Introduction
Gastric cancer, one of the major malignant tumor
types, is the fourth most common and the second leading cause of
cancer mortality around the world, with ~1,000,000 new cases and
700,000 mortalities every year (1,2). In
the past two decades, the morbidity and mortality of gastric cancer
have gradually been declining around the world (2). However, in East Asia, it remains the
second leading cause of cancer-associated mortality and the most
common type of gastroenteric tumor (3). Increasing numbers of studies have
demonstrated that many risk factors are involved in the occurrence
and development of gastric cancer, such as genetic alterations, the
Helicobacter pylori infection, smoking, alcohol drinking,
habitual excessive intake of salt, and environmental factors
(4). Although great effort has
been made in various therapeutic strategies that combined surgery
resection, radiotherapy and chemotherapy, the overall prognosis
remains unsatisfactory (5).
Therefore, an improved understanding of the mechanisms underlying
the carcinogenesis and progression of gastric cancer would
facilitate the development of novel therapeutic treatments for this
disease.
MicroRNAs (miRNAs) are a large group of 19- to
25-nucleotide, endogenous, short RNA molecules (6). miRNAs negatively regulate the
expression of their target genes at the post-transcriptional and/or
translational level by directly binding to the 3′-untranslated
regions (UTRs) of their target genes, resulting in degradation of
the target genes or inhibition of protein translation (7,8).
Numerous studies demonstrate that miRNAs exert a critical role in
various types of biological processes, including cell
proliferation, the cell cycle, invasion, migration, metastasis and
differentiation (9,10).
Furthermore, the dysregulation of miRNAs has been
reported in a diverse range of human cancers, such as colorectal
(11) and cervical (12) cancers, hepatocellular (13) and renal cell (14) carcinoma, and gastric cancer
(15) amongst others. Increasing
evidence has demonstrated that miRNAs function as tumor suppressors
or oncogenes in the carcinogenesis and progression of numerous
types of cancer. For example, miR-524-5p inhibited gastric cancer
proliferation and invasion by negative regulation of oncogenes,
matrix metalloproteinase (MMP)-2 and MMP-9 (16). miR-3646 acted as an oncogene in
breast cancer, through promoting cell proliferation, migration and
invasion via directly targeting the p53/p21/cyclin-dependent kinase
1/cyclin B1 pathway (17).
In the current study, the expression level,
biological functions and molecular mechanism of miR-455 were
investigated in gastric cancer.
Materials and methods
Clinical samples
A total of 57 gastric cancer and matched,
non-tumorous gastric tissue samples were collected from patients of
the The Affiliated Huai'an Hospital of Xuzhou Medical University
(Huai'an, China) and The Second People's Hospital of Huai'an
(Huai'an, China) between 2011 and 2014. None of the gastric cancer
patients received chemotherapy or radiotherapy treatments prior to
surgery. All tissue samples were immediately frozen in liquid
nitrogen and stored at −80°C. The seventh edition of UICC tumor
node metastasis (TNM) classification system was used as clinical
staging criteria (18). The
present study was approved by the Ethics Committee of The
Affiliated Huai'an Hospital of Xuzhou Medical University and The
Second People's Hospital of Huai'an, and written informed consent
was obtained from all gastric cancer patients.
Cell culture
Five gastric cancer cell lines (SGC-7901, BGC-823,
AGS, MKN-1 and MKN-45) and a normal gastric epithelium cell line
(GES-1) were purchased from the Shanghai Institute of Biochemistry
and Cell Biology (Shanghai, China) and cultured in Gibco Dulbecco's
modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with Gibco 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere
consisting of 5% CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissue samples and cells
using Invitrogen TRIzol (Thermo Fisher Scientific, Inc.). A SYBR
PrimeScript miRNA RT PCR kit (Takara Biotechnology Co., Ltd.,
Dalian, China) was used to determine the miR-455 expression level
and U6 served as an internal control. To quantify IGF-1R mRNA
expression, total RNA was used to synthesize cDNA using a
PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.),
followed by qPCR using SYBR Premix Ex Taq (Takara Biotechnology
Co., Ltd.). The amplification reaction was performed using the
following cycling conditions: An initial predenaturation step for 5
min at 95°C, followed by 40 cycles of denaturation at 95°C for 30
sec and annealing at 65°C for 45 sec. The primer sequences were as
follows: Forward, 5′-ACACTCCAGCTGGGTATGTGCCTT-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′ for miR-455; forward,
5′-CTCGCTTCGGCAGCACATATACT-3′ and reverse,
5′-ACGCTTCACGAATTTGCGTGTC-3′ for U6; forward,
5′-CCGCTGCCAGAAAATGTGCCCA-3′ and reverse,
5′-TGTCGTTGTCAGGCGCGCTG-3′ for IGF-1R; and forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′ for GAPDH. RT-qPCR was performed in
triplicate on an AB7300 thermo-recycler (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The relative expression of miR-455 and
IGF-1R mRNA were determined using the 2−ΔΔCq cycle
threshold method (19).
Oligonucleotides and cell
transfection
miR-455 mimics and negative control miRNA mimics
(NC) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai,
China). Small interfering RNAs (siRNAs) specifically for IGF-1R and
the control siRNA (IGF-1R siRNA and NC siRNA) were obtained from
the Chinese Academy of Sciences (Changchun, China). Cells were
transfected with these oligonucleotides using Invitrogen
Lipofectamine 2000 Thermo Fisher Scientific, Inc.). Following
transfection for 6–8 h, the culture medium containing Lipofectamine
2000 and oligonucleotides was discarded and replaced with fresh
culture medium.
MTT assay
Following the 24-h transfection, the transfected
cells were collected and re-seeded in 96-well plates at a density
of 2,000 cells per well. Cells were then incubated at 37°C in 5%
CO2 atmosphere. At different time points (24, 48, 72 and
96 h), an MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
was performed according to the manufacturer's instructions.
Briefly, 20 µl MTT solution (5 mg/ml) was added to each well and
subsequently incubated at 37°C for 4 h. The culture medium
containing MTT solution was discarded and 200 µl DMSO solution
(Sigma-Aldrich; Merck KGaA) was added to dissolve the formazan
precipitates. The absorbance at a wavelength of 490 nm was measured
using an automatic multi-well spectrophotometer (BioTek
Instruments, Inc., Winooski, VT, USA).
Tranwell migration and invasion
assay
Following a 48-h transfection, the transfected cells
were harvested, washed with Gibco phosphate-buffered saline (PBS;
Thermo Fisher Scientific, Inc.) three times and resuspended in
FBS-free culture medium. For the Transwell migration assay,
5×104 cells were plated into the top side of
polycarbonate Transwell chambers (8 µm; Costar; Thermo Fisher
Scientific, Inc.). For the Transwelll invasion assay,
5×104 cells were plated into the upper chambers coated
with Matrigel (BD Biosciences, San Jose, CA, USA). The lower
chambers were filled with DMEM containing 20% FBS. The chambers
were incubated at 37°C in an atmosphere of 5% CO2 for 48
h. Migratory and invasive cells on the lower membranes were fixed,
stained with 0.5% crystal violet, washed with PBS three times and
counted in five random fields (magnification, ×200) per Transwell
chamber using an IX71 inverted microscope (Olympus Corporation,
Tokyo, Japan).
Protein extraction and western
blotting
Following transfection for 72 h, cells were washed
with ice-cold PBS and lysed in RIPA buffer (Beyotime Institute of
Biotechnology, Haimen, China) in the presence of protease inhibitor
cocktail. The concentration of total protein was quantified using
the BCA Protein Assay kit (Beyotime Institute of Biotechnology).
Proteins (30 µg) were separated on 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto polyvinylidene fluoride membrane at 100 V for 1.5
h. Subsequent to blocking in 5% non-fat milk in Tris-buffered
saline containing Tween-20 for 1 h at room temperature, the
membranes were probed with mouse anti-human monoclonal IGF-1R
(1:1,000 dilution; cat. no. sc-81464; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) and mouse anti-human monoclonal GAPDH
(1:1,000 dilution; cat. no. sc-137179; Santa Cruz Biotechnology,
Inc.) at 4°C overnight, followed by incubation with the goat
anti-mouse horseradish peroxidase-conjugated secondary antibodies
(1:5,000 dilution; cat. no. sc-2005; Santa Cruz Biotechnology,
Inc.) for 1 h at room temperature. The signals were visualized
using the ECL Plus Detection kit (Pierce; Thermo Fisher Scientific,
Inc.).
Bioinformatic analysis and luciferase
reporter assay
Bioinformatic analysis was performed to investigate
the potential target genes of miR-455 using three algorithm
databases: TargetScan (http://www.targetscan.org/), PicTar (http://pictar.bio.nyu.edu) and miRanda (http://www.sanger.ac.uk).
Luciferase reporter assay was performed to
investigate whether IGF-1R was a direct target gene of miR-455.
Luciferase report vectors, PGL3-IGF-1R-3′UTR Wt and
PGL3-IGF-1R-3′UTR Mut, were synthesized by Shanghai GenePharma Co.,
Ltd. HEK293T cells, purchased from Shanghai Institute of
Biochemistry and Cell Biology, were seeded into 24-well plates at a
density of 30–40%, and transfected with PGL3-IGF-1R-3′UTR Wt or
PGL3-IGF-1R-3′UTR Mut, along with miR-455 mimics or NC using
Lipofectamine 2000. At 48 h post-transfection, transfected cells
were collected, washed with PBS, and subjected to luciferase
reporter assay using a Dual-Luciferase Reporter Assay System
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's instructions. All samples were run in
triplicate.
Statistical analysis
Data are presented as the mean ± standard deviation,
and were compared using a Student's t-test or one-way analysis of
variance followed by the Student-Newman-Keuls post hoc test using
SPSS version 13.0 (SPSS, Inc., Chicago, IL, USA). Spearman's
correlation analysis was used to analyze the association between
miR-455 and IGF-1R mRNA expression levels. Two-tailed P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-455 was downregulated in gastric
cancer tissue samples and cell lines
miR-455 expression was detected in gastric cancer
tissue samples and their matched non-tumorous gastric tissue
samples using RT-qPCR. The expression level of miR-455 in the
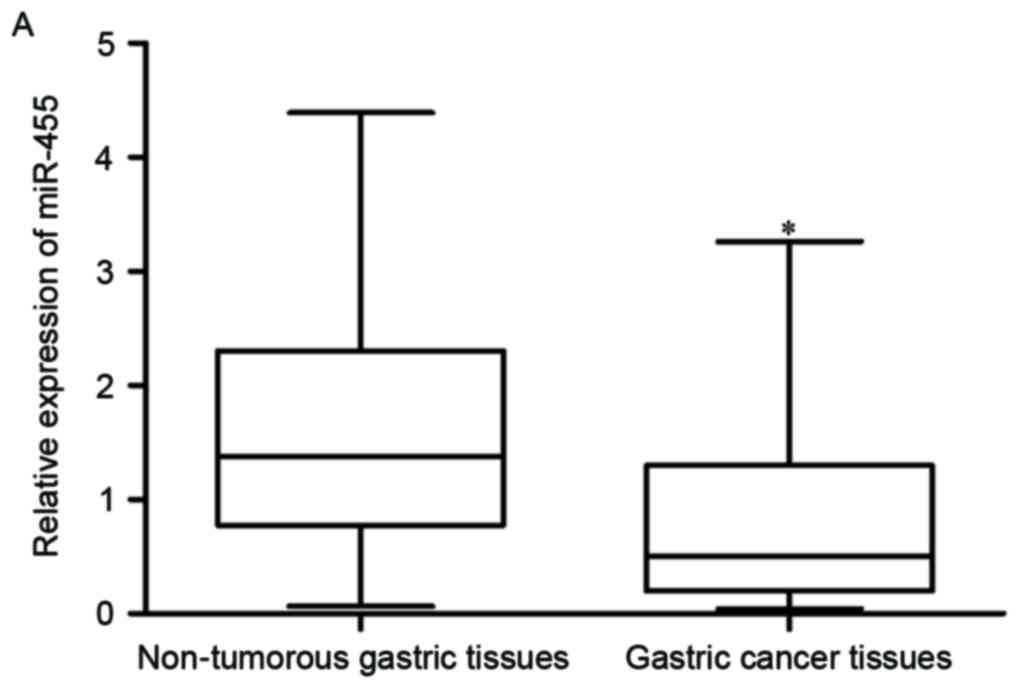
gastric cancer tissue samples was significantly lower than that in
the matched non-tumorous gastric tissue samples (Fig. 1A; P<0.05).
In addition, the association between miR-455
expression and the clinicopathological features of gastric cancer
were evaluated. As shown in Table
I, reduced miR-455 expression was significantly associated with
the clinical stage (P=0.003), lymph node metastasis (P=0.008) and
tumor invasion (P=0.032). However, there was no correlation between
miR-455 expression and age (P=0.962), sex (P=0.607), tumor size
(P=0.314) or age (P=0.578).
 | Table I.Association between miR-455 expression
and clinicopathological features of gastric cancer. |
Table I.
Association between miR-455 expression
and clinicopathological features of gastric cancer.
|
|
| miR-455
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathologic
feature | Cases (n) | Low | High | P-value |
|---|
| Age, years |
|
|
| 0.962 |
|
<60 | 23 | 13 | 10 |
|
| ≥60 | 34 | 19 | 15 |
|
| Sex |
|
|
| 0.607 |
| Male | 39 | 21 | 18 |
|
|
Female | 18 | 11 | 7 |
|
| Tumor size, cm |
|
|
| 0.314 |
|
<3 | 42 | 26 | 16 |
|
| ≥3 | 15 | 6 | 9 |
|
| Differentiation |
|
|
| 0.578 |
| Well
and moderate | 25 | 13 | 12 |
|
| Poor
and signet | 32 | 19 | 13 |
|
| Clinical stage |
|
|
| 0.003 |
|
I-II | 20 | 6 | 14 |
|
|
III-IV | 37 | 26 | 11 |
|
| Lymph node
metastasis |
|
|
| 0.008 |
| No | 17 | 5 | 12 |
|
|
Yes | 40 | 27 | 13 |
|
| Invasion |
|
|
| 0.032 |
| T1 +
T2 | 11 | 3 | 8 |
|
| T3 +
T4 | 46 | 29 | 17 |
|
Subsequently, miR-455 expression levels in the
gastric cancer cell lines were compared with the normal gastric
epithelium cell line. As presented in Fig. 1B, miR-455 was downregulated in all
five gastric cancer cell lines (SGC-7901, BGC-823, AGS, MKN-1 and
MKN-45) compared with that in GES-1 (P<0.05).
miR-455 inhibited gastric cancer cell
proliferation, migration and invasion
Given the downregulation of miR-455 in gastric
cancer, it was hypothesized that miR-455 may be involved in the
progression of cervical cancer. Firstly, BGC-823 and MKN-45 cells
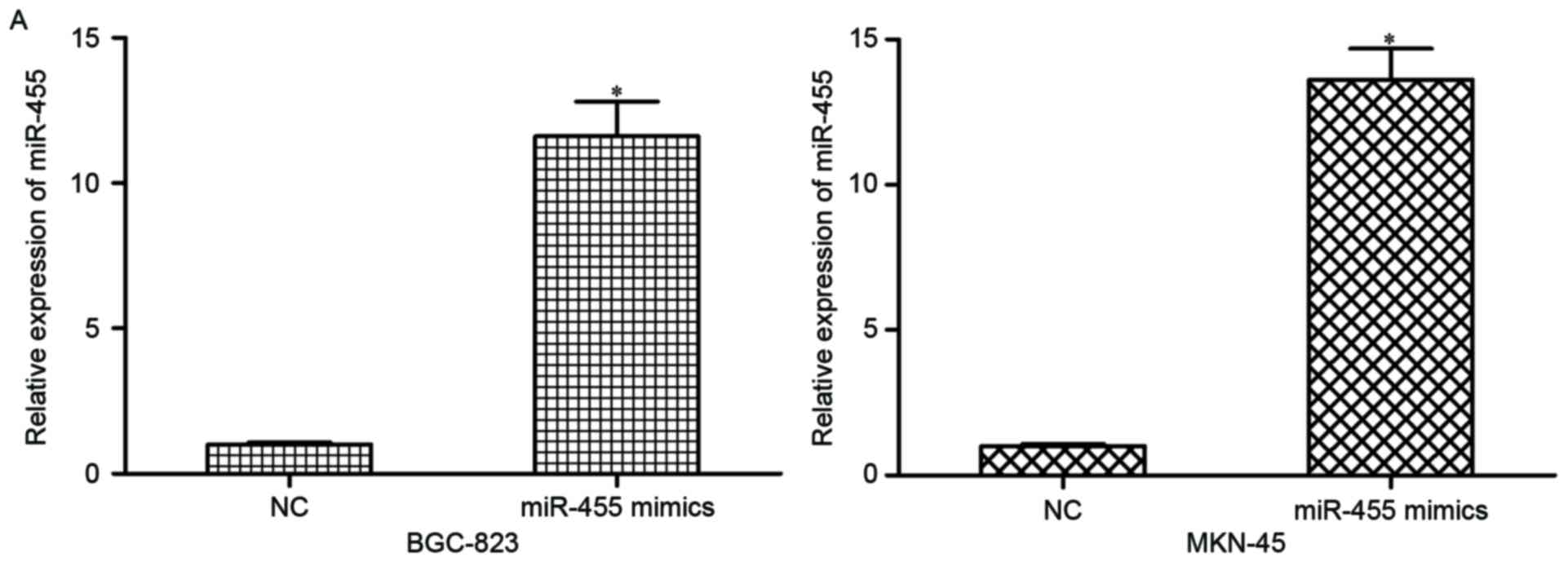
were transfected with miR-455 mimics or NC. At 48 h
post-transfection, results of RT-qPCR indicated that miR-455 was
markedly upregulated in miR-455 mimic-transfected BGC-823 and
MKN-45 cells (Fig. 2A; P<0.05).
Subsequently, MTT assay, and Transwell migration and invasion
assays were used to analyzed the effects of miR-455 on cell
proliferation, migration and invasion in gastric cancer. BGC-823
and MKN-45 cells transfected with miR-455 mimics exhibited
decreased proliferation (Fig. 2B;
P<0.05), migration and invasion (Fig. 2C; P<0.05) compared with NC.
IGF-1R was a direct target of
miR-455
Bioinformatic analysis with three algorithm
databases indicated that IGF-1R was a potential target of miR-455
(Fig. 3A). A luciferase reporter
assay was used to confirm whether the 3′-UTR of IGF-1R was directly
targeted by miR-455. HEK293T cells were co-transfected with
luciferase reporter vectors and miR-455 mimics or NC. As presented
in Fig. 3B, miR-455 overexpression
significantly decreased the luciferase activities of the
PGL3-IGF-1R-3′UTR Wt. However, there was no significant difference
between the miR-455 mimics and NC groups co-transfected with
PGL3-IGF-1R-3′UTR Mut.
 | Figure 3.miR-455 directly targeted the 3′-UTR
of IGF-1R to decrease its expression. (A) Bioinformatics analysis
demonstrated that miR-455 bound to the 3′-UTR of IGF-1R at the
3,782–3,788th base site. (B) Luciferase reporter assay was
performed in HEK293T cells injected with PGL3-IGF-1R-3′UTR Wt or
PGL3-IGF-1R-3′UTR Mut, together with miR-455 mimics or NC.
*P<0.05 vs. NC. Data are presented as the mean ± standard
deviation. (C) IGF-1R mRNA expression level in gastric cancer
tissue samples and their matched non-tumorous gastric tissue
samples was analyzed using reverse transcription-quantitative
polymerase chain reaction. Data are presented as the mean ±
standard deviation. *P<0.05 vs. non-tumorous gastric tissues.
(D) Spearman's correlation analysis revealed the significant
inverse correlation between miR-455 and IGF-1R mRNA in gastric
cancer tissues. reverse transcription-quantitative polymerase chain
reaction and western blotting were performed to detect IGF-1R (E)
mRNA and (F) protein expression in BGC-823 and MKN-45 cells
following transfection with miR-455 mimics or NC, respectively.
Data are presented as the mean ± standard deviation. *P<0.05 vs.
NC. mir-455, microRNA-455; 3′-UTR, 3′-untranslated region; IGF-1R,
insulin-like growth factor 1 receptor; Wt, wild-type; Mut,
mutation; NC, negative control miRNA mimics. |
The expression levels of IGF-1R mRNA in gastric
cancer tissue samples and their matched non-tumorous gastric tissue
samples were observed using qRT-PCR. As demonstrated in Fig. 3C, IGF-1R mRNA was expressed at
higher levels in gastric cancer tissue samples when compared with
the matched non-tumorous gastric tissue samples (P<0.05).
Spearman's correlation analysis revealed a significant inverse
correlation between miR-455 and IGF-1R mRNA (r=−0.8408) in gastric
cancer tissue samples (Fig. 3D;
P<0.001).
To investigate whether miR-455 regulates the IGF-1R
mRNA and protein expression levels in gastric cancer cells, RT-qPCR
and western blotting were performed. The results demonstrated that
IGF-1R mRNA (Fig. 3E; P<0.05)
and protein (Fig. 3F; P<0.05)
expression levels were suppressed by upregulation of miR-455 in the
BGC-823 and MKN-45 cells. These findings indicate that miR-455 may
directly target IGF-1R in gastric cancer cells via interaction with
the binding sites in the 3′UTR of IGF-1R.
Knockdown of IGF-1R inhibited cell
proliferation, migration and invasion in cervical cancer
To evaluate the effects of IGF-1R on gastric cancer,
IGF-1R siRNA or NC siRNA was injected into BGC-823 and MKN-45
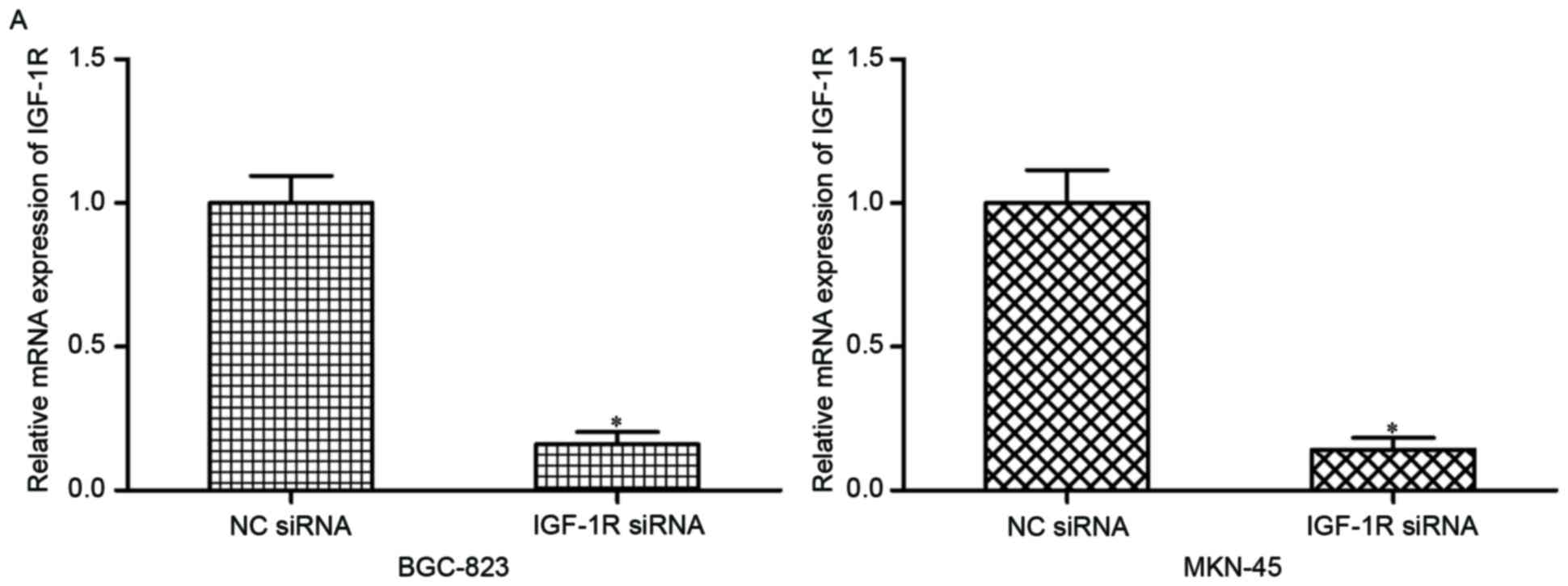
cells. Following transfection, RT-qPCR and western blotting were
conducted and IGF-1R was demonstrated to be downregulated at the
mRNA (Fig. 4A; P<0.05) and
protein (Fig. 4B; P<0.05)
levels in the BGC-823 and MKN-45 cells transfected with IGF-1R
siRNA. Consistent with the results of the current study,
downregulation of IGF-1R mimics the effects of miR-455
overexpression on BGC-823 and MKN-45 cell proliferation (Fig. 4C; P<0.05), migration and
invasion (Fig. 4D; P<0.05).
Discussion
In the current study, the miR-455 expression level
was measured in gastric cancer tissue samples and five gastric
cancer cell lines, and the expression levels of miR-455 were found
to be reduced in the gastric cancer tissue samples and cell lines
when compared with the matched non-tumorous gastric tissue samples
and normal gastric epithelium cell line, respectively. In addition,
a low miR-455 expression level was identified to be significantly
associated with the clinical stage, lymph node metastasis and tumor
invasion in gastric cancer. These findings indicate that a low
miR-455 expression level was contributed to the progression of this
disease. In addition, rapid growth and metastasis are the leading
cause of gastric cancer-associated mortalities. Therefore, the
roles of miR-455 on cell proliferation, migration and invasion of
gastric cancer were evaluated in the present study. Functional
experiments demonstrated that enforced miR-455 expression
significantly suppressed gastric cancer cell proliferation,
migration and invasion in vitro. Notably, IGF-1R was
identified as a direct and functional target of miR-455, which was
confirmed by a series of experiments. These findings elucidated the
decreased expression levels of miR-455 in gastric cancer and
detailed roles of miR-455 in gastric cancer, and further
contributed to establishing effective therapeutic targets for the
treatment of gastric cancer.
The expression levels of miR-455 vary in different
diseases. Li et al (20)
reported that miR-455 was significantly downregulated in non-small
cell lung cancer tissue samples and cell lines. A study by Chai
et al (21) demonstrated
that miR-455 expression was lower in colorectal cancer tissue
samples when compared with matched normal tissue samples. Cheng
et al (22) demonstrated
that the expression level of miR-455 was greater in oral squamous
cancer tissue samples than in the normal tissue samples. In
addition, its expression was also upregulated in oral squamous
cancer cell lines in comparison with that in normal keratinocyte
cell lines (22). In addition to
in tumors, miR-455 was identified to be upregulated in human
cartilage from patients with osteoarthritis compared with that in
healthy control subjects (23).
These findings indicate that miR-455 expression is diverse and
tissue-dependent.
The roles of miR-455 have primarily been evaluated
in human cancers. For example, in non-small cell lung cancer,
restoration of miR-455 expression repressed cell growth and
motility by downregulation of zinc finger E-box binding homeobox 1
(20). In colorectal cancer,
upregulation of miR-455 suppressed proliferation and invasion of
colorectal cancer cells by directly targeting Raf-1 proto-oncogene,
serine/threonine kinase (21). In
oral squamous cancer, reduced expression of miR-455 decreased the
cell anchorage-independent growth and proliferative ability by
inhibition of ubiquitin conjugating enzyme E2 B (22). In addition, in cardiac hypertrophy,
overexpression of miR-455 decreased myocardial fibrosis and
inhibited apoptosis by targeting calreticulin, indicating a
potential therapeutic strategy to reverse pressure-induced cardiac
hypertrophy and prevent maladaptive cardiac remodeling (24). Furthermore, miR-455 was found to be
involve in brown adipogenesis via regulation of differentiation and
thermogenesis via hypoxia inducible factor 1 a subunit, an
AMP-activated protein kinase-PPARG coactivator 1a signaling pathway
(25). These studies demonstrated
that miR-455 significantly contributes to these diseases, and may
therefore serve as a potential therapeutic target for their
treatment.
miRNAs perform their biological roles by binding to
the 3′-UTRs of their target genes and decreasing gene expression by
degrading the target mRNA or repressing its translation. It is
therefore important to investigate the potential target genes
involved in miR-455-mediated tumor suppressive roles in gastric
cancer. In the current study, IGF-1R was demonstrated to be a novel
direct and functional downstream target of miR-455, and IGF-1R
expression was negatively modulated by miR-455 by direct
interaction with the 3′-UTR of IGF-1R. This finding was supported
by a series of experiments. Firstly, bioinformatic analysis
indicated that the 3′-UTR of IGF-1R contained potential binding
sites of miR-455. Secondly, IGF-1R was identified as a direct
target of miR-455 using a luciferase reporter assay. Thirdly, the
expression and association between miR-455 and IGF-1R mRNA
expression in gastric cancer were analyzed, and it was demonstrated
that their expression levels were inversely correlated in tumor
tissue samples. Additionally, the expression levels of IGF-1R mRNA
and protein were downregulated by miR-455 overexpression in gastric
cancer cells. Finally, IGF-1R knockdown imitated the effects of
miR-455 overexpression on gastric cancer cell proliferation,
migration and invasion. These findings indicate that IGF-1R was a
novel direct target of miR-455 in gastric cancer, and, to the best
of our knowledge, this is the first study to investigate the
miR-455/IGF-1R interaction.
IGF-1R, a transmembrane tyrosine kinase receptor of
the insulin receptor family, contains two extracellular α subunits
with the ligand-binding site and two transmembrane β subunits with
intracellular tyrosine kinase activity (26). In gastric cancer, IGF-1R was
significantly upregulated in tumor tissue samples (27,28).
The expression of IGF-1R was correlated with the TNM staging, lymph
node metastasis and distant metastasis (29). Furthermore, gastric cancer patients
exhibiting low IGF-1R expression levels demonstrated longer overall
survival than those with high IGF-1R expression (30). Additionally, functional experiments
have indicated that IGF-1R knockdown decreased gastric cancer cell
growth, motility and invasion and enhanced cell apoptosis (31). All these studies demonstrated that
IGF-1R is upregulated in gastric cancer, and act as an oncogene in
tumorigenesis and tumor development. Combined with the current
results, miR-455/IGF-1R based targeted therapy may present as an
effective therapeutic treatment for gastric cancer.
In conclusion, the present study demonstrated that
miR-455 acted as a tumor suppressor in gastric cancer by inhibiting
cell proliferation, migration and invasion. Furthermore, it was
identified that miR-455 exerted its functions by directly targeting
IGF-1R, thus providing further insights into the molecular
mechanisms of gastric cancer carcinogenesis and progression.
However, the effects of miR-455 on growth and metastasis in gastric
cancer cells in vivo were not examined. This will be
investigated in future research.
Acknowledgements
The present study was supported by the Key Project
of Research and Development of Huai'an (grant no. HAS2015009).
References
|
1
|
Yang L: Incidence and mortality of gastric
cancer in China. World J Gastroenterol. 12:17–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herszényi L and Tulassay Z: Epidemiology
of gastrointestinal and liver tumors. Eur Rev Med Pharmacol Sci.
14:249–258. 2010.PubMed/NCBI
|
|
4
|
Choi AH, Nelson RA, Merchant SJ, Kim JY,
Chao J and Kim J: Rates of lymph node metastasis and survival in
T1a gastric adenocarcinoma in Western populations. Gastrointest
Endosc. 83:1184–1192.e1. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takahashi T, Saikawa Y and Kitagawa Y:
Gastric cancer: Current status of diagnosis and treatment. Cancers
(Basel). 5:48–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hutvágner G and Zamore PD: A microRNA in a
multiple-turnover RNAi enzyme complex. Science. 297:2056–2060.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kwak PB, Iwasaki S and Tomari Y: The
microRNA pathway and cancer. Cancer Sci. 101:2309–2315. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shah NR and Chen H: MicroRNAs in
pathogenesis of breast cancer: Implications in diagnosis and
treatment. World J Clin Oncol. 5:48–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng D, Zhao S, Tang H, Zhang D, Sun H,
Yu F, Jiang W, Yue B, Wang J, Zhang M, et al: MicroRNA-20a-5p
promotes colorectal cancer invasion and metastasis by
downregulating Smad4. Oncotarget. 7:45199–45213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang Z, Song Q, Zeng R, Li J, Li J, Lin
X, Chen X, Zhang J and Zheng Y: MicroRNA-218 inhibits EMT,
migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical
cancer. Oncotarget. 7:45622–45636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han SY, Han HB, Tian XY, Sun H, Xue D,
Zhao C, Jiang ST, He XR, Zheng WX, Wang J, et al: MicroRNA-33a-3p
suppresses cell migration and invasion by directly targeting PBX3
in human hepatocellular carcinoma. Oncotarget. 7:42461–42473. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ
and Li Y: miR-630 functions as a tumor oncogene in renal cell
carcinoma. Arch Med Sci. 12:473–478. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu C, Li M, Zhang L, Bi Y, Wang P, Li J
and Jiang X: MicroRNA-205 suppresses the invasion and
epithelial-mesenchymal transition of human gastric cancer cells.
Mol Med Rep. 13:4767–4773. 2016.PubMed/NCBI
|
|
16
|
Liu GH, Liu YH, Yang Z, Zhu AL and Zhao
CL: MicroRNA-524-5p suppresses the growth and invasive abilities of
gastric cancer cells. Oncol Lett. 11:1926–1932. 2016.PubMed/NCBI
|
|
17
|
Tao S, Liu YB, Zhou ZW, Lian B, Li H, Li
JP and Zhou SF: MiR-3646 promotes cell proliferation, migration,
and invasion via regulating G2/M transition in human breast cancer
cells. Am J Transl Res. 8:1659–1677. 2016.PubMed/NCBI
|
|
18
|
Kwon SJ: Evaluation of the 7th UICC TNM
staging system of gastric cancer. J Gastric Cancer. 11:78–85. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li YJ, Ping C, Tang J and Zhang W:
MicroRNA-455 suppresses non-small cell lung cancer through
targeting ZEB1. Cell Biol Int. 40:621–628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chai J, Wang S, Han D, Dong W, Xie C and
Guo H: MicroRNA-455 inhibits proliferation and invasion of
colorectal cancer by targeting RAF proto-oncogene
serine/threonine-protein kinase. Tumour Biol. 36:1313–1321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng CM, Shiah SG, Huang CC, Hsiao JR and
Chang JY: Upregulation of miR-455-5p by the TGF-β-SMAD signalling
axis promotes the proliferation of oral squamous cancer cells by
targeting UBE2B. J Pathol. 240:38–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
MicroRNA-455 important in chondrogenesis.
Bonekey Rep. 1:1652012.PubMed/NCBI
|
|
24
|
Wu C, Dong S and Li Y: Effects of
miRNA-455 on cardiac hypertrophy induced by pressure overload. Int
J Mol Med. 35:893–900. 2015.PubMed/NCBI
|
|
25
|
Zhang H, Guan M, Townsend KL, Huang TL, An
D, Yan X, Xue R, Schulz TJ, Winnay J, Mori M, et al: MicroRNA-455
regulates brown adipogenesis via a novel HIF1an-AMPK-PGC1α
signaling network. EMBO Rep. 16:1378–1393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu Q, Gong JP, Li J, Zhong SL, Chen WX,
Zhang JY, Ma TF, Ji H, Lv MM, Zhao JH and Tang JH: Down-regulation
of miRNA-452 is associated with adriamycin-resistance in breast
cancer cells. Asian Pac J Cancer Prev. 15:5137–5142. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pavelić K, Kolak T, Kapitanović S,
Radosević S, Spaventi S, Kruslin B and Pavelić J: Gastric cancer:
The role of insulin-like growth factor 2 (IGF 2) and its receptors
(IGF 1R and M6-P/IGF 2R). J Pathol. 201:430–438. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gryko M, Kiśluk J, Cepowicz D, Zińczuk J,
Kamocki Z, Guzińska-Ustymowicz K, Pryczynicz A, Czyżewska J, Kemona
A and Kędra B: Expression of insulin-like growth factor receptor
type 1 correlate with lymphatic metastases in human gastric cancer.
Pol J Pathol. 65:135–140. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao Y, Cui J, Xi H, Shen W, Zhang K, Li J,
Liang W, Hu C, Wei B and Chen L: Association of prognosis with
insulin-like growth factor receptor type I expression in gastric
cancer patients: A meta-analysis. Zhonghua Wei Chang Wai Ke Za Zhi.
18:1051–1055. 2015.(In Chinese). PubMed/NCBI
|
|
30
|
Matsubara J, Yamada Y, Nakajima TE, Kato
K, Hamaguchi T, Shirao K, Shimada Y and Shimoda T: Clinical
significance of insulin-like growth factor type 1 receptor and
epidermal growth factor receptor in patients with advanced gastric
cancer. Oncology. 74:76–83. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ge J and Chen Z, Huang J, Yuan W, Den Z
and Chen Z: Silencing insulin-like growth factor-1 receptor
expression inhibits gastric cancer cell proliferation and invasion.
Mol Med Rep. 11:633–638. 2015.PubMed/NCBI
|