Introduction
Osteosarcoma (OS) represents the most frequent
malignant primary bone tumor arising from primitive bone-forming
mesenchymal cells in children and adolescents (1). It occurs predominantly in metaphyseal
regions of the long bone, with the distal femur and proximal tibia
accounting for ~50% of all OS cases (2). There is evidence that OS is caused by
genetic and epigenetic changes, and environmental factors, which
inhibit mesenchymal stem cells differentiating into osteoblasts
(3). As a result of the progress
made in diagnostic techniques, surgery, neoadjuvant radiotherapy
and chemotherapy, the prognosis for patients with OS has improved
(4). However, the presence of
advanced phenotypes at the time of diagnosis, distant metastasis,
and resistance to chemotherapy and/or therapy remain major causes
of treatment failure (5). The
molecular mechanisms underlying the initiation and progression of
OS remain to be elucidated. Therefore, a comprehensive
understanding of the carcinogenesis and progression of OS is
required to investigate novel therapeutic strategies for this
disease.
MicroRNAs (miRNAs) are an abundant group of
endogenous, non-protein-coding, single stranded RNA molecules of
~18–25 nucleotides in length (6).
miRNAs regulate gene expression at the transcriptional and
post-transcriptional levels through direct interaction with the
3′untranslated regions (3′UTRs) of their target genes via
base-pairing, which leads to translational repression or mRNA
degradation (7). Computational
estimations suggest that there are ~1,000 miRNAs in the human
genome, which regulate one third of human protein-encoding genes
(8,9). Through this mechanism, miRNAs are
involved in the regulation of a wide range of physiological and
pathological processes, including cell proliferation, cell cycle,
apoptosis, survival, differentiation, invasion and metastasis
(10–12). The dysregulation of miRNAs has been
reported in several types of human cancer, in which these
aberrantly expressed miRNAs are involved in tumor tumorigenesis,
growth, metastasis, chemotherapy resistance and radiation
resistance (13,14). miRNAs can function either as
tumor-suppressors or oncogenes in different types of human cancer
depending on the characteristics of their target genes (15). Therefore, the identification of
cancer-associated miRNAs may provide therapeutic targets for
cancer.
The present study is the first, to the best of our
knowledge, to report that miR-302a was significantly downregulated
in OS tissues and cell lines. It also provided the first evidence
that miR-302a targeted ADAM9 to inhibit cell proliferation,
migration and invasion in OS.
Materials and methods
Patient samples
A total of 75 OS tissues and pair-matched
non-tumorous tissues were collected from patients diagnosed with OS
at the Department of Orthopedics, 89th Hospital of PLA (Weifang,
China) between February 2013 and December 2015. No patients had
received chemotherapy and/or radiotherapy prior to surgery. All
specimens were frozen in liquid nitrogen and stored at −80°C. The
present study was approved by the Ethics Committee of the 89
Hospital of PLA and written informed consent was also obtained from
each patient.
Cell culture and transfection
OS cell lines (MG63, HOS, U2OS, SAOS-2) and a human
normal osteoblastic cell line (hFOB 1.19) were purchased from
America Type Culture Collection (Manassas, VA, USA), and grown in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin in a
37°C humidified 5% CO2 incubator. The hsa-miR-302a
mimics and its negative control mimics (NC) were synthesized by
GenePharma Co., Ltd. (Shanghai, China). ADAM9 small interfering
(si)RNA and negative control siRNA (NC siRNA) were purchased from
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The AMAM9 siRNA
sequence was 5′-GGAATGGCATTGTGGGAA-3′ and the NC siRNA sequence was
5′-TTCTCCGAACGTGTCACGTTT-3′. The cells were seeded in 6-well plates
at a density of 8×105 cells/well, and transfected with
oligonucleotides at room temperature using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) when the cells were
grown to 60–70% density.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
RNA was isolated using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). The expression of miR-302a was
detected using TaqMan microRNA assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.), with U6 as an internal control. To
determine mRNA expression levels, total RNA was reverse transcribed
into cDNA using an M-MLV Reverse Transcription system (Promega
Corporation, Madison, WI, USA). SYBR Green PCR Master mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to measure the
mRNA expression of ADAM9, and data was normalized to β-actin. This
reaction included 2 µl cDNA (100 ng), 2 µl forward primer, 2 µl
reverse primer, 10 µl SYBR® Green PCR Master Mix and 4
µl ddH2O. The thermocycling conditions for qPCR were as
follows: 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 1 min. The primers were designed as follows: miR-302a,
5′-CGTGGATGTACTTGCTTTGAA-3′ (forward) and
5′-TCACCAAAACATGGAAGCAC-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); ADAM9,
5′-CATTGAGGGAGGGACTTCTGGT-3′ (forward) and
5′-ATGGGAACTGCTGAGGTTGCTT-3′ (reverse); and β-actin,
5′-TGACGTGGACATCCGCAAAG-3′ (forward) and 5′-CTGGAAGGTGGACAGCGAGG-3′
(reverse). The relative expression of miR-302a and ADAM9 were
calculated using the 2−∆∆Cq method (16).
Cell proliferation assay
A Cell counting kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) assay was used to investigate
the effect of miR-302a on OS cell proliferation. A total of 3,000
transfected OS cells were seeded into 96-well plates and incubated
at 37°C in an atmosphere of 5% CO2 for 1, 2, 3 and 4
days. At these time points, 10 µl CCK-8 solution was added into
each well and cultured for additional 2 h. The absorbance at 450 nm
was detected using an automatic multi-well spectrophotometer
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Each assay was
performed in triplicate and repeated three times.
Cell migration and invasion assay
Transwell chambers (8-µm pore size; BD Biosciences,
San Jose, CA, USA), precoated with or without Matrigel (BD
Biosciences) were used to the perform cell migration and invasion
assays, respectively. In brief, a total of 3×104
transfected OS cells suspended in 200 µl FBS-free culture medium
were seeded in the upper Transwell chambers, and 500 µl culture
medium containing 10% FBS was added to the lower chambers.
Following incubation at 37°C in an atmosphere of 5% CO2
for 48 h, cotton swabs were used to remove the nonmigrated and
noninvaded cells in the upper chamber. The migrated and invaded
cells were fixed with 4% paraformaldehyde for 10 min, stained with
0.5% crystal violet for 30 min, and washed with PBS (Gibco; Thermo
Fisher Scientific, Inc.). The cell numbers were counted in five
randomly selected fields of each Transwell chambers under an
inverted microscope (Olympus, Tokyo, Japan).
Luciferase reporter assay
The luciferase reporter vectors, pMir-ADAM9-3′UTR
wild-type (WT) and pMir-ADAM9-3′UTR mutant-type (MUT), were
synthesized by GenePharma, Co., Ltd. Using Lipofectamine 2000,
HEK293T cells (American Type Culture Collection, Manassas, VA, USA)
were transfected with luciferase reporter vectors, in addition to
miR-302a mimics or NC. Following incubation for 48 h at 37°C in an
atmosphere of 5% CO2, luciferase activities were
detected using a Dual-Luciferase Reporter Assay system (Promega
Corporation). Renilla luciferase activities were normalized to
firefly luciferase activities. Each assay was performed in
triplicate.
Bioinformatics analysis
Bioinformatics analysis was conducted to predict the
candidate genes of miR-302a using TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/getDownloads.do).
Protein extraction and western blot
analysis
Total protein was extracted using RIPA buffer
(Beyotime Institute of Biotechnology, Haimen, China) supplemented
with protease inhibitors and phosphate inhibitors. The
concentration of total protein was detected using a BCA protein
assay (Pierce; Thermo Fisher Scientific, Inc.). Equal quantities
(20 µg) of protein were separated by 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and then transferred
onto polyvinylidene difluoride membranes (EMD Millipore, Bedford,
MA, USA). Subsequently, the membranes were blocked with 5% non-fat
skimmed milk powder in Tris-buffered saline (TBS), and incubated
with mouse anti-human monoclonal ADAM9 antibody (1:1,000 dilution;
cat. no. sc-377233; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and mouse anti-human monoclonal GADPH antibody (1:1,000
dilution; cat. no. sc-166574; Santa Cruz Biotechnology, Inc.), at
4°C overnight. Following washing with TBS containing 0.5% Tween,
the membranes were probed with goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (1:1,000 dilution; cat.
no. sc-2005; Santa Cruz Biotechnology, Inc.) at room temperature
for 1 h. Protein bands were visualized using enhanced
chemiluminescence, according to the manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± standard deviation.
The differences between groupswere compared using Student's t-test
or one-way analysis of variance using SPSS 13.0 statistical
software (SPSS, Inc, Chicago, IL, USA). Student-Newman-Keuls test
was utilized to compare between two groups in experiments involving
multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-302a in OS tissues
and cell lines
The present study first investigated whether
miR-302a was dysregulated in OS tissues, compared with pair-matched
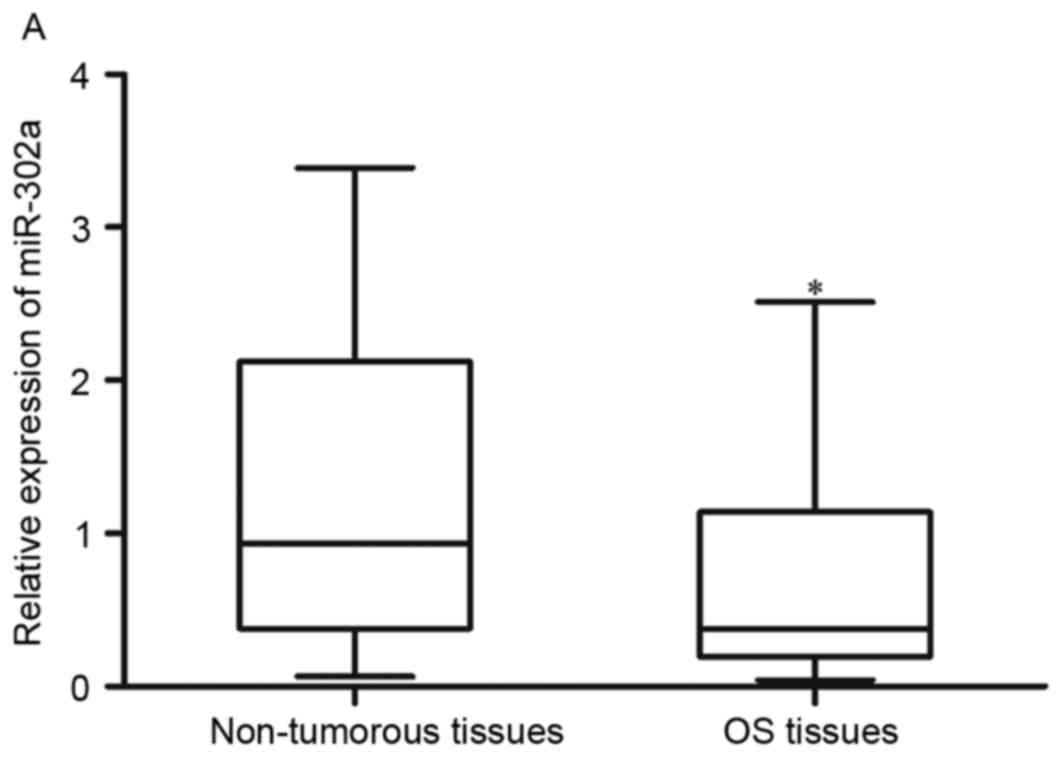
non-tumorous tissues, using RT-qPCR analysis. The results showed
that the expression of miR-302a was significantly lower in the OS
tissues, compared with the pair-matched non-tumorous tissues
(Fig. 1A; P<0.05).
RT-qPCR was also performed to detect the expression
of miR-302a in OS cell lines, including the MG63, HOS, U2OS, SAOS-2
cell lines, and a human normal osteoblastic cell line (FOB 1.19).
Comparison with the expression in the FOB 1.19 cells, the
expression of miR-302a was significantly lower in all the examined
OS cell lines (Fig. 1B;
P<0.05). These data suggested that miR-302a was downregulated in
OS tissues and cell lines.
Downregulation of miR-302a is
associated with aggressive tumor progression in patients with
OS
The correlations between the expression of miR-302a
and clinicopathological characteristics were analyzed. As shown in
Table I, the expression levels of
miR-302a were significantly correlated with tumor-node-metastasis
(TNM) stage (P=0.001) and metastasis (P=0.030) in OS. However, no
significant associations were found between the expression of
miR-302a and the other clinicopathological characteristics,
including age, gender, tumor size, and tumor location
(P>0.05).
 | Table I.Correlation between the expression of
miR-302a and clinicopathological characteristics of in patients
with osteosarcoma. |
Table I.
Correlation between the expression of
miR-302a and clinicopathological characteristics of in patients
with osteosarcoma.
|
|
| Expression of
miR-302a |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
feature | Cases (n) | Low | High | P-value |
|---|
| Age (years) |
|
|
| 0.576 |
|
<19 | 55 | 29 | 26 |
|
|
≥19 | 20 | 12 | 8 |
|
| Gender |
|
|
| 0.213 |
|
Male | 39 | 24 | 15 |
|
|
Female | 36 | 17 | 19 |
|
| Tumor size
(cm) |
|
|
| 0.620 |
|
<10 | 31 | 18 | 13 |
|
|
≥10 | 44 | 23 | 21 |
|
| Tumor location |
|
|
| 0.837 |
|
Tibia/femur | 61 | 33 | 28 |
|
|
Elsewhere | 14 | 8 | 6 |
|
| TNM stage |
|
|
| 0.001 |
|
I-II | 35 | 12 | 23 |
|
|
III | 40 | 29 | 11 |
|
| Metastasis |
|
|
| 0.030 |
|
Present | 36 | 15 | 21 |
|
|
Absent | 39 | 26 | 13 |
|
Expression of miR-302a in OS cells
following transfection
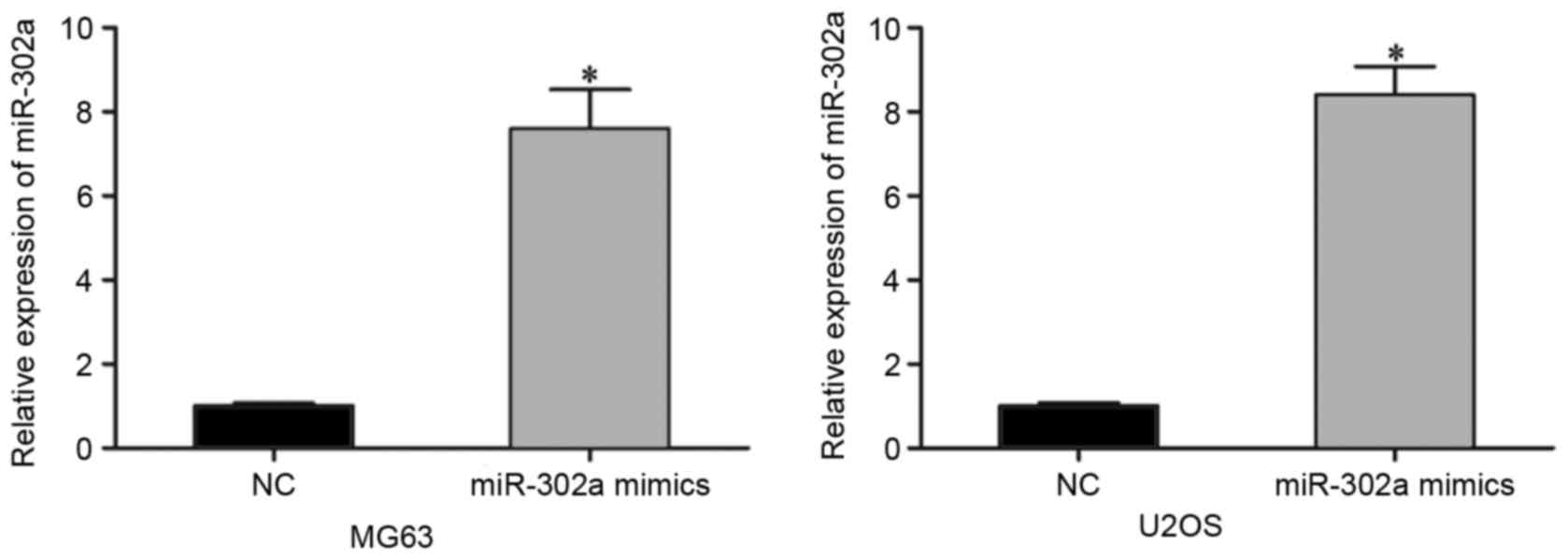
To examine the functions of miR-302a in OS, the
miR-302a mimics or NC mimic was injected into MG63 and U2OS cells,
for relatively lower expression levels of miR-302a. Following
transfection for 48 h, RT-qPCR analysis was performed to evaluate
its transfection efficiency. As shown in Fig. 2, miR-302a was significantly
increased in the MG63 and U2OS cells transfected with miR-302a
mimics (P<0.05).
Effects of miR-302a on the
proliferation, migration and invasion of OS cells
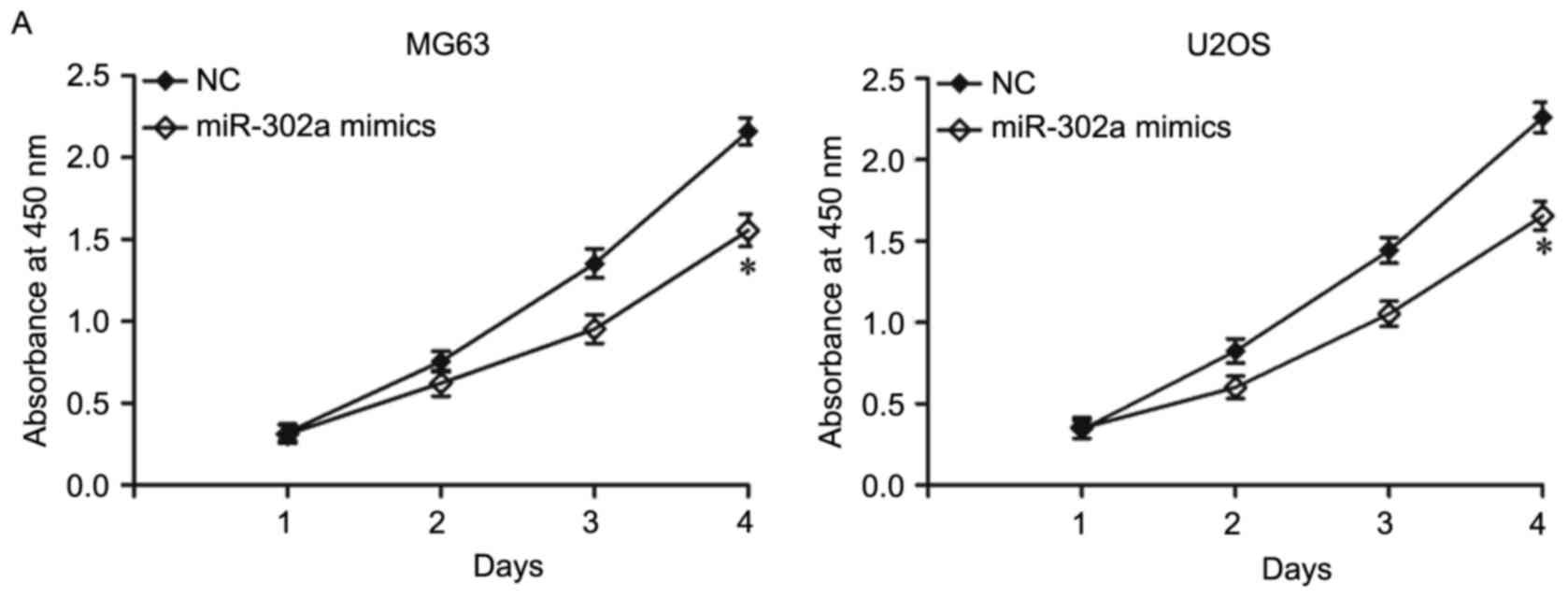
To examine the effects of miR-302a on the
proliferation of OS cells, cell proliferation assays were performed
using the CCK-8 method. As shown in Fig. 3A, the overexpression of miR-302a
inhibited MG63 and U2OS cell proliferation (P<0.05). As the
expression level of miR-302a is correlated with metastasis in
patients with OS, the present study examined whether ectopic
miR-302a affected the migration and invasion capacities in OS. To
confirm this hypothesis, cell migration and invasion assays were
used, which revealed that the migration and invasion abilities were
significantly decreased in the MG63 and U2OS cells transfected with
miR-302a mimics, compared with the cells transfected with NC
(Fig. 3B; P<0.05). These data
suggested that miR-302a acted as a tumor suppressor in OS.
Identification of ADAM9 as a direct
target of miR-302a
To investigate the molecular mechanism by which
miR-302a inhibited OS cell growth and metastasis, bioinformatics
analysis was performed using TargetScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/getDownloads.do).
The analysis showed that ADAM9 was among the list of predicted
miR-302a targets (Fig. 4A).
The correlation between miR-302a and ADAM9 was
further examined by evaluating the mRNA and protein expression
levels of ADAM9 in MG63 and U2OS cells following transfection with
miR-302a mimics. The results of the RT-qPCR analysis showed that
the restoration of miR-302a in MG63 and U2OS cells had no
regulatory effect on the mRNA expression of ADAM9 (Fig. 4B; P>0.05). However, the results
of western blot analysis revealed that the overexpression of
miR-302a downregulated the protein expression of ADAM9 in MG63 and
U2OS cells (Fig. 4C; P<0.05).
These results indicated that miR-302a regulated the expression of
ADAM9 at the post-transcriptional level in OS.
Luciferase reporter assays were also performed to
determine whether ADAM9 was a direct target gene of miR-302a.
HEK293T cells were transfected with pMir-ADAM9-3′UTR WT or
pMir-ADAM9-3′UTR MUT, in addition to miR-302a mimics or NC. As
shown in Fig. 4D, the
overexpression of miR-302a decreased the luciferase activity of
pMir-ADAM9-3′UTR WT (P<0.05), but not that of pMir-ADAM9-3′UTR
MUT. Taken together, these results demonstrated that ADAM9 was a
direct target gene of miR-302a.
Expression of ADAM9 is inversely
correlated with miR-302a in OS tissues
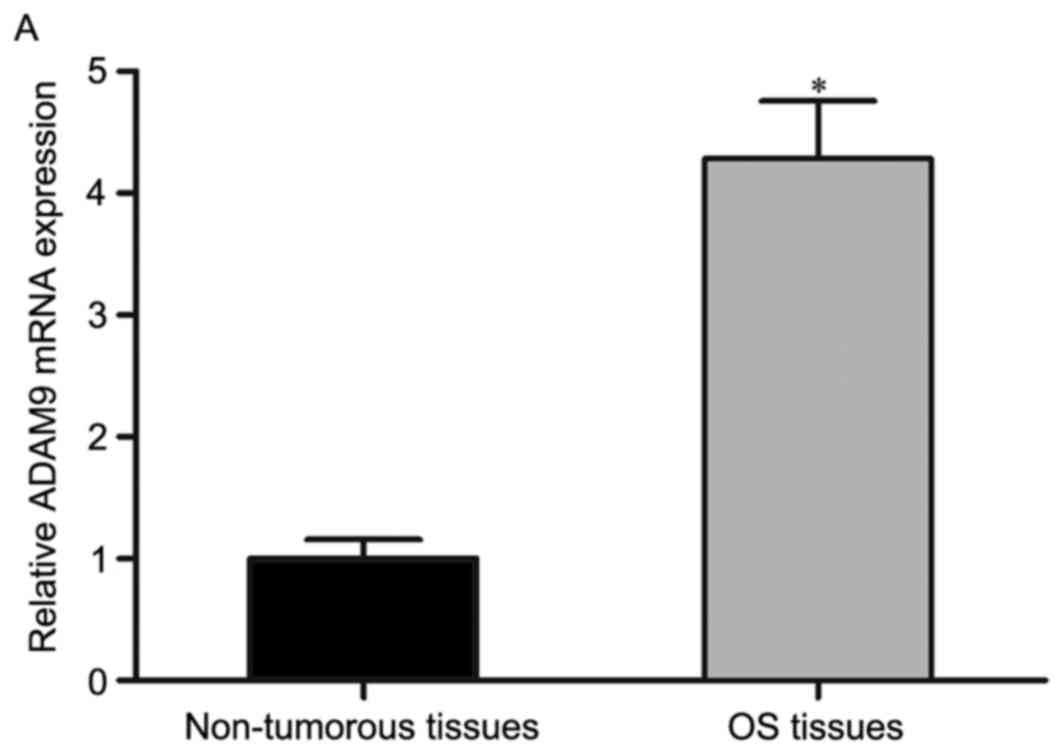
As ADAM9 was identified as a direct target gene of
miR-302a in OS, the present study measured its expression in OS
tissues and pair-matched non-tumorous tissues using RT-qPCR
analysis. It was found that the mRNA levels of ADAM9 were
significantly upregulated in OS tissues, compared with those in
pair-matched non-tumorous tissues (Fig. 5A; P<0.05). Spearman's
correlation analysis revealed that ADAM9 was inversely correlated
with the expression of miR-302a in OS tissues (Fig. 5B; r=−0.6043; P<0.001).
Knockdown of ADAM9 inhibits cell
proliferation, migration and invasion in OS
To investigate the roles of ADAM9 in OS,
loss-of-function experiments were performed. MG63 and U2OS cells
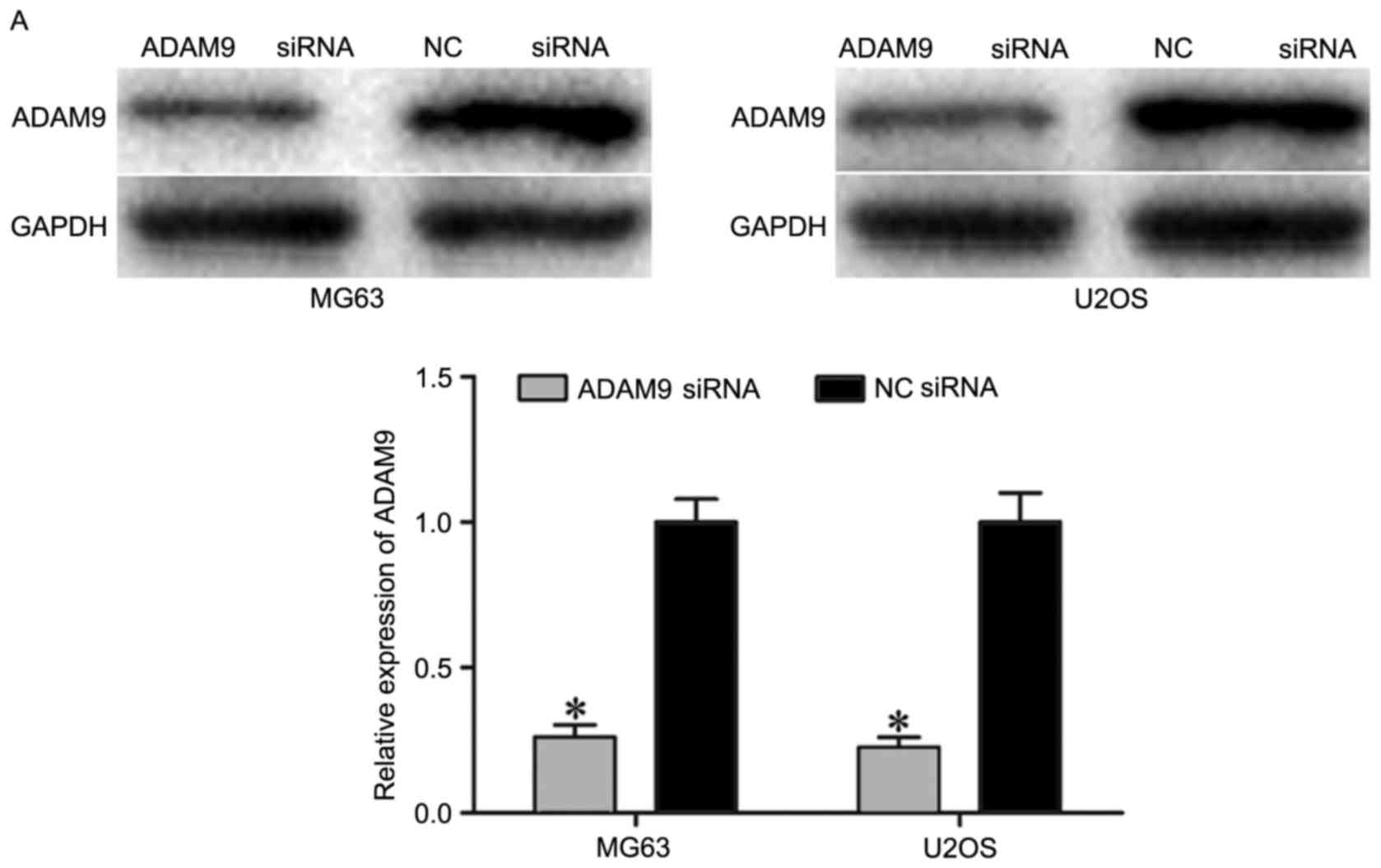
were transfected with ADAM9 siRNA or NC siRNA. As shown in Fig. 6A, ADAM9 siRNA markedly decreased
the expression of ADAM9 in MG63 and U2OS cells (P<0.05).
Following transfection, cell proliferation, cell migration and
invasion assays were performed, which demonstrated that the
knockdown of ADAM9 significantly suppressed MG63 and U2OS cell
proliferation (Fig. 5B;
P<0.05), migration and invasion (Fig. 5C; P<0.05). The results showed
that the effects of ADAM9 knockdown were similar with those of
miR-302a in OS cells, suggesting that ADAM9 was a direct and
functional target of miR-302a in OS.
Discussion
Previous studies have provided evidence for the
dysregulation of miR-302a in various types of human cancer. For
example, in colorectal cancer, the expression of miR-302a was found
to be lower in tumor cell lines, compared with that in normal colon
epithelium cells (17). Zhang
et al (18) reported that,
compared with normal prostate tissues, the expression level of
miR-302a was reduced in prostate cancer tissues, and even lower in
tumor tissues with a Gleason score ≥8. Liang et al (19,20)
demonstrated that miR-302a was significantly downregulated in
metastatic breast cancer tissues and cell lines, and in irradiated
breast cancer cell lines. Guo et al (21) showed that miR-302a was
significantly downregulated in ovarian cancer tissues and cell
lines, compared with normal cervical tissues and normal cells,
respectively. The reduced expression levels of miR-302a in tumor
tissues has also been shown to correlate with TNM stage in patients
with ovarian cancer (21).
However, until now, there have been no reports on the expression
level of miR-302a in OS. The present study initially measured the
expression of miR-302a in OS tissues and pair-matched non-tumorous
tissues using RT-qPCR analysis. The results showed that miR-302a
was markedly downregulated in OS tissues. Consistent with the
expression pattern in tissues, OS cell lines also showed reduced
expression of miR-302a compared with the human normal osteoblastic
cell line. The statistical analysis showed that low expression
levels of miR-302a were correlated with TNM stage and metastasis in
patients with OS. These data suggested that miR-302a may be
involved in the progression of several types of human cancer.
Previous studies have reported significant roles of
miR-302a in cancer progression. Wei et al (17) found that the overexpression of
miR-302a suppressed cell proliferation and invasion in colorectal
cancer. In prostate cancer, functional investigations revealed that
the restoration of miR-302a inhibited prostate cancer cells growth
in vitro and in vivo, and enhanced G1/S cell cycle
arrest (18). In breast cancer,
the upregulation of miR-302a decreased tumor cell invasion
abilities and metastasis in vitro and in vivo
(19). In addition, the ectopic
expression of miR-302a improved the radiosensitivity of breast
cancer cells to radiation therapy in vitro and in
vivo (20). In ovarian cancer,
the induced expression of miR-302a has been found to repress cell
proliferation, induce cell cycle arrest at the G1 phase and
decrease transition from the G1 phase to the S phase (21). However, the functions of miR-302 in
OS have not been reported previously. In the present study, the
downregulation of miR-302a in OS suggested that miR-302a may act as
a potential tumor suppressor in OS. Functional experiments showed
that the overexpression of miR-302a suppressed OS cell
proliferation, migration and invasion in vitro, suggesting
that the decreased expression of miR-302a in OS may promote OS cell
growth and metastasis.
At the molecular level, several target genes of
miR-302a have been identified, including mitogen-activated protein
kinase (MAPK) in colorectal cancer (17), AKT in prostate cancer (18), C-X-C chemokine receptor type 4
(19), breast cancer resistance
protein (22), MAPK kinase 1
(23) in breast cancer, and
syndecan-1 in ovarian cancer (21). In the present study, the results
demonstrated that ADAM9 may be one of the direct targets of
miR-302a in OS. Members of the ADAM family have been demonstrated
to contribute to several biological functions, including
fertilization, adhesion, migration and proteolysis (24,25).
ADAM9, a member of the ADAM family, was found to be significantly
upregulated in non-small cell lung cancer (26), colon cancer (27), gastric cancer (28) and prostate cancer (29). Several studies have reported that
ADAM9 is important in cancer cell growth, invasion and metastasis
(28,30–32).
A study by Jiang et al (33) found that ADAM9 is produced and
secreted by human osteoblasts and acts as one of the insulin-like
growth factor binding protein-5 proteases, which is important in
regulating bone formation. ADAM9 was also reported to be
upregulated in OS tissues and cell lines, compared with normal
tissues and cells (33). These
findings indicated that ADAM9 may be involved in the initiation and
progression of OS, and may be investigated as an effective
therapeutic target for OS.
In conclusion, the present study showed that
miR-302a was significantly downregulated in OS. Its reduced
expression was correlated with TNM stage and metastasis in OS.
Functionally, the upregulation of miR-302a inhibited the
proliferation, migration and invasion of OS cells by directly
targeting ADAM9, thus being involved in the carcinogenesis and
progression of OS. The results of the present study suggested that
miR-302a may be a therapeutic target in the treatment of patients
with OS.
References
|
1
|
Ando K, Heymann MF, Stresing V, Mori K,
Rédini F and Heymann D: Current therapeutic strategies and novel
approaches in osteosarcoma. Cancers (Basel). 5:591–616. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He H, Ni J and Huang J: Molecular
mechanisms of chemoresistance in osteosarcoma (Review). Oncol Lett.
7:1352–1362. 2014.PubMed/NCBI
|
|
3
|
Sun L, Li Y, Zhang J, Li H, Li B and Ye Z:
Prognostic value of pathologic fracture in patients with high grade
localized osteosarcoma: A systemic review and meta-analysis of
cohort studies. J Orthop Res. 33:131–139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Broadhead ML, Clark JC, Myers DE, Dass CR
and Choong PF: The molecular pathogenesis of osteosarcoma: A
review. Sarcoma. 2011:9592482011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Poletajew S, Fus L and Wasiutynski A:
Current concepts on pathogenesis and biology of metastatic
osteosarcoma tumors. Ortop Traumatol Rehabil. 13:537–545. 2011.(In
English, Polish). View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carrington JC and Ambros V: Role of
microRNAs in plant and animal development. Science. 301:336–338.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu J: Control of protein synthesis and
mRNA degradation by microRNAs. Curr Opin Cell Biol. 20:214–221.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nouraee N and Mowla SJ: miRNA therapeutics
in cardiovascular diseases: Promises and problems. Front Genet.
6:2322015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bertoli G, Cava C and Castiglioni I:
MicroRNAs: New biomarkers for diagnosis, prognosis, therapy
prediction and therapeutic tools for breast cancer. Theranostics.
5:1122–1143. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aigner A: MicroRNAs (miRNAs) in cancer
invasion and metastasis: Therapeutic approaches based on
metastasis-related miRNAs. J Mol Med (Berl). 89:445–457. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rottiers V and Näär AM: MicroRNAs in
metabolism and metabolic disorders. Nat Rev Mol Cell Biol.
13:239–250. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cho WC: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chistiakov DA and Chekhonin VP:
Contribution of microRNAs to radio- and chemoresistance of brain
tumors and their therapeutic potential. Eur J Pharmacol. 684:8–18.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fan W, Huang J, Xiao H and Liang Z:
MicroRNA-22 is downregulated in clear cell renal cell carcinoma,
and inhibits cell growth, migration and invasion by targeting PTEN.
Mol Med Rep. 13:4800–4806. 2016.PubMed/NCBI
|
|
17
|
Wei ZJ, Tao ML, Zhang W, Han GD, Zhu ZC,
Miao ZG, Li JY and Qiao ZB: Up-regulation of microRNA-302a
inhibited the proliferation and invasion of colorectal cancer cells
by regulation of the MAPK and PI3K/Akt signaling pathways. Int J
Clin Exp Pathol. 8:4481–4491. 2015.PubMed/NCBI
|
|
18
|
Zhang GM, Bao CY, Wan FN, Cao DL, Qin XJ,
Zhang HL, Zhu Y, Dai B, Shi GH and Ye DW: MicroRNA-302a suppresses
tumor cell proliferation by inhibiting AKT in prostate cancer. PLoS
One. 10:e01244102015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang Z, Bian X and Shim H: Inhibition of
breast cancer metastasis with microRNA-302a by downregulation of
CXCR4 expression. Breast Cancer Res Treat. 146:535–542. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang Z, Ahn J, Guo D, Votaw JR and Shim
H: MicroRNA-302 replacement therapy sensitizes breast cancer cells
to ionizing radiation. Pharm Res. 30:1008–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo T, Yu W, Lv S, Zhang C and Tian Y:
MiR-302a inhibits the tumorigenicity of ovarian cancer cells by
suppression of SDC1. Int J Clin Exp Pathol. 8:4869–4880.
2015.PubMed/NCBI
|
|
22
|
Wang Y, Zhao L, Xiao Q, Jiang L, He M, Bai
X, Ma M, Jiao X and Wei M: miR-302a/b/c/d cooperatively inhibit
BCRP expression to increase drug sensitivity in breast cancer
cells. Gynecol Oncol. 141:592–601. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao L, Wang Y, Jiang L, He M, Bai X, Yu L
and Wei M: MiR-302a/b/c/d cooperatively sensitizes breast cancer
cells to adriamycin via suppressing P-glycoprotein (P-gp) by
targeting MAP/ERK kinase kinase 1 (MEKK1). J Exp Clin Cancer Res.
35:252016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Blobel CP: ADAMs: Key components in EGFR
signalling and development. Nat Rev Mol Cell Biol. 6:32–43. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Edwards DR, Handsley MM and Pennington CJ:
The ADAM metalloproteinases. Mol Aspects Med. 29:258–289. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang J, Qi J, Chen N, Fu W, Zhou B and He
A: High expression of a disintegrin and metalloproteinase-9
predicts a shortened survival time in completely resected stage I
non-small cell lung cancer. Oncol Lett. 5:1461–1466.
2013.PubMed/NCBI
|
|
27
|
Li J, Ji Z, Qiao C, Qi Y and Shi W:
Overexpression of ADAM9 promotes colon cancer cells invasion. J
Invest Surg. 26:127–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim JM, Jeung HC, Rha SY, Yu EJ, Kim TS,
Shin YK, Zhang X, Park KH, Park SW, Chung HC and Powis G: The
effect of disintegrin-metalloproteinase ADAM9 in gastric cancer
progression. Mol Cancer Ther. 13:3074–3085. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sung SY, Kubo H, Shigemura K, Arnold RS,
Logani S, Wang R, Konaka H, Nakagawa M, Mousses S, Amin M, et al:
Oxidative stress induces ADAM9 protein expression in human prostate
cancer cells. Cancer Res. 66:9519–9526. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang L, Gong F and Cui Y: RNAi-mediated A
disintegrin and metalloproteinase 9 gene silencing inhibits the
tumor growth of non-small lung cancer in vitro and in vivo. Mol Med
Rep. 12:1197–1204. 2015.PubMed/NCBI
|
|
31
|
Martin AC, Cardoso AC, Selistre-de-Araujo
HS and Cominetti MR: Recombinant disintegrin domain of human ADAM9
inhibits migration and invasion of DU145 prostate tumor cells. Cell
Adh Migr. 9:293–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen CM, Hsieh YH, Hwang JM, Jan HJ, Hsieh
SC, Lin SH and Lai CY: Fisetin suppresses ADAM9 expression and
inhibits invasion of glioma cancer cells through increased
phosphorylation of ERK1/2. Tumour Biol. 36:3407–3415. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang L, He A, Zhang Q and Tao C: miR-126
inhibits cell growth, invasion, and migration of osteosarcoma cells
by downregulating ADAM-9. Tumour Biol. 35:12645–12654. 2014.
View Article : Google Scholar : PubMed/NCBI
|