Introduction
The endothelial tyrosine kinase receptor (TIE2) is
required for vascular remodeling and the maintenance of mammalian
vessel integrity (1–3). Angiopoietin-1 (ANG-1) is an
angiogenic protein which binds and activates the endothelial TIE2
receptor. The ANG-1/TIE2 signaling cascade plays a crucial role in
remodeling and maturation/stabilization of the embryonic
vasculature (4,5). Both ANG-1 and TIE2 are widely
expressed in the adult vasculature, illustrating their importance
in the maintenance of vascular homeostasis. The angiogenic growth
factors vascular endothelial growth factor (VEGF), basic fibroblast
growth factor (bFGF) and ANG-1 are upregulated in endothelial
vessels during inflammatory processes such as wound healing,
atherosclerosis and arthritis (6–9).
ANG-1 and ANG-2 can be used as clinically informative biomarkers of
disease severity and outcome in severe sepsis (10,11).
Interleukin-Iβ (IL-1β) and tumor necrosis factor α (TNF-α) can
induce the expression of both VEGF and bFGF (12,13),
and Brown et al demonstrated that the expression of ANG-1 is
also stimulated by TNF-α (14).
Recent studies have shown that microRNAs (miRNAs), short 22
nucleotide non-coding RNAs, can modulate gene expression by binding
to the 3′-untranslated regions (UTRs) of their target mRNAs via
complementary base pairing, to repress translation or direct
sequence-specific degradation of the mRNAs. miRNAs are increasingly
being proven to play an important function in the control of
inflammatory responses, and have also been associated with the
proliferation, migration, adhesion, stimuli response and retraction
of vascular cells (15,16). The present study was undertaken to
investigate whether the expression of ANG-1 is regulated by
specific miRNAs during inflammatory processes. Chen et al,
have shown that the functional variant in the 3′-UTR of
angiopoietin-1 might reduce stroke risk by interfering with the
binding efficiency of miR-211 (17). Here, we provide evidence to show
that expression of ANG-1 and the migratory ability of EA.hy926
cells are reduced in response to miR-204/211. Additionally, we
demonstrate that miR-204/211 are downregulated in EA.hy926
endothelial cells after exposure to lipopolysaccharide (LPS).
Materials and methods
Cell culture and LPS exposure
The EA.hy926 endothelial cell line (18), derived from immortalized human
umbilical vein endothelial cells (HUVECs), was cultured in DMEM
containing 10% fetal bovine serum (FBS), 2 mM glutamine, HAT (final
concentrations of 100 mM hypoxanthine, 0.4 mM aminopterin and 16 mM
thymidine) and antibiotics at 37°C in a 5% CO2
atmosphere. The growth media was replaced every 3–5 days. For
transfection and LPS exposure experiments, the cells were seeded in
24-well plates and used at 80% confluence. The cells were
stimulated with 1 µg/ml of LPS for different periods of time (0, 1,
2, 4, 6, 12, 18 and 24 h), washed once with PBS, and then
harvested.
Plasmid construction
Luciferase reporter constructs were generated by
introducing the ANG-1 3′-UTR, carrying a putative miR-204/211
binding site, into the vector pGL3 control (Promega, Madison, WI,
USA). The 3′-UTR sequence was amplified by PCR using the primers
ANG-1-UTR-Sense (5′-CGGGGTACCGCGCAATGTCAGAAGCGATTATG-3′) and
ANG-1-UTR-Antisense (5′-GGAAGATCTGTAGTTTGAAGCACAGCAAGC-3′) to
introduce KpnI and BglII restriction sites
(underlined). The PCR product was cloned into pGL3 control and the
resultant plasmid (pGL3C-WT) was used as a template to produce the
mutant plasmid pGL3C-MUT. Site-directed mutagenesis of the
miR-204/211 binding site was performed using the Quik-Change
Site-Directed Mutagenesis kit (Stratagene, Heidelberg, Germany).
Specifically, the bases AAA, complementary to UUU in the seed
sequence (UUUCCCUU) of miR-204/211, were mutated to TTT; the three
base pairs were mutated without introducing or removing other
nucleotides in the binding site. All plasmid DNA was purified using
the QIAfilter plasmid kit (Qiagen, Hilden, Germany) and confirmed
to have the correct sequence by sequencing (Takara, Dalian, China).
miR-NC (empty vector), miR-204 (miRBase accession number:
MI0000247) and miR-211 (miRBase accession number: MI0000708)
overexpressing retroviral vector carrying were constructed as
described by Huang et al (19,20).
Luciferase reporter assay
To analyze the role of the ANG-1 mRNA 3′UTR,
EA.hy926 cells were seeded into 24-well plates and transfected with
100 ng of pGL3C-WT/pGL3C-Mut or co-transfected with
pGL3C-WT/pGL3C-Mut and miR-204/211 overexpressing plasmids. The
cells were also co-transfected with the control plasmid pRL-TK
Renilla (Promega). At 24 h after transfection, the firefly
and Renilla luciferase activities of the cell lysates were
assayed using the Dual Luciferase Reporter Assay system kit
(Promega) with a Modulus Luminometer (Turner Biosystems, Sunnyvale,
CA, USA). The relative firefly luciferase activities (RLU) were
calculated by normalizing for transfection efficiency against
Renilla luciferase activity. All data are the mean ± SD of
at least three independent experiments.
Western blot analysis
Cells were lysed using RIPA buffer containing 1%
NP-40, 0.1% SDS, 10 mM Tris-HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl
and protease inhibitor cocktail, and then subsequently sonicated.
Protein concentrations were determined using the BCA protein assay
kit (Pierce, Rockford, IL, USA) according to the manufacturer's
protocol. Equal amounts of protein were resolved by SDS-PAGE and
immunoblotted using an anti-ANG-1 antibody and secondary antibody,
anti-goat IgG-HRP (Santa Cruz Biotechnology, Santa Cruz, CA). The
signals were detected using the ECL Advance Western Blotting
Detection kit (Amersham Biosciences, Piscataway, NJ, USA); an
anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody
(Santa Cruz Biotechnology, Santa Cruz, CA). was used as a loading
control.
Quantitative reverse
transcription-polymerase chain reaction
After treatment with LPS or transfection with
plasmids, the cells were harvested and total RNA was isolated using
the miRVana miRNA Isolation Kit (Ambion, Austin, TX, USA). The
expression of miR-204/211 was quantified using quantitative reverse
transcription-PCR (qRT-PCR). The specific miR-204/211 RT primer
sequence was
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGCAW-3′ (W=A or
T); the PCR primer sequences for miR-204/211 were
5′-GGGCTTCCCTTTGTCATCCT-3′ and 5′-GTGCAGGGTCCGAGGT-3′. U6 snRNA was
used as an internal control (5′-CGCTTCGGCAGCACATATAC-3′ and
5′-CAGGGGCCATGCTAATCTT-3′). The expression of ANG-1 mRNA was
quantified by qRT-PCR using the primers sense
5′-TTTCCTCGCTGCCATTCTGACTC-3′ and antisense
5′-TATGGATGTCAATGGGGGAGGTT-3′; GAPDH was amplified as an internal
control using the primers sense 5′-CAAAGTTGTCATGGATGACC-3′ and
antisense 5′-CCATGGAGAAGGCTGGGG-3′.
Assay of mRNA stability
ANG-1 mRNA stability was determined by treating
EA.hy926 cells with the transcription inhibitor
5,6-dichloro-1-β-D-ribobenzimidazole (DRB). The cells were treated
with 1 µg/ml LPS for different periods of time (0, 1, 2, 4 and 6 h)
and then treated with 50 uM DRB for 24 h to inhibit transcription.
Total RNA was collected at the indicated time points, and the
expression of ANG-1 mRNA was determined by qPCR and expressed as a
percentage of the initial mRNA level after LPS treatment. The
half-life of ANG-1 mRNA was calculated using decay curves.
Wound-scratch assay
The wound-scratch assay was used to investigate cell
migration. Briefly, EA.hy926 cells in the log phase were seeded in
6-well plates, cultured for 24 h until approaching 100% confluence
and then transfected with miR-NC, miR-204 or miR-211-expressing
retroviruses. At 24 h post-transfection, the monolayers were
wounded using 10 µl pipette tips to produce 300 µm wide wounds. The
unattached cells were washed away with three washes of PBS, and the
monolayers were cultured in media containing 0.1% FBS. The wounds
were photographed immediately and 12 h after wounding by
phase-contrast microscopy at 10× magnification, and the distance
migrated was calculated.
Statistical analysis
Each experiment was performed at least three times;
paired or unpaired Student's t-tests were used for statistical
analysis; significant P-values (P<0.05) are indicated by
asterisks in the figures.
Results
LPS induces ANG-1 mRNA and protein
expression
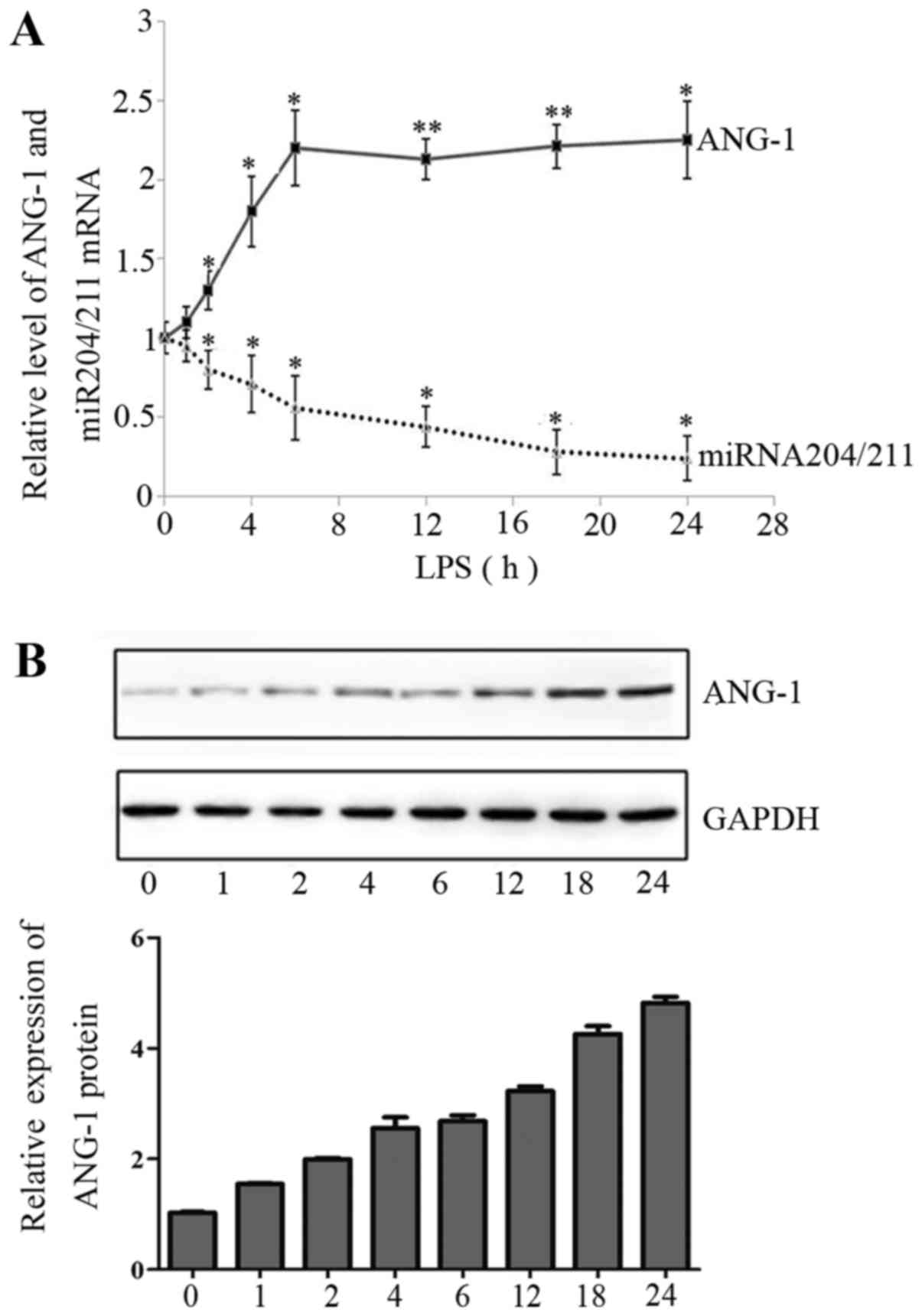
To confirm whether ANG-1 is upregulated as an
inflammatory response, we investigated the regulation of ANG-1 in
EA.hy926 endothelial cells. Firstly, we analyzed the levels of
ANG-1 mRNA in EA.hy926 cells stimulated with 1 µg/ml LPS for 1, 2,
4, 6, 12, 18 and 24 h by qRT-PCR. We observed a rapid, significant
increase in ANG-1 mRNA expression after 6 h LPS treatment (about
2.2-fold), which was followed by a slight increase over the
remainder of the time course (Fig.
1A). Western blotting was used to determine whether LPS-induced
ANG-1 mRNA expression was reflected at the protein level. A
time-dependent increase in ANG-1 protein expression was observed,
with a maximum at 6 h; however, ANG-1 protein expression did not
markedly increase after this time (Fig. 1B).
LPS reduces expression of
miR-204/211
We analyzed the time-course of miR-204/211
expression in response to LPS treatment. Quantitative RT-PCR
demonstrated an inverse correlation between the expression of
miR-204/211 and ANG-1 mRNA in LPS-treated EA.hy926 cells (Fig. 1A), indicating that miR-204/211 may
mediate LPS-induced expression of ANG-1 mRNA.
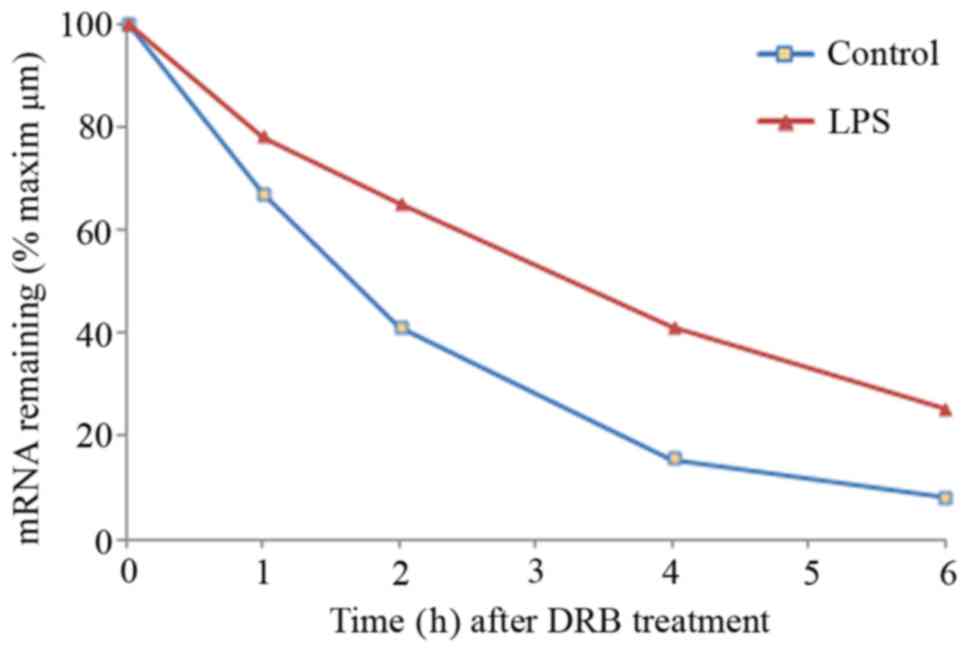
LPS treatment stabilizes ANG-1
mRNA
The mechanisms which regulate the transcription of
ANG-1 have been extensively researched (21), whereas relatively little is known
about the post-transcriptional regulation of ANG-1 during
inflammation. Thus, we analyzed the post-transcriptional regulation
of ANG-1 during LPS stimulation. To determine if LPS increased the
steady-state expression level of ANG-1 mRNA by increasing the
stability of ANG-1 mRNA, we treated EA.hy926 cells with PBS or LPS
for 6 h, the time point which led to the maximal induction of ANG-1
mRNA (Fig. 1A). Then, DRB, an
inhibitor of the TFIIH-associated CTD kinase was added to inhibit
ongoing transcription. Total RNA was isolated after 0, 1, 2, 4 and
6 h, and the levels of ANG-1 mRNA were quantified by qRT-PCR. ANG-1
mRNA was highly unstable in PBS-treated EA.hy926 cells, with a
half-life of less than 2 h (Fig.
2). However, LPS treatment increased the half-life of ANG-1
mRNA to approximately 3.5 h. Previous studies have demonstrated
that LPS induces the transcription of ANG-1 mRNA (22); however, the response to the
transcription inhibitor DRB suggested that LPS could contribute to
the stabilization of ANG-1 mRNA.
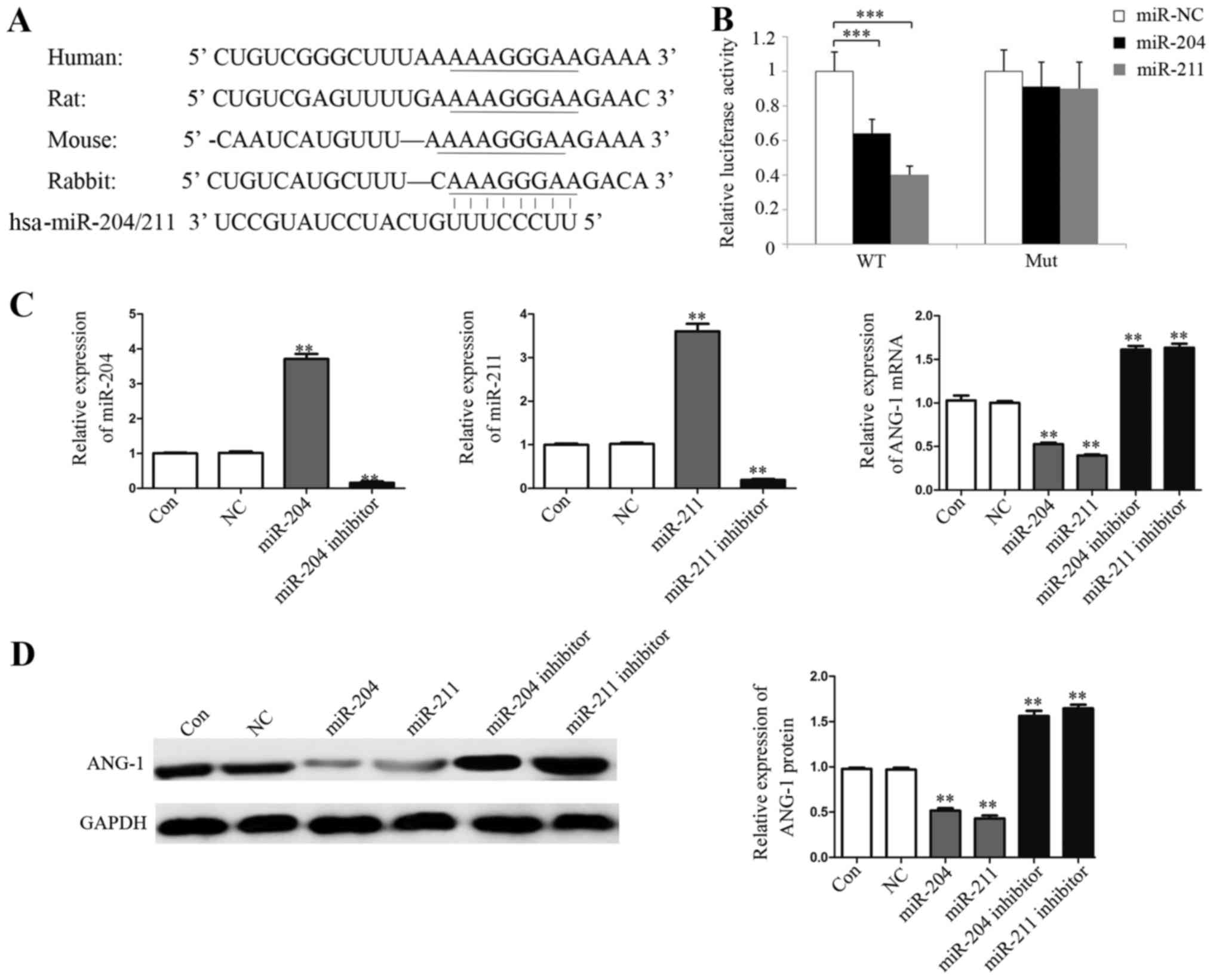
Overexpression of miR-204/211
downregulate ANG-1 expression
As mRNA stability is primarily regulated by the
binding of miRNAs to the 3′UTR of mRNAs, we analyzed the 3′UTR
sequence of ANG-1 mRNA and found some adenylate uridylate elements
(AREs), a sequence typical of instable mRNA transcripts. The
bioinformatic programs TargetScan, PicTar and miRanda were used to
predict which miRNAs may target the conserved region of the ANG-1
3′UTR, not other genes such as VEGF, bFGF and ANG-2, which
indicates that the function of miRNA204/211 on ANG-1 si specific.
Several miRNAs, including miR-204/211, miR-1244, miR-153 and
miR-448 were identified to potentially bind the 3′UTR of ANG-1
mRNA, and we hypothesized that ANG-1 may be regulated by these
miRNAs. However, manipulation of miR-1244, miR-153 and miR-448
resulted in no meaningful changes in the cell line following
treatment with LPS, therefore it was hypothesized that they were
not be involved in the interaction between ANG-1 and miRNA-204/211
MiR-204/211 was singled out for further analysis (Fig. 3A), followed by filtering out those
sites that do not appear to be conserved in multiple species with
the optimal free energy. A dual-luciferase reporter system was
employed to validate whether ANG-1 is a direct target gene of
miR-204/211. We cloned a 200 bp region of the ANG-1 3′UTR
containing the predicted miRNA target site of miR-204/211 (wild
type; pGL3C-WT) or the mutated miR-204/211 target site (mutant,
pGL3C-Mut) into the luciferase reporter vector pGL3 control.
miR-204 or miR-211 were overexpressed in EA.hy926 cells transiently
transfected with pGL3C-WT and pGL3C-Mut, and the relative
luciferase activities were measured. Overexpression of miR-204 and
miR-211 significantly suppressed the luciferase activity of
pGL3C-WT by 36.4 and 60.9%, respectively (P<0.001), whereas
overexpression of miR-204 and miR-211 did not significantly affect
the luciferase activity of pGL3C-Mut (Fig. 3B). This illustrated that both
miR-204 and miR-211 can directly target the 3′UTR of ANG-1. The
effects of miR-204 and miR-211 on the endogenous expression levels
of ANG-1 were subsequently examined in EA.hy926 cells.
Overexpression of miR-204/211 reduced the expression of ANG-1 mRNA
by 49.1 and 66.3%, respectively, and the effect can be reversed by
adding inhibitors of miR-204/211 (Fig.
3C). Western blotting indicated that overexpression of
miR-204/211 in EA.hy926 cells also reduced ANG-1 protein expression
by at least 50% (Fig. 3D).
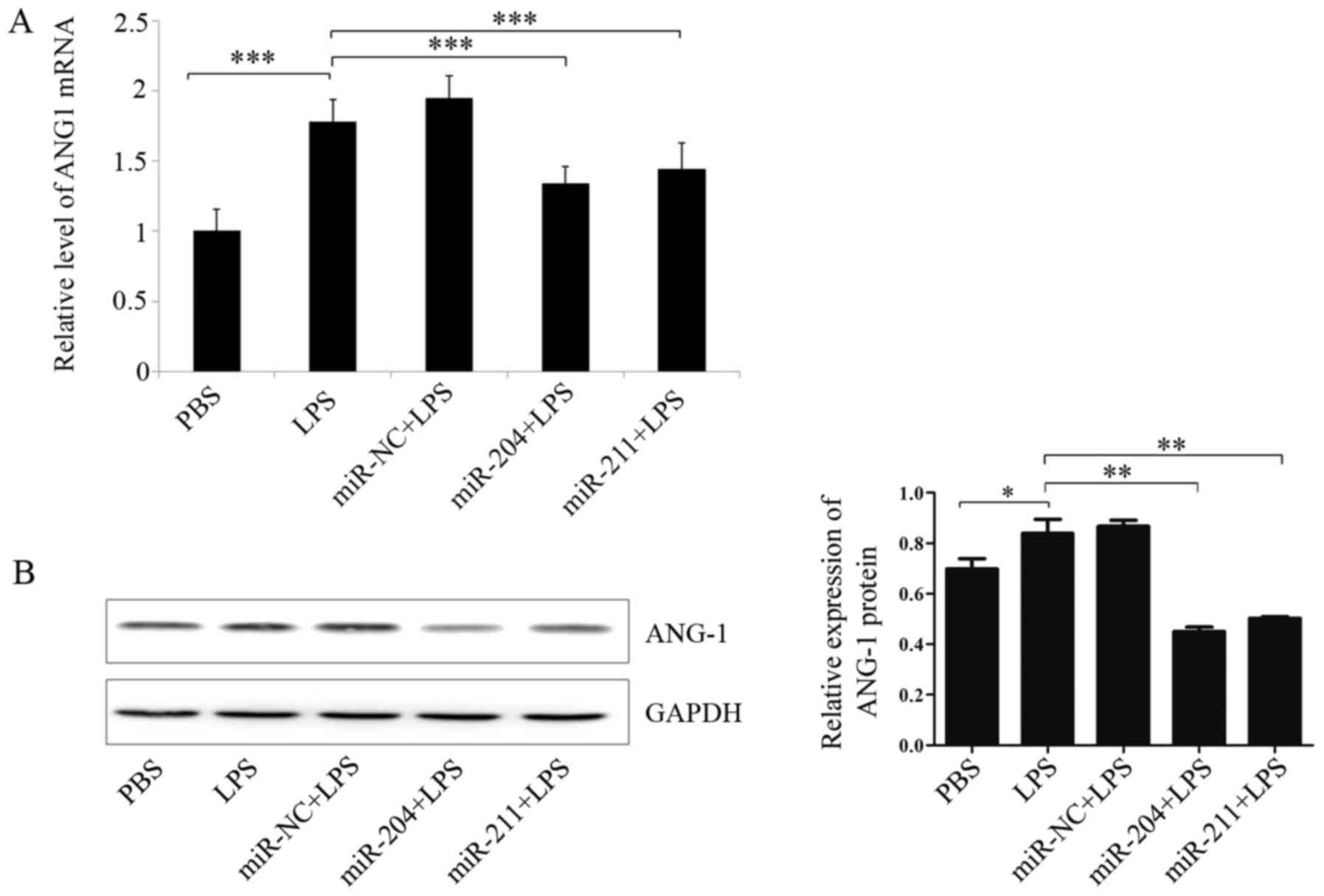
Overexpression of miR-204/211 partly
reverses the LPS-induced ANG-1 expression
To determine if gain-of-function of miR-204/211
could decrease ANG-1 protein levels induced by LPS, we treated the
EA.hy926 cells with PBS or LPS, and then retrovirally
over-expressed miR-204/211 gene or control vector. Quantitative
RT-PCR and western blotting results demonstrated that LPS induced
the expression of ANG-1, while, ANG-1 mRNA (Fig. 4A) and protein levels (Fig. 4B) were significantly reduced in the
microRNA overexpressing cells. The results indicated that
miR-204/211 could partly reverses the LPS-induced ANG-1
expression.
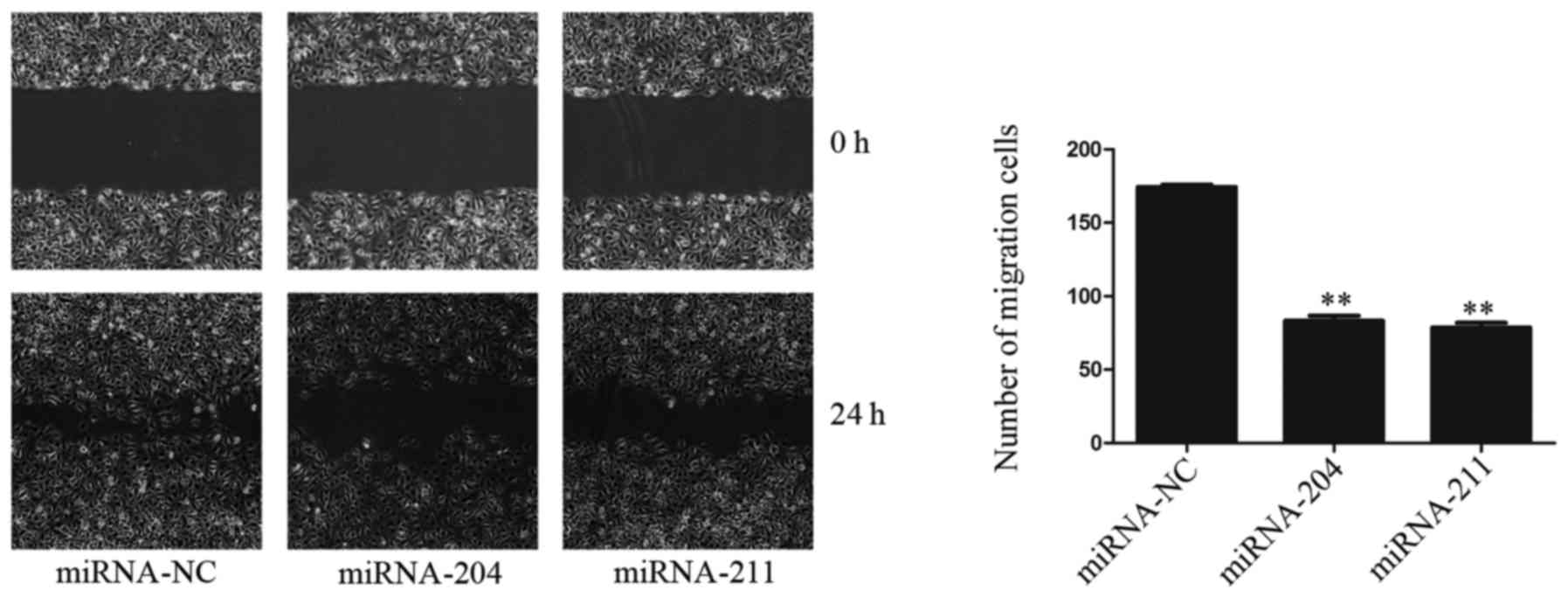
Overexpression of miR-204/211 reduces
the migration of EA.hy926 cells in vitro
As ANG-1 is also implicated in cell migration, we
examined the effect of miR-204/211 on the migration of EA.hy926
cells in vitro. miR-204/211 were overexpressed in confluent
EA.hy926 cell monolayers, and a wound was formed by scratching with
pipette tip. Cell migration was monitored at different time points
by light microscopy. After 12 h, the cells in the miR-NC group had
migrated 258±26 µm. In contrast, the miR-204/211-expressing cells
migrated more slowly (161±36 and 174±31 µm, respectively; Fig. 5). This data clearly demonstrated
that overexpression of miR-204/211 markedly reduced the migration
of EA.hy926 cells.
Discussion
miRNAs are short noncoding RNAs which can regulate a
variety of biological processes via regulating the expression of
multiple target genes. In the present study, we demonstrated that
LPS treatment downregulated the expression of miR-204/211 in
EA.hy926 endothelial cells, which upregulated the expression of
ANG-1 mRNA. Inflammatory diseases such as rheumatoid arthritis
(RA), chronic granulomatous response and certain benign tumors
often are accompanied by intense angiogenesis (23–25).
During angiogenesis, endothelial cell behavior is modulated by
various cytokines. A number of inflammatory cytokines, including
TNF-α, IL-1 and VEGF are released by inflamed tissues (26). ANG-1 promotes the stabilization of
vascular networks and branching morphogenesis, without stimulating
endothelial cell proliferation (27). As ANG-1 is a critical mediator of
the later stages of blood vessel development and vessel maturation,
activation of ANG-1 transcription may also be required during the
vascular endothelial cell response associated with inflammatory
diseases. Results from studies have shown miRNAs are important
regulators of innate immune responses (28). mir-155 is reported to be
substantially upregulated in murine macrophages activated with
polyriboinosinic:polyribocytidylic acid [poly (I:C)] or IFN-β
(29). mir-204 was significantly
up-regulated in T cells activated by anti-CD3 antibodies in
vitro. miR-204 and probably miR-211 are downregulated in
inflammation-associated gastric cancer (30). Furthermore, recently data indicated
miR-204/211 involved in maintaining epithelial barrier function and
cell physiology (31). LPS acts as
a classical imitator of gram-negative bacterial infection, which
induces the innate and adaptive immune responses and has the
capacity to activate endothelial cells (32). In this study, we demonstrated that
ANG-1 mRNA expression increased in EA.hy926 endothelial cells
exposed to LPS. As post-transcriptional regulation contributes to
gene expression during chronic inflammation and cancer (16), we sought to probe the
post-transcriptional mechanisms which potentially regulate ANG-1.
The transcription inhibitor DRB was used to determine the stability
ANG-1 mRNA. After exposure to LPS, the half-life of ANG-1 mRNA
increased from 2 to 3.5 h, indicating that ANG-1 mRNA was
stabilized in LPS-treated EA.hy926 endothelial cells. We identified
a highly conserved 8-mer miR-204/211 binding site in the 3′-UTR of
ANG-1 mRNA. miR-204 and miR-211 are closely related homologues.
Wang et al reported that miR-204 and miR-211 are highly
expressed in human fetal retinal pigment epithelium (hfRPE) cells
and are involved in maintaining epithelial barrier function and
normal cell physiology (31).
miR-204/211 also act as important endogenous negative regulators of
runt-related transcription factor 2 (Runx2), which inhibits
osteogenesis and promotes adipogenesis of mesenchymal progenitor
cells and bone marrow stromal cells (BMSCs) (20). Additionally, the expression of
miR-204 is elevated in activated T cells in vitro (33). These results indicate that
miR-204/211 are expressed in epithelial cells and may also play a
role in inflammatory processes. The use of reporter constructs is
an established method to verify potential mRNA destabilization
mechanisms. Luciferase assays using an ANG-1 3′UTR reporter gene
and point mutation of the miR-204/211 binding sites revealed that
miR-204/211 can bind to the ANG-1 3′UTR and post-transcriptionally
regulate ANG-1. Furthermore, we showed that LPS treatment could
reduce the abundance of miR-204/211 by approximately 40–60% in
EA.hy926 endothelial cells. Therefore, we conclude that LPS
negatively modulates miR-204/211, which results in increased
expression of ANG-1. ANG-1 has been shown to participate in the
endothelial cell migration and proliferation after vascular injury.
In the present study, we demonstrate that the overexpression of
miR-204/211 dramatically decreased migration of EA.hy926
endothelial cells. These results suggest that downregulation of
miR-204/211 may contribute to the repair process of endothelial
cell after vascular injury. In the future, further experiments are
planned to complete the hypothesis including infecting EA.h926
cells with different amount of miRNA 204/211 retroviral vector,
then checking ANG-1 mRNA and protein levels at different timepoints
Jennewein et al discovered that the NF-κB-dependent
downregulation of peroxisome proliferator-activated receptor 1
(PPAR-1) mRNA expression in response to LPS was due, at least in
part, to NF-κB-dependent induction of miR-27b (34). Therefore, it is possible that
miR-204/211 may be regulated via the similar NF-κB-dependent
mechanism. It is our expectation that the precise mechanisms which
regulate ANG-1 will be identified in the near future, which will
enable a better understanding of the molecular basis of
ANG-1-mediated inflammatory disease.
Acknowledgments
This study was funded by the Guangdong Natural
Science Fund Project, grant number: 2016A030310174.
References
|
1
|
Hansen TM, Singh H, Tahir TA and Brindle
NP: Effects of angiopoietins-1 and −2 on the receptor tyrosine
kinase tie2 are differentially regulated at the endothelial cell
surface. Cell Signal. 22:527–532. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Augustin HG, Koh GY, Thurston G and
Alitalo K: Control of vascular morphogenesis and homeostasis
through the angiopoietin-tie system. Nat Rev Mol Cell Biol.
10:165–177. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen L, Li J, Wang F, Dai C, Wu F, Liu X,
Li T, Glauben R, Zhang Y, Nie G, et al: Tie2 expression on
macrophages is required for blood vessel reconstruction and tumor
relapse after chemotherapy. Cancer Res. 76:6828–6838. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Erber R, Thurnher A, Katsen AD, Groth G,
Kerger H, Hammes HP, Menger MD, Ullrich A and Vajkoczy P: Combined
inhibition of VEGF and PDGF signaling enforces tumor vessel
regression by interfering with pericyte-mediated endothelial cell
survival mechanisms. FASEB J. 18:338–340. 2004.PubMed/NCBI
|
|
5
|
Jackson KA, Majka SM, Wang H, Pocius J,
Hartley CJ, Majesky MW, Entman ML, Michael LH, Hirschi KK and
Goodell MA: Regeneration of ischemic cardiac muscle and vascular
endothelium by adult stem cells. J Clin Invest. 107:1395–1402.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen L, Endler A, Uchida K, Horiguchi S,
Morizane Y, Iijima O, Toi M and Shibasaki F: Int6/eif3e silencing
promotes functional blood vessel outgrowth and enhances wound
healing by upregulating hypoxia-induced factor 2alpha expression.
Circulation. 122:910–919. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abdel-Malak NA, Mofarrahi M, Mayaki D,
Khachigian LM and Hussain SN: Early growth response-1 regulates
angiopoietin-1-induced endothelial cell proliferation, migration,
and differentiation. Arterioscler Thromb Vasc Biol. 29:209–216.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Clavel G, Bessis N, Lemeiter D, Fardellone
P, Mejjad O, Ménard JF, Pouplin S, Boumier P, Vittecoq O, Le Loët X
and Boissier MC: Angiogenesis markers (VEGF, soluble receptor of
VEGF and angiopoietin-1) in very early arthritis and their
association with inflammation and joint destruction. Clin Immunol.
124:158–164. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chan W, Ismail H, Mayaki D, Sanchez V,
Tiedemann K, Davis EC and Hussain SN: Fibulin-5 regulates
angiopoietin-1/tie-2 receptor signaling in endothelial cells. PLoS
One. 11:e01569942016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Conroy AL, Phiri H, Hawkes M, Glover S,
Mallewa M, Seydel KB, Taylor TE, Molyneux ME and Kain KC:
Endothelium-based biomarkers are associated with cerebral malaria
in malawian children: A retrospective case-control study. PLoS One.
5:e152912010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Paulus P, Jennewein C and Zacharowski K:
Biomarkers of endothelial dysfunction: Can they help us deciphering
systemic inflammation and sepsis? Biomarkers. 16 Suppl 1:S11–S21.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ben-Av P, Crofford LJ, Wilder RL and Hla
T: Induction of vascular endothelial growth factor expression in
synovial fibroblasts by prostaglandin e and interleukin-1: A
potential mechanism for inflammatory angiogenesis. FEBS Lett.
372:83–87. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paleolog EM, Young S, Stark AC, McCloskey
RV, Feldmann M and Maini RN: Modulation of angiogenic vascular
endothelial growth factor by tumor necrosis factor alpha and
interleukin-1 in rheumatoid arthritis. Arthritis Rheum.
41:1258–1265. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brown C, Gaspar J, Pettit A, Lee R, Gu X,
Wang H, Manning C, Voland C, Goldring SR, Goldring MB, et al: Ese-1
is a novel transcriptional mediator of angiopoietin-1 expression in
the setting of inflammation. J Biol Chem. 279:12794–12803. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao
W and Cui Q: An analysis of human microRNA and disease
associations. PLoS One. 3:e34202008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khabar KS: Post-transcriptional control
during chronic inflammation and cancer: A focus on AU-rich
elements. Cell Mol Life Sci. 67:2937–2955. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen J, Yang T, Yu H, Sun K, Shi Y, Song
W, Bai Y, Wang X, Lou K, Song Y, et al: A functional variant in the
3′-UTR of angiopoietin-1 might reduce stroke risk by interfering
with the binding efficiency of microRNA 211. Hum Mol Genet.
19:2524–2533. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Edgell CJ, McDonald CC and Graham JB:
Permanent cell line expressing human factor VIII-related antigen
established by hybridization. Proc Natl Acad Sci USA. 80:3734–3737.
1983; View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kitamura T, Koshino Y, Shibata F, Oki T,
Nakajima H, Nosaka T and Kumagai H: Retrovirus-mediated gene
transfer and expression cloning: Powerful tools in functional
genomics. Exp Hematol. 31:1007–1014. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang J, Zhao L, Xing L and Chen D:
MicroRNA-204 regulates Runx2 protein expression and mesenchymal
progenitor cell differentiation. Stem Cells. 28:357–364.
2010.PubMed/NCBI
|
|
21
|
Qin J, Chen X, Xie X, Tsai MJ and Tsai SY:
COUP-TFII regulates tumor growth and metastasis by modulating tumor
angiogenesis. Proc Natl Acad Sci USA. 107:3687–3692. 2010;
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mofarrahi M, Nouh T, Qureshi S, Guillot L,
Mayaki D and Hussain SN: Regulation of angiopoietin expression by
bacterial lipopolysaccharide. Am J Physiol Lung Cell Mol Physiol.
294:L955–L963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bodolay E, Koch AE, Kim J, Szegedi G and
Szekanecz Z: Angiogenesis and chemokines in rheumatoid arthritis
and other systemic inflammatory rheumatic diseases. J Cell Mol Med.
6:357–376. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szekanecz Z, Pakozdi A, Szentpetery A,
Besenyei T and Koch AE: Chemokines and angiogenesis in rheumatoid
arthritis. Front Biosci (Elite Ed). 1:44–51. 2009.PubMed/NCBI
|
|
25
|
Santos IC, Silbiger VN, Higuchi DA, Gomes
MA, Barcelos LS, Teixeira MM, Lopes MT, Cardoso VN, Lima MP, Araujo
RC, et al: Angiostatic activity of human plasminogen fragments is
highly dependent on glycosylation. Cancer Sci. 101:453–459. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jackson JR, Bolognese B, Kircher CH,
Marshall LA and Winkler JD: Modulation of angiogenesis in a model
of chronic inflammation. Inflamm Res. 46 Suppl 2:S129–S130. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Papapetropoulos A, Fulton D, Mahboubi K,
Kalb RG, O'Connor DS, Li F, Altieri DC and Sessa WC: Angiopoietin-1
inhibits endothelial cell apoptosis via the Akt/survivin pathway. J
Biol Chem. 275:9102–9105. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Taganov KD, Boldin MP and Baltimore D:
MicroRNAs and immunity: Tiny players in a big field. Immunity.
26:133–137. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
O'Connell RM, Taganov KD, Boldin MP, Cheng
G and Baltimore D: MicroRNA-155 is induced during the macrophage
inflammatory response. Proc Natl Acad Sci USA. 104:1604–1609. 2007;
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lam EK, Wang X, Shin VY, Zhang S, Morrison
H, Sun J, Ng EK, Yu J and Jin H: A microRNA contribution to
aberrant Ras activation in gastric cancer. Am J Transl Res.
3:209–218. 2011.PubMed/NCBI
|
|
31
|
Wang FE, Zhang C, Maminishkis A, Dong L,
Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S and Miller SS:
MicroRNA-204/211 alters epithelial physiology. FASEB J.
24:1552–1571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pioli PA, Weaver LK, Schaefer TM, Wright
JA, Wira CR and Guyre PM: Lipopolysaccharide-induced IL-1 beta
production by human uterine macrophages up-regulates uterine
epithelial cell expression of human beta-defensin 2. J Immunol.
176:6647–6655. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pedersen I and David M: MicroRNAs in the
immune response. Cytokine. 43:391–394. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jennewein C, von Knethen A, Schmid T and
Brune B: MicroRNA-27b contributes to lipopolysaccharide-mediated
peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA
destabilization. J Biol Chem. 285:11846–11853. 2010. View Article : Google Scholar : PubMed/NCBI
|