nuclei were counterstained with
Mayer's haematoxylin for 20 sec. The level of background staining
by was observed to be negligible in the absence of antibodies (data
not shown). Slides were analyzed with a light microscope (Olympus
BX53; Olympus Corporation, Tokyo, Japan). Immunostaining of the
nuclei and the cytoplasm in target tissues (in a 250×250-µm area)
with 20x objective lens was evaluated in samples selected at
random, and analysed in triplicate.
A pathologist (S.S.) assessed the staining level by
observation of the sections under a light microscope. The following
scoring was employed: Negative staining=0; weak positive
staining=1; strong positive staining=2. The difference between 1
and 2 was arbitrarily by the pathologist.
Immunofluorescence and confocal
microscopic examination of the subcellular localization of
ATBF1-A
The human colon cancer cell line HCT116 (ref. no.
CCL-274; American Type Culture Collection, Manassas, VA, USA) was
cultured in McCoy's 5A medium (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) supplemented with 10% fetal bovine serum and 1%
ampicillin and streptomycin (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Cell line authentication by profiling of short
tandem repeats was performed by the Japanese Collection of Research
Bioresources Cell Bank (Osaka, Japan), on the 25th February
2014.
A total of 4 heamagglutinin (HA)-tagged ATBF1
expression vectors were employed for the purposes of the current
study, including HA-ATBF1 (1–3703 amino acids), ATBF1-2.7N (1–892
amino acids), ATBF1-7.2M (893–3290 amino acids) and ATBF1-1.2C
(3291–3703 amino acids). The plasmid vectors were used as described
previously (12) and transfected
into HCT116 cells (1.25×105 cells/well) using
Lipofectamine LTX with PLUS Reagent, (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Immunofluorescence staining was performed using anti-HA-tag
antibody at room temperature for 1 h (1:200; no. 561; MBL
International Co., Woburn, MA, USA) as the primary antibody, and
Alexa fluor 594-labelled goat anti-rabbit IgG (H+L) antibody at
room temperature for 1 h (1:200; no. A11012; Invitrogen; Thermo
Fisher Scientific, Inc.) as the secondary antibody. All slides were
counterstained with DAPI (0.1 mg/ml) at room temperature for 5 min
(no. D523; Dojindo Molecular Technologies, Inc., Kumamoto, Japan).
Stained cells were observed and 10 fields of view were analysed for
each sample under a confocal laser microscope (Nikon A1 Confocal
Microscope; Nikon Instech, Co., Ltd., Tokyo, Japan). The results
were analysed using NIS Elements Microscope Imaging Software
(version 4.20; Nikon Corporation, Tokyo, Japan).
Statistical analysis
Descriptive statistics and simple analyses were
performed using R software (version 3.1.1; The R Foundation,
Vienna, Austria). A χ2-test was used to compare the
expression of ATBF1 using D1-120 and MB44 antibodies between cancer
tissues and benign polyps, as well as between well-differentiated
(tub 1) and moderately differentiated (tub 2) tubular
adenocarcinomas. Univariate regression analysis was used to measure
the association between depth of tumour invasion and ATBF
expression. P<0.05 was considered to indicate a statistically
significant difference.
Results
ATBF1 expression in the colonic
mucosa
As indicated in Table
II, no ATBF1 expression was observed in normal colonic mucosa
tissues using all four of the anti-ATBF1 antibodies. ATBF1
expression was almost completely absent from hyperplastic polyps
and serrated adenomas, with positive expression detected in the
nuclei of 25% hyperplastic polyps and 18% serrated adenomas using
the MB44 antibody (Table II).
Tubular adenomas were not observed to express ATBF1, however, a
number of tubulovillous adenomas stained positive for the middle
portion of ATBF1 (using D1-120 and MB44 antibodies) in the
cytoplasm and nucleus (Table II).
In particular, 41% of nuclei in tubulovillous adenomas were
positive for MB44 staining. The expression of ATBF1 in the nucleus
and cytoplasm, as detected using the D1-120 and MB44 antibodies,
was significantly higher in adenocarcinoma tissues when compared
with benign polyps and adenomas (P<0.001 for D1-120 and MB44
antibodies; Table II). Notably,
all poorly differentiated adenocarcinomas were observed to express
ATBF1 using the MB34, MB44 and MB49 antibodies, with 100% nuclear
expression detected using the MB44 antibody (Table II). Comparisons between tub 1 and
tub 2 tubular adenocarcinomas revealed significantly higher ATBF1
expression in tub 2 compared with tub 1 adenocarcinomas using the
D1-120 and MB44 antibodies (P<0.01; Table II).
 | Table II.Rate of positive staining for ATBF1
fragments in different colon samples using four types of anti-ATBF1
polyclonal antibody. |
Table II.
Rate of positive staining for ATBF1
fragments in different colon samples using four types of anti-ATBF1
polyclonal antibody.
|
| Anti-ATBF1
antibody |
|---|
|
|
|
|---|
|
| MB34 (%) | D1-120 (%) | MB44 (%) | MB49 (%) |
|---|
|
|
|
|
|
|
|---|
| Colon tissue | Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm |
|---|
| Normal mucosa
(n=80) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hyperplastic polyp
(n=4) | 0 | 0 | 0 | 0 | 25 | 0 | 0 | 0 |
| Serrated adenoma
(n=11) | 0 | 0 | 0 | 0 | 18 | 0 | 0 | 9 |
| Tubular adenoma
(n=13) | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 0 |
| Tubulovillous
adenoma (n=17) | 0 | 0 | 12 | 29 | 41 | 12 | 0 | 24 |
| Tubular
adenocarcinoma |
| tub1
(n=38) | 0 | 58 | 29a | 26a |
53b |
42b | 0 | 55 |
| tub2
(n=19) | 0 | 84 | 63a | 84a |
74b |
74b | 0 | 84 |
| Poorly
differentiated adenocarcinoma (n=3) | 0 | 100 | 33a | 67a | 100b | 100b | 0 | 100 |
| Mucinous
adenocarcinoma (n=6) | 17 | 50 | 50a | 50a |
83b |
33b | 17 | 33 |
Correlation between ATBF1 expression
and colon cancer invasion
The next aim of the present study was to examine the
association between ATBF1 expression (and/or localization) and
cancer invasion in tub 1 and tub 2 cancer tissues. As indicated in
Table III, N-terminal (MB34) and
C-terminal (MB49) fragments of ATBF1 were highly expressed in the
cytoplasm. The expression of N-terminal and C-terminal fragments
were statistically significantly correlated with the cancer
invasion depth (MB34 in tub1: P=0.004; MB49 in tub1: P=0.006).
However, nuclear expression of N-terminal and C-terminal fragments
was not detected. Furthermore, nuclear expression of the middle
region of the ATBF1 fragment (detected using D1-120) was
significantly correlated with invasion to the deep layer of the
colon wall (D1-120 in tub2 nucleus, P=0.004) (Table III).
 | Table III.Rate of positive staining for ATBF1
fragments in tubular adenocarcinoma of the colon using four types
of anti-ATBF1 polyclonal antibodies. |
Table III.
Rate of positive staining for ATBF1
fragments in tubular adenocarcinoma of the colon using four types
of anti-ATBF1 polyclonal antibodies.
|
| Anti-ATBF1
antibody |
|---|
|
|
|
|---|
|
| MB34 (%) | D1-120 (%) | MB44 (%) | MB49 (%) |
|---|
|
|
|
|
|
|
|---|
| Tubular
adenocarcinoma | Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm |
|---|
| tub1 |
| m
(n=11) | 0 |
27a | 36 | 18 | 46 | 27 | 0 |
27c |
| sm
(n=22) | 0 |
63a | 14 | 23 | 45 | 45 | 0 |
59c |
| mp
(n=5) | 0 | 100a | 80 | 60 | 100 | 60 | 0 | 100c |
| tub2 |
| m
(n=2) | 0 | 50 |
0b | 100 | 50 | 50 | 0 | 50 |
| sm
(n=4) | 0 | 50 |
25b | 50 | 25 | 50 | 0 | 50 |
| mp
(n=13) | 0 | 100 | 85b | 92 | 92 | 85 | 0 | 100 |
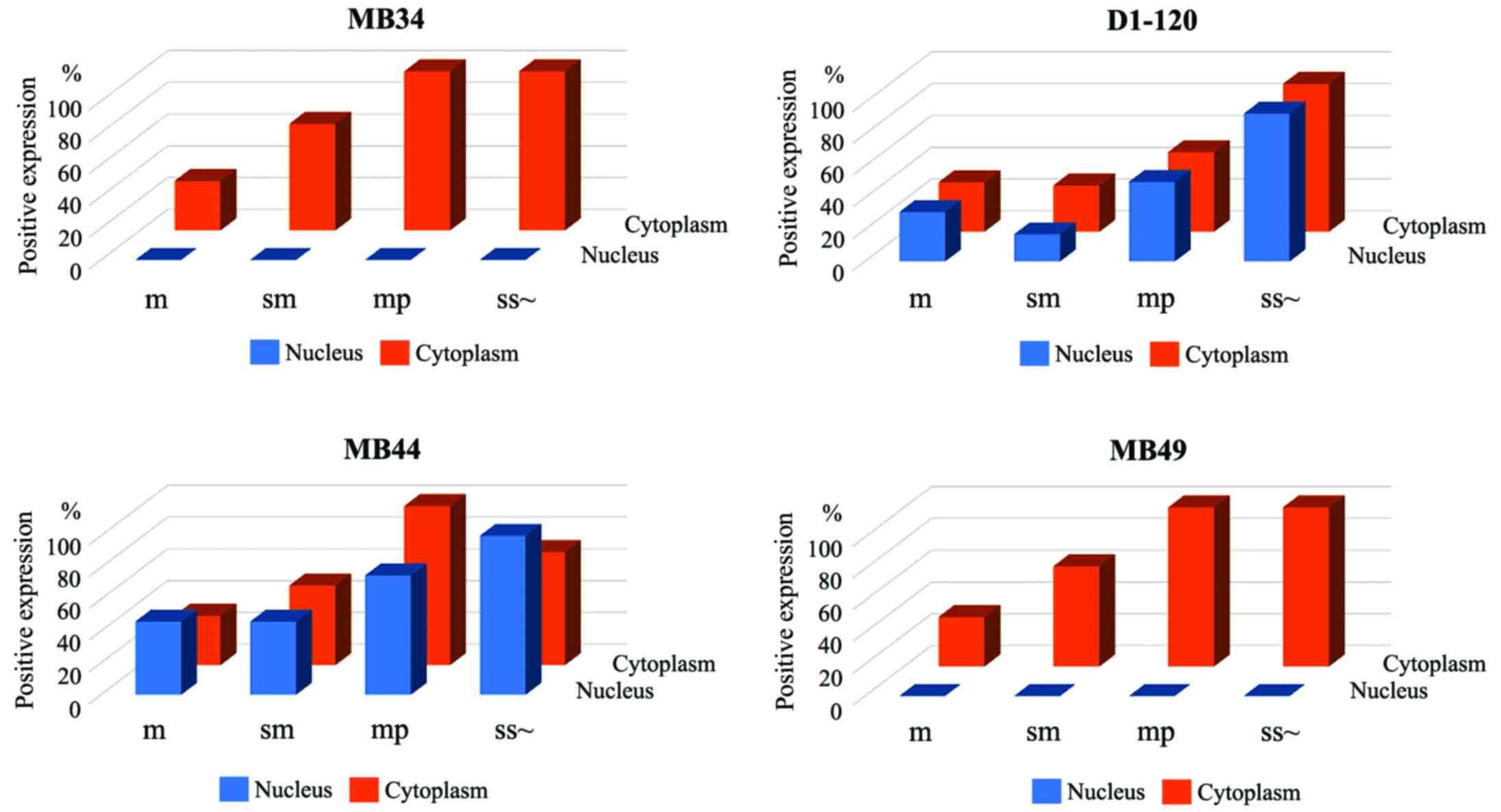
The rate of positive nuclear and cytoplasmic
staining for each of the four antibodies in tub1 and tub2 colon
cancers is shown in Fig. 2.
Expression of the middle region of ATBF1 was increased in the
cytoplasm and nucleus with increasing depth of invasion, whereas,
increases in the expression of the N- and C-terminal regions of
ATBF1 were observed in the cytoplasm (Fig. 2). The depth of invasion was
assigned the following values: Mucosa, 0; submucosa, 1;
muscularis propria, 2; subserosa, 3. Univariate regression
analysis revealed significant correlations between the depth of
tumour invasion and cytoplasmic ATBF1 expression detected using
MB34 (P=0.002), D1-120 (P=0.001), MB44 (P=0.022) and MB49 (P=0.002)
antibodies. Furthermore, significant correlations were observed
between the depth of tumour invasion and nuclear expression when
using D1-120 (P<0.001) and MB44 (P=0.002) antibodies, but not
when using MB34 (P=1.00) and MB49 (P=1.00) antibodies.
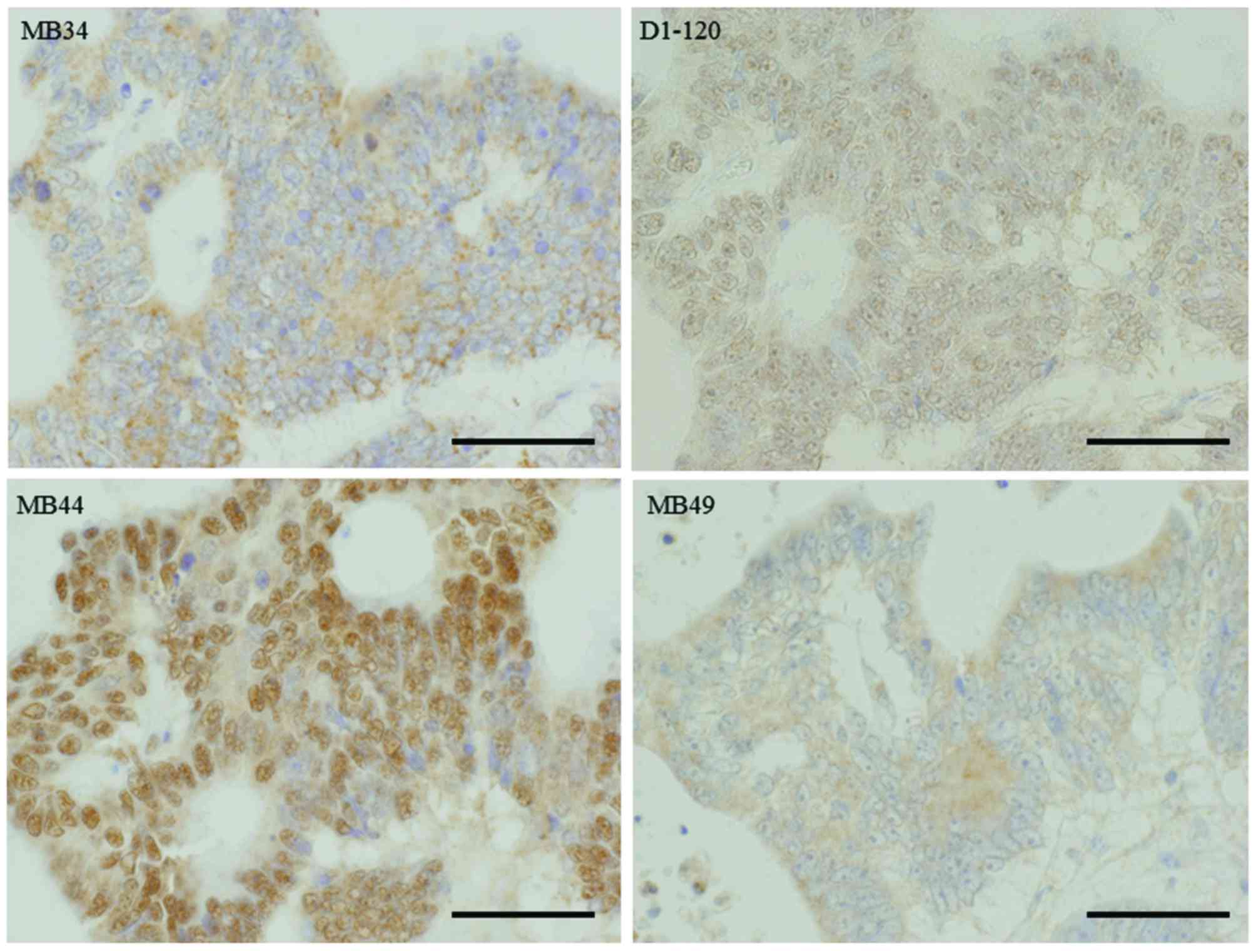
Representative images of immunohistochemical staining of colon
cancer specimens using the four anti-ATBF1 antibodies are shown in
Fig. 3.
Putative role of the middle region of
ATBF1 in nuclear translocation
In order to confirm the mechanism underlying ATBF1
nuclear translocation, the HCT116 colon cancer cells were
transfected with the four HA-tagged ATBF1 expression vectors, and
subcellular localization was evaluated using confocal microscopy
(Fig. 4). As shown in Fig. 4A, the HA-ATBF1 (full-length ATBF1)
and ATBF1-7.2M expression vectors encompass 3 nuclear localization
signals (NLS), namely NLS1387, NLS2947 and NLS2987, whereas the
ATBF1-2.7N and ATBF1-1.2C expression vectors do not encompass any
NLS. Confocal microscopy analysis revealed that HA-ATBF1 and
ATBF1-7.2M translocated to the nucleus, whereas ATBF1-2.7N and
ATBF1-1.2C were localized in the cytoplasm (Fig. 4B). Based on these observations, the
authors speculate that the ATBF1 N- and C-terminal fragments
detected by the MB34 and MB49 antibodies may have been cleaved
post-transcriptionally, and were therefore unable to translocate to
the nucleus as they lack NLS. By contrast, the full-length and
middle regions of ATBF1, detected by the D1-120 and MB44
antibodies, were able to translocate to the nucleus due to the
presence of NLS.
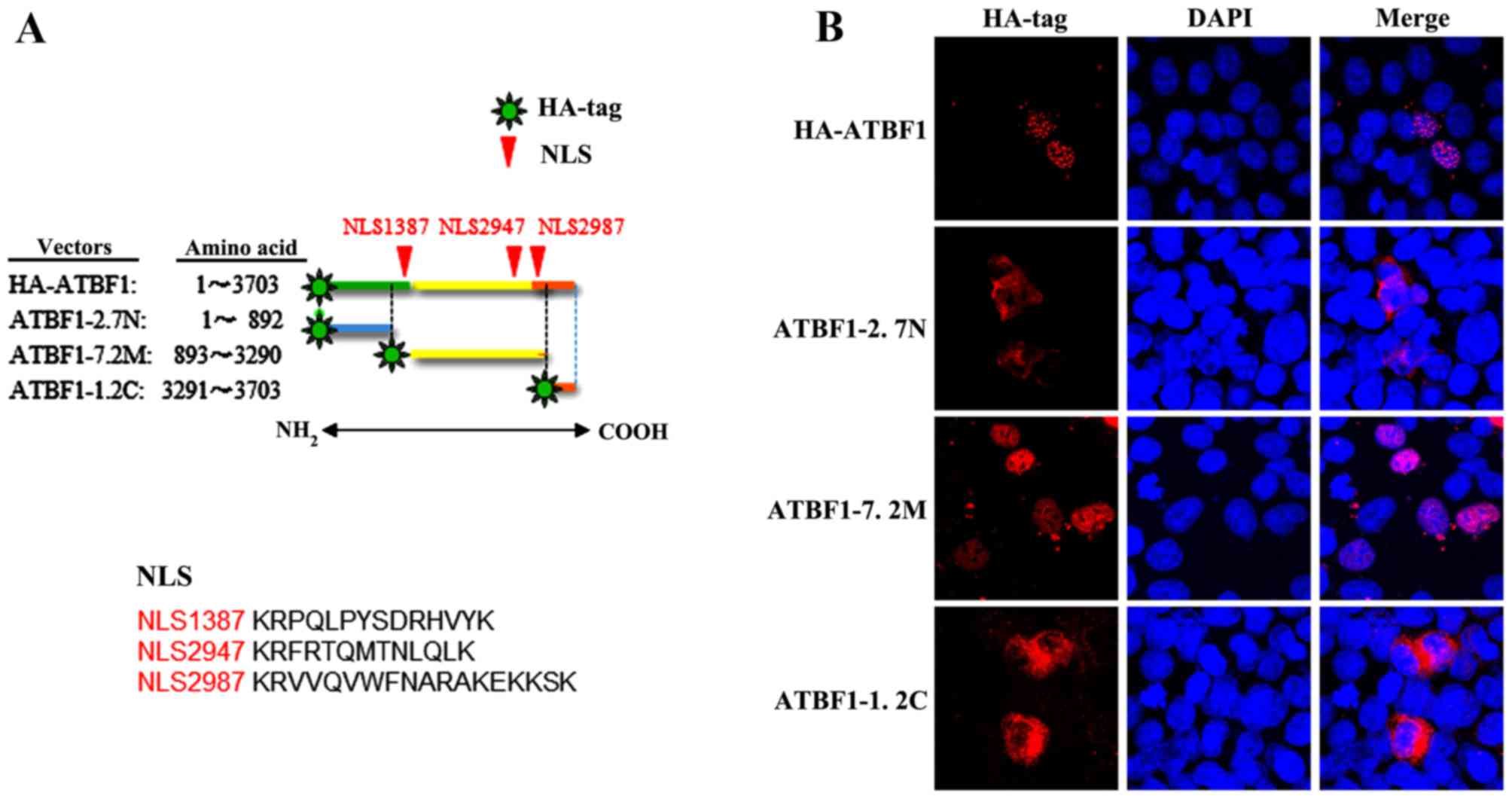 | Figure 4.(A) Details of the following four
ATBF1 expression vector constructs: The HA-ATBF1 vector, which
contains the full-length ATBF1 sequence; the ATBF1-2.7N vector,
which contains the N-terminal fragment of ATBF1 (amino acids 1–892)
and does not possess any NLS; the ATBF1-7.2M vector, which contains
the middle region of ATBF1 (amino acids 893–3,290), and possesses
three NLS; and the ATBF1-1.2C vector, which contains the C-terminal
fragment of ATBF1 (amino acids 3,291–3,703) and does not possess
any NLS. (B) Confocal microscopy images of HCT116 cells transfected
with the four HA-tagged ATBF1 expression vectors. Transfection with
the full-length ATBF1 vector (HA-ATBF1) and the vector containing
the middle region of the ATBF1 protein (ATBF1-7.2M) were detected
in the nucleus, whereas the N-terminal (ATBF1-2.7N) and C-terminal
(ATBF1-1.2C transfection) fragment vectors were detected in the
cytoplasm. ATBF1, AT motif binding factor 1; HA, haemagglutinin;
NLS, nuclear localization signal. |
Discussion
In the present study a positive association between
ATBF1 expression and the step-wise process of carcinogenesis in the
colonic mucosa was observed, and the level of ATBF1 expression
increased with the extent of tumour invasion to the colonic wall.
In addition, the N- and C-terminal regions of ATBF1 were
demonstrated to not translocate to the nucleus. This suggests that
the ATBF1 protein is cleaved and the N- and C-terminal fragments of
ATBF1 are released from the remainder of ATBF1. These fragments
remain in the cytoplasm as they lack NLS. A previous study reported
that nuclear translocation of ATBF1 is associated with TGF-β signal
transduction and/or RUNX3 (9). In
addition, sumoylation is involved in the nuclear localization of
ATBF1 (13). The authors of the
present study are currently investigating the key enzyme that
cleaves ATBF1, as an additional factor that may contribute to its
subcellular localization.
As reported previously, ATBF1 is a novel tumour
suppressor protein that interacts with p53 and Myb (14,15).
Together with the findings of the present study, these observations
indicate that increased ATBF1 expression may be associated with
increased levels of p53 or Myb in colon cancer cells.
A number of ATBF1 mutations have been
reported in prostate cancer (7),
however, there is limited information regarding ATBF1 gene
mutations in colon cancer. There are two potential reasons for the
increased expression of ATBF1 in colon cancer during the process of
carcinogenesis. Firstly, as ATBF1 is a tumour suppressor, its
expression may increase in an attempt to inhibit cell cycle
progression and suppress cancer cell growth (7,16).
Secondly, it is possible that abnormal ATBF1 proteins, which lack
the normal function of ATBF1, exhibit a dominant negative effect,
which leads to cancer development (12). Future studies should therefore
investigate genetic mutations of ATBF1 in colon cancer. The
molecular mechanism of the regulation of ATBF1 gene
expression has been reported previously (17). ATBF1 contains several DNA binding
domains including four homeodomains and twenty-three zinc finger
domains (4). In addition to these
DNA binding domains ATBF1 has distinct protein-protein interaction
domains to bind Myb (14), p53
(15) and protein inhibitor of
activated STAT3 (PIAS3) (18).
These protein-protein interactions suggest ATBF1 may function as a
suppressive factor for tumor progression. Without these co-factors,
ATBF1 does not function as a tumor-suppressor. For example, ATBF1
activates p21(Waf1/Cip1) to halt the cell cycle only when it
interacts with p53 (15). The
fragmentation of ATBF1 may interfere with the full length ATBF1 and
prevent the effective suppression of malignancy in cancer cells.
The lack of ATBF1 expression has been reported in the majority of
malignancies (12) and embryonic
carcinomas (16). Notably,
improved clinical prognosis has been observed with the expression
of ATBF1 fragments compared with no expression (12). However, it is not known whether the
ATBF1 fragments are responsible, or whether unknown mechanisms
besides ATBF1 serve an oncosuppressive role in these cases.
The authors of the present study hypothesize that
fragments of ATBF1 may function as dominant negative factors for
the same target genes. ATBF1 was initially identified as a
suppressor of the AFP gene. A previous study demonstrated
that a fragment of ATBF1 was unable to suppress AFP
(19). The cytoplasmic expression
of ATBF1 fragments in hepatocellular carcinomas may not suppress
AFP. ATBF1 contains three nuclear localization signals
(NLSs) (12). NLSs are distinct
motifs from the DNA binding domains on the ATBF1 (4). The ATBF1 fragments localized in the
cytoplasm that contain DNA binding domains, but not the NLSs, will
have no chance to interact with DNA because the target DNA is
exclusively localized in the nucleus. Nuclear localization of ATBF1
is an important factor in the improved prognosis of patients with
bladder carcinomas (12). However,
it is possible that ATBF1 fragments present in the cytoplasm
interact with PIAS3 to inhibit the signal transduction of signal
transducer and activator of transcription (STAT)3 (18); suppression of the STAT3
inflammatory reaction may limit the progression of malignant
tumors. Janus kinase (JAK)-STAT3 signaling promotes cancer
progression through inflammation, obesity, stem cells and the
pre-metastatic niche (20). The
JAKs and STAT proteins, particularly STAT3, are among the most
promising new targets for cancer therapy (20).
In conclusion, the results of the present study
demonstrate that ATBF1 expression increases during the malignant
progression of colon cancer. In addition, the nuclear localization
of ATBF1 is regulated by three NLS located in the middle region of
ATBF1. The C- and N-terminal fragments of ATBF1, which lack NLS,
remain in the cytoplasm of colon cancer cells. Cleavage of ATBF1
increases the level of cytoplasmic ATBF1 fragments. Alterations in
the subcellular localization of ATBF1, due to its fragmentation,
are associated with malignant features of colon cancer.
Acknowledgements
The authors of the present study are grateful to Dr
Satoshi Osaga and Mrs. Yukimi Ito (Nagoya City University Graduate
School of Medical Sciences, Nagoya, Japan) for their assistance
with the statistical analyses and for their technical assistance,
respectively. In addition, the authors would like to thank Mr. Yuji
Fujinawa, Mr. Hiroyuki Ozawa and Mr. Osamu Yamamoto (Niigata Rosai
Hospital, Joetsu, Japan) for their help with the
immunohistochemical staining procedures.
References
|
1
|
Morinaga T, Yasuda H, Hashimoto T,
Higashio K and Tamaoki T: A human alpha-fetoprotein
enhancer-binding protein, ATBF1, contains four homeodomains and
seventeen zinc fingers. Mol Cell Biol. 11:6041–6049. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ido A, Miura Y and Tamaoki T: Activation
of ATBF1, a multiple-homeodomain zinc-finger gene, during neuronal
differentiation of murine embryonal carcinoma cells. Dev Biol.
163:184–187. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yasuda H, Mizuno A, Tamaoki T and Morinaga
T: ATBF1, a multiple-homeodomain zinc finger protein, selectively
down-regulates AT-rich elements of the human alpha-fetoprotein
gene. Mol Cell Biol. 14:1395–1401. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miura Y, Tam T, Ido A, Morinaga T, Miki T,
Hashimoto T and Tamaoki T: Cloning and characterization of an ATBF1
isoform that expresses in a neuronal differentiation-dependent
manner. J Biol Chem. 270:26840–26848. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kataoka H, Miura Y, Joh T, Seno K, Tada T,
Tamaoki T, Nakabayashi H, Kawaguchi M, Asai K, Kato T and Itoh M:
Alpha-fetoprotein producing gastric cancer lacks transcription
factor ATBF1. Oncogene. 20:869–873. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cho YG, Song JH, Kim CJ, Lee YS, Kim SY,
Nam SW, Lee JY and Park WS: Genetic alterations of the ATBF1 gene
in gastric cancer. Clin Cancer Res. 13:4355–4359. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun X, Frierson HF, Chen C, Li C, Ran Q,
Otto KB, Cantarel BL, Vessella RL, Gao AC, Petros J, et al:
Frequent somatic mutations of the transcription factor ATBF1 in
human prostate cancer. Nat Genet. 37:407–412. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mori Y, Kataoka H, Miura Y, Kawaguchi M,
Kubota E, Ogasawara N, Oshima T, Tanida S, Sasaki M, Ohara H, et
al: Subcellular localization of ATBF1 regulates MUC5AC
transcription in gastric cancer. Int J Cancer. 121:241–247. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mabuchi M, Kataoka H, Miura Y, Kim TS,
Kawaguchi M, Ebi M, Tanaka M, Mori Y, Kubota E, Mizushima T, et al:
Tumor suppressor, AT motif binding factor 1 (ATBF1), translocates
to the nucleus with runt domain transcription factor 3 (RUNX3) in
response to TGF-beta signal transduction. Biochem Biophys Res
Commun. 398:321–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nishio E, Miura Y, Kawaguchi M and Morita
A: Nuclear translocation of ATBF1 is a potential prognostic marker
for skin cancer. Acta Dermatovenerol Croat. 20:239–245.
2012.PubMed/NCBI
|
|
11
|
Li M, Zhao D, Ma G, Zhang B, Fu X, Zhu Z,
Fu L, Sun X and Dong JT: Upregulation of ATBF1 by progesterone-PR
signaling and its functional implication in mammary epithelial
cells. Biochem Biophys Res Commun. 430:358–363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kawaguchi M, Hara N, Bilim V, Koike H,
Suzuki M, Kim TS, Gao N, Dong Y, Zhang S, Fujinawa Y, et al: A
diagnostic marker for superficial urothelial bladder carcinoma:
Lack of nuclear ATBF1 (ZFHX3) by immunohistochemistry suggests
malignant progression. BMC Cancer. 16:8052016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun X, Li J, Dong FN and Dong JT:
Characterization of nuclear localization and SUMOylation of the
ATBF1 transcription factor in epithelial cells. PLoS One.
9:e927462014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaspar P, Dvoráková M, Králová J, Pajer P,
Kozmik Z and Dvorak M: Myb-interacting protein, ATBF1, represses
transcriptional activity of Myb oncoprotein. J Biol Chem.
274:14422–14428. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Miura Y, Kataoka H, Joh T, Tada T, Asai K,
Nakanishi M, Okada N and Okada H: Susceptibility to killer T cells
of gastric cancer cells enhanced by Mitomycin-C involves induction
of ATBF1 and activation of p21 (Waf1/Cip1) promoter. Microbiol
Immunol. 48:137–145. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jung CG, Kim HJ, Kawaguchi M, Khanna KK,
Hida H, Asai K, Nishino H and Miura Y: Homeotic factor ATBF1
induces the cell cycle arrest associated with neuronal
differentiation. Development. 132:5137–5145. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim TS, Kawaguchi M, Suzuki M, Jung CG,
Asai K, Shibamoto Y, Lavin MF, Khanna KK and Miura Y: The ZFHX3
(ATBF1) transcription factor induces PDGFRB, which activates ATM in
the cytoplasm to protect cerebellar neurons from oxidative stress.
Dis Model Mech. 3:752–762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nojiri S, Joh T, Miura Y, Sakata N, Nomura
T, Nakao H, Sobue S, Ohara H, Asai K and Ito M: ATBF1 enhances the
suppression of STAT3 signaling by interaction with PIAS3. Biochem
Biophys Res Commun. 314:97–103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ninomiya T, Mihara K, Fushimi K, Kayashi
Y, Hashimoto-Tamaoki T and Tamaoki T: Regulation of the
alpha-fetoprotein gene by the isoforms of ATBF1 transcription
factor in human hepatoma. Hepatology. 35:82–87. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signaling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|