Introduction
Antioxidant of bamboo leaves (AOB) was approved as a
natural food additive by the Chinese Ministry of Health in 2007.
AOB may be used as a food antioxidant, preservative or flavoring in
numerous types of foods. AOB has several types of bioactive
components including flavonoids, lactones, and phenolic acids,
however, it predominantly consists of four representative
flavonoids (orientin, isoorientin, vitexin, and isovitexin). AOB is
obtained from bamboo leaves and has been a focus of research due to
its antioxidative activity (1).
However, the dose-dependent toxicity of AOB and its impact on
animal reproductive and developmental function remain unclear
(2). The working principle of the
genechip technique is based on hybridization between target DNA/RNA
extracted from cell lines or tissues and complementary short
DNA-nucleotide oligomers grafted to the solid surface of the chip
(3,4). Genechip has been widely used in
functional genomics and investigation of pathogenic mechanisms.
Mouse embryonic fibroblasts (MEF) are a type of undifferentiated
cell that have the potential for infinite proliferation and
totipotential differentiation (5,6).
MEFs have been successfully applied in a variety of biological
mechanism and toxicological studies (7,8).
However, the effects of AOB on the reproductive toxicity of MEFs
have not been reported. In the present study, MEF cells were used
to detect the impact of different concentrations of AOB on MEF
proliferation. Additionally, the gene expression of MEF cells was
analyzed to explore the molecular mechanism through which AOB may
affect the proliferation and apoptosis of MEFs.
The present study aimed to investigate the impact of
AOB on the expression of reproduction-associated proteins. These
findings may provide a broader understanding of the role of AOB in
the activation of the ERK and Wnt signaling pathways.
Materials and methods
Preparation of MEFs
A total of 8 pregnant ICR mice (6 weeks old; weight,
26±5 g) were purchased from Zhejiang Academy of Medical Sciences
(MIS20034; Zhejiang, China). All mice had free access to water and
food and were maintained at 24°C in a humidity-controlled room with
a 12–12 h light-dark cycle. Mice were sacrificed at day 13.5 of
gestation by cervical dislocation. The body was placed into aseptic
conditions following disinfection by immersion for 3–5 min in 75%
ethanol. The uterine horns were dissected, briefly rinsed in PBS
3–5 times, and each embryo was separated from its placenta and
embryonic sac. The uterus was cut open along the uterine membrane
to remove the embryo that was covered by the membrane envelope, the
embryos were washed with PBS and placed into a clean Petri dish.
The tissue was finely minced using a sterile razor blade in order
to facilitate pipetting. A total of 1 ml 0.05% trypsin/0.02% EDTA
was added and cells were dissociated by pipetting up and down
thoroughly and incubated for 5–10 min at 37°C. The supernatant was
aspirated and the cells were centrifuged at low-speed (300 × g) at
4°C for 5 min; the supernatant was subsequently removed and the
cell pellet resuspended in PBS. Cells were counted, plated in MEF
medium (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at a density
of 35,000 cells/cm2 in 6-well plates and incubated at
37°C with 5% CO2 for 24 h. The medium was replaced with
fresh MEF medium and the cells were cultured at 37°C with 5%
CO2. Cells were continuously passaged or frozen for
future usage when they had reached 90–95% confluence. The protocol
of the present study was approved by the Ethics Committee of
Zhejiang University (Hangzhou, China; approval no. ZU20150324).
Cell viability (MTT) assay
AOB was provided as a kind gift from Professor Ying
Zhang (Zhejiang University). Cells were seeded at 1×105
cells/ml in a 24-well plate with final AOB concentrations of 0,
100, 200, 300, 400, 500 to 800 µg/ml, and cultured at 37°C with 5%
CO2 for 72 h. Following incubation, the medium was
removed and supplemented with MTT reagent (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Following incubation at 37°C
for 4 h, the medium was removed and 100 µl DMSO was added to each
well and agitated for 15 min at 37°C. The cell viability percentage
was assessed via spectrophotometry at 570 nm using an ELx800
Absorbance Reader (BioTek Instruments, Inc., Winooski, VA, USA).
Based on the results of this experiment, a concentration of 400
µg/ml AOB was used for all subsequent experiments.
Flow cytometric analysis for apoptosis
quantification
Flow cytometric analysis was used to detect the
apoptotic rate of cells processed with a fluorescein isothiocyanate
(FITC) Apoptosis Detection kit (BD Biosciences, San Jose, CA, USA).
A total of 1×106 cells were harvested and washed twice
with cold PBS. Cells were subsequently washed and incubated in
buffer containing 5 µl FITC-Annexin V and 5 µl propidium iodide.
Apoptosis quantification was performed using an Annexin V-FITC
cellular apoptosis assay reagent kit (BD Biosciences), according to
the manufacturer's protocol. Cells were subsequently collected
using a FACSCalibur flow cytometer and analyzed using CellQuest
Software (version 3.3, BD Biosciences).
Microarray gene expression and data
analysis
The RNA samples from each group were sent to Genergy
Biotechnology Co., Ltd. (Shanghai, China) for Illumina MouseWG-6
v2.0 Expression BeadChip microarray analysis (Illumina, Inc., San
Diego, CA, USA). The quantile measure was used to normalize the
different arrays and identify differentially-expressed genes. A
fold change ≥|2| and P<0.05 was considered to indicate
significant differential expression of genes.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA (1 µg) was reverse-transcribed
to cDNA using an AccuPower RT PreMix kit (Bioneer Corporation,
Daejeon, Korea), with incubations performed at 37°C for 15 min
followed by 85°C for 10 sec. Gene specific primers for the 20 mRNAs
and β-actin are summarized in Table
I, β-actin served as an internal control for gene expression
normalization. PCR amplifications were performed using Takara
master mix (Takara Biotechnology Co., Ltd., Dalian, China). For
each PCR, 1 µl template cDNA, equivalent to ~100 ng total RNA, was
mixed with 12.5 µl 2X SYBR-Green PCR Master mix and 0.4 µM each of
the forward and reverse primers in a final volume of 20 µl under
the following conditions: Initial enzyme activation at 95°C for 10
min, amplification for 40 cycles (95°C for 30 sec and 60°C for 60
sec), followed by a dissociation curve analysis. The relative RNA
level was calculated using the 2ΔΔCq method (9).
 | Table I.Primer sequences for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primer sequences for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Primer sequence
5′-3′ | Tm | Length (bp) |
|---|
| β-actin | F:
GTGACGTTGACATCCGTAAAGA | 60.3 | 245 |
|
| R:
GCCGGACTCATCGTACTCC | 61.6 |
|
| Cabyr | F:
AGAGGATCACCTTGGGGTACA | 61.7 | 98 |
|
| R:
CGAAGCGACAGATGGTGGTC | 62.9 |
|
| Pcsk4 | F:
TTGCTGGGTCTTACAAGCTACT | 60.4 | 106 |
|
| R:
ACTGGTGAATAGAACAGGGCT | 60.1 |
|
| Myl2 | F:
CCCTGAAGTCGAGGAGCTG | 60.9 | 114 |
|
| R:
CTGCTGCACCTCTAAGCGA | 61.7 |
|
| Kng2 | F:
GAAGCGTCTCACTCCCGAAG | 62.3 | 93 |
|
| R:
GAAGAAAACGTCGCGCTACT | 60.7 |
|
| Cpz | F:
AGAGGATCACCTTGGGGTACA | 61.7 | 98 |
|
| R:
CGAAGCGACAGATGGTGGTC | 62.9 |
|
| B230120H23Rik | F:
CAAACCTCAATGTGTCTCTTTGC | 60.2 | 97 |
|
| R:
AGAGTAAAGCCTATCTCGCTGT | 60.4 |
|
| Pla2g4b | F:
CTTCCCTCATCCTCCTGCTAC | 61.1 | 145 |
|
| R:
ACAAACTGGGTAAAGGTGATGG | 60.2 |
|
| Rap1a | F:
CCAAAGCGGAGTCTCGCAT | 62.4 | 125 |
|
| R:
GCCTAGCATCTTGCTTAGCTC | 60.6 |
|
| Dhrs9 | F:
TTGCTCCGGTAACAGCAGTG | 62.4 | 105 |
|
| R:
GTGGTCGCTTGTGTAGAAGGA | 61.7 |
|
| Pdgfb | F:
ATGCTGGGAAAGTCATGGAAG | 60.0 | 201 |
|
| R:
CGTGTTCTGGTCACGAGAGA | 61.2 |
|
| Mapk12 | F:
ACCTGCACCCGATTCACAG | 62.0 | 112 |
|
| R:
TGGCAGCATACTCCTGACCA | 62.8 |
|
| Smc3 | F:
TCTGATCCGCTGTACTCTCCT | 61.6 | 60 |
|
| R:
AGGCGGCAATTCAACATCCA | 62.5 |
|
| Cxxc4 | F:
GAGAATTTCAAGTCGTGGCGA | 60.6 | 193 |
|
| R:
CAGGTTTTCCAGTATGTGCTCC | 60.9 |
|
| Fzd1 | F:
TTGGTTCGTCATAAGGCATCAC | 62.8 | 94 |
|
| R:
TGTTGGCAAAGGCCATAATATCT | 61.5 |
|
| Sfrp1 | F:
GGTTGGGAGAATCGTGACTGC | 63.0 | 299 |
|
| R:
TAGACACACGTCGCCTCTTCA | 62.9 |
|
| Lrp6 | F:
ATGCAGTACATTGGAGAAGGTG | 60.0 | 138 |
|
| R:
CGTCTCTCGGCTGCCTATTT | 61.7 |
|
| Ntrk2 | F:
TGCCCATCATTTCATTCATCCTT | 60.3 | 232 |
|
| R:
AAAAGCGGTTTCTCACTCTCC | 60.2 |
|
| Pla2g4e | F:
TGGTGTCCTTTATAGCCTCCTG | 59.2 | 157 |
|
| R:
CATCTCCTGTACCTTCAAGTTGTG | 59.3 |
|
| Fgf10 | F:
TGGTGTCCTTTATAGCCTCCTG | 58.7 | 124 |
|
| R:
CATCTCCTGTACCTTCAAGTTGTG | 58.7 |
|
| Pdgfra | F:
TGGTGTCCTTTATAGCCTCCTG | 61.2 | 136 |
|
| R:
CATCTCCTGTACCTTCAAGTTGTG | 60.3 |
|
Western blot analysis
Total proteins were extracted separately from
experimental groups and control using radioimmunoprecipitation
assay lysis buffer (Beyotime Institute of Biotechnology, Jiangsu,
China). A total of 60 µg of protein was separated by 10% SDS-PAGE.
The proteins were subsequently transferred to polyvinylidene
difluoride membranes and subjected to western blotting. Membranes
were blocked in Tris-buffered saline and Tween-20 [150 mM NaCl, 10
mM Tris-HCl (pH 7.5) and 0.1% Tween-20] containing 5% (w/v) milk at
room temperature for 1.5 h, and incubated with the primary and
secondary antibodies. Membranes were incubated with the primary
antibodies overnight at 4°C. the following antibodies were used:
Rabbit anti-phosphorylated extracellular signal-regulated kinase
(p-ERK; 1:1,000 dilution, cat. no. NKC20314), β-catenin (1:1,000
dilution, cat. no. NKC31478), transcription factor SOX-17 (SOX17;
1:1,000 dilution, cat. no. LUY220473), calcium-binding tyrosine
phosphorylation-regulated protein (CABYR; 1:1,000 dilution, cat.
no. ZYS01775), cholesterol side-chain cleavage enzyme mitochondrial
(P450scc; 1:1,000 dilution, cat. no. JX012004) and β-actin (1:2,000
dilution, cat. no. KC015541) (all from Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Membranes were subsequently incubated with a
horseradish peroxidase-labeled goat anti-rabbit secondary antibody
(1:5,000 dilution; cat. no. MB005, Beijing Solarbio Science and
Technology, Co., Ltd., Beijing, China) at 37°C for 2 h. Protein
signals were detected using Electrochemiluminescence Plus Western
Blotting Detection system (GE Healthcare Bio-Sciences, Pittsburgh,
PA, USA). Target bands were analyzed for gray levels using ImageJ
1.37 (National Institutes of Health, Bethesda, MD, USA).
Bioinformatic analysis
The functional annotation of
differentially-expressed genes was performed according to the Gene
Ontology (GO) databases (http://www.geneontology.org). The GO category was
classified using hypergeometric distribution, and the Benjamini and
Hochberg (BH) false discovery rate (FDR) algorithm was used to
adjust the resulting P-values. GO enrichment was considered to be
significant if the FDR values were ≤0.05. Pathway analysis was used
to identify the significant pathways of the
differentially-expressed genes according to the Kyoto Encyclopedia
of Genes and Genomes (KEGG; www.genome.jp/kegg). Similarly, hypergeometric
distribution, followed by BH multiple testing correction, was
performed to select the significant pathways, and the threshold of
significance was defined as P=0.05.
Statistical analysis
All data were analyzed using SPSS 17.0 software
(SPSS, Inc., Chicago, IL, USA). Experiments were performed in
triplicate. Data are expressed as the mean ± standard deviation.
One-way analysis of variance was performed to compare the
differences among the groups. Pairwise comparisons among multiple
sets of data were analyzed using the least significant difference
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
AOB effectively inhibits MEF cell
proliferation
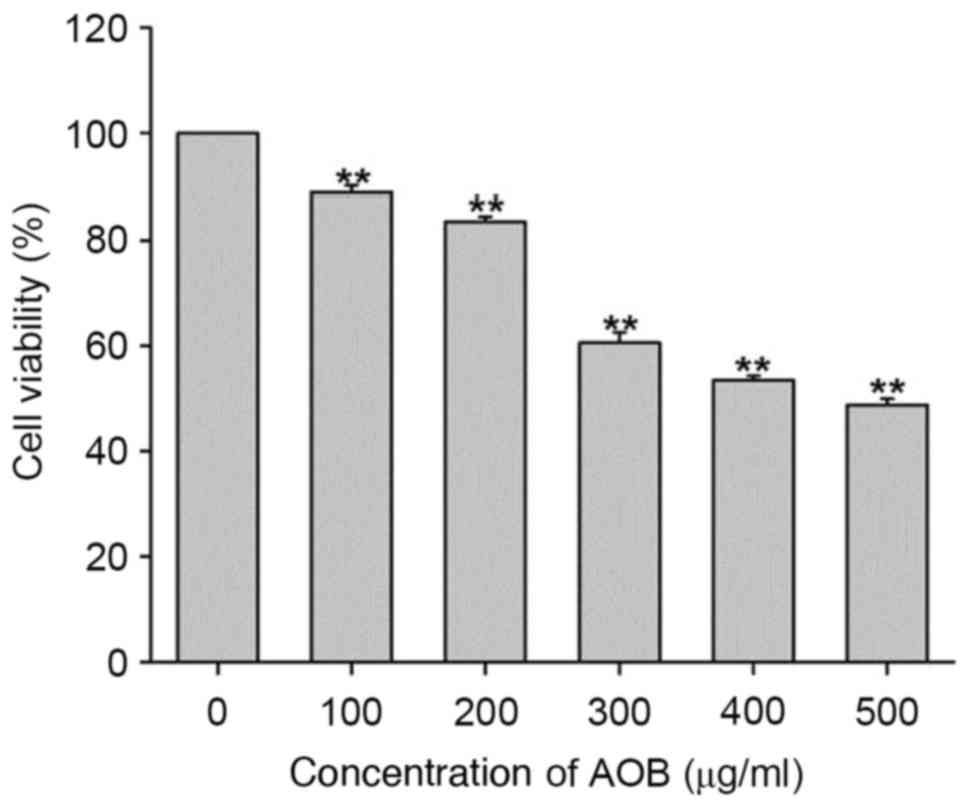
In order to measure the cytotoxic effects of AOB,
MEF cells were treated with increasing concentrations of AOB for 72
h. Subsequently, cell viability was measured using an MTT assay.
The results of the present study indicated that AOB inhibited the
cell growth of MEF cells more effectively at increased
concentrations (Fig. 1). The
IC50 value of AOB for MEF cells was 429.8 µg/ml.
Compared with the control group, ~50% of MEF cells were killed by a
400 µg/ml concentration of AOB. Therefore, the concentration of 400
µg/ml was selected for the follow-up experiments.
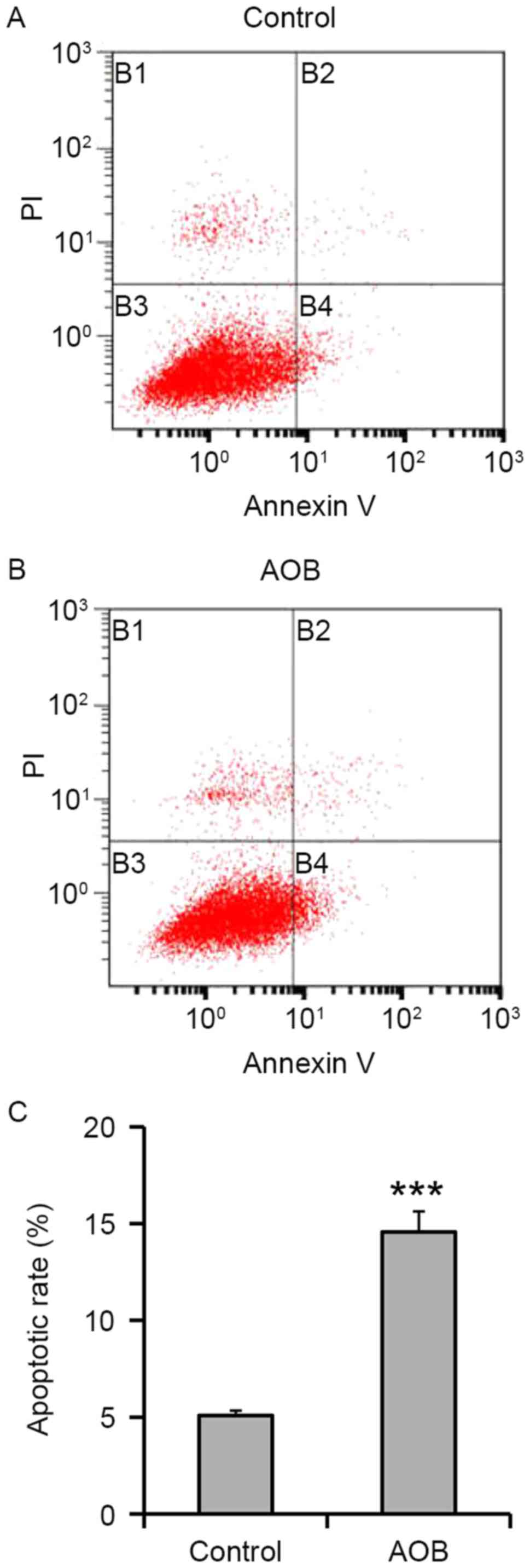
Treatment with AOB results in
increased apoptosis
In order to address the cytotoxic effects of the
selected treatment, 400 µg/ml AOB-treated cells were analyzed for
their apoptosis phenotype. According to the results, treatment with
400 µg/ml AOB in MEF cells for 72 h significantly increased the
apoptotic population to 14.57±1.06% (P<0.001; Fig. 2), whereas the control group
contained an apoptotic population of 5.09±0.26%.
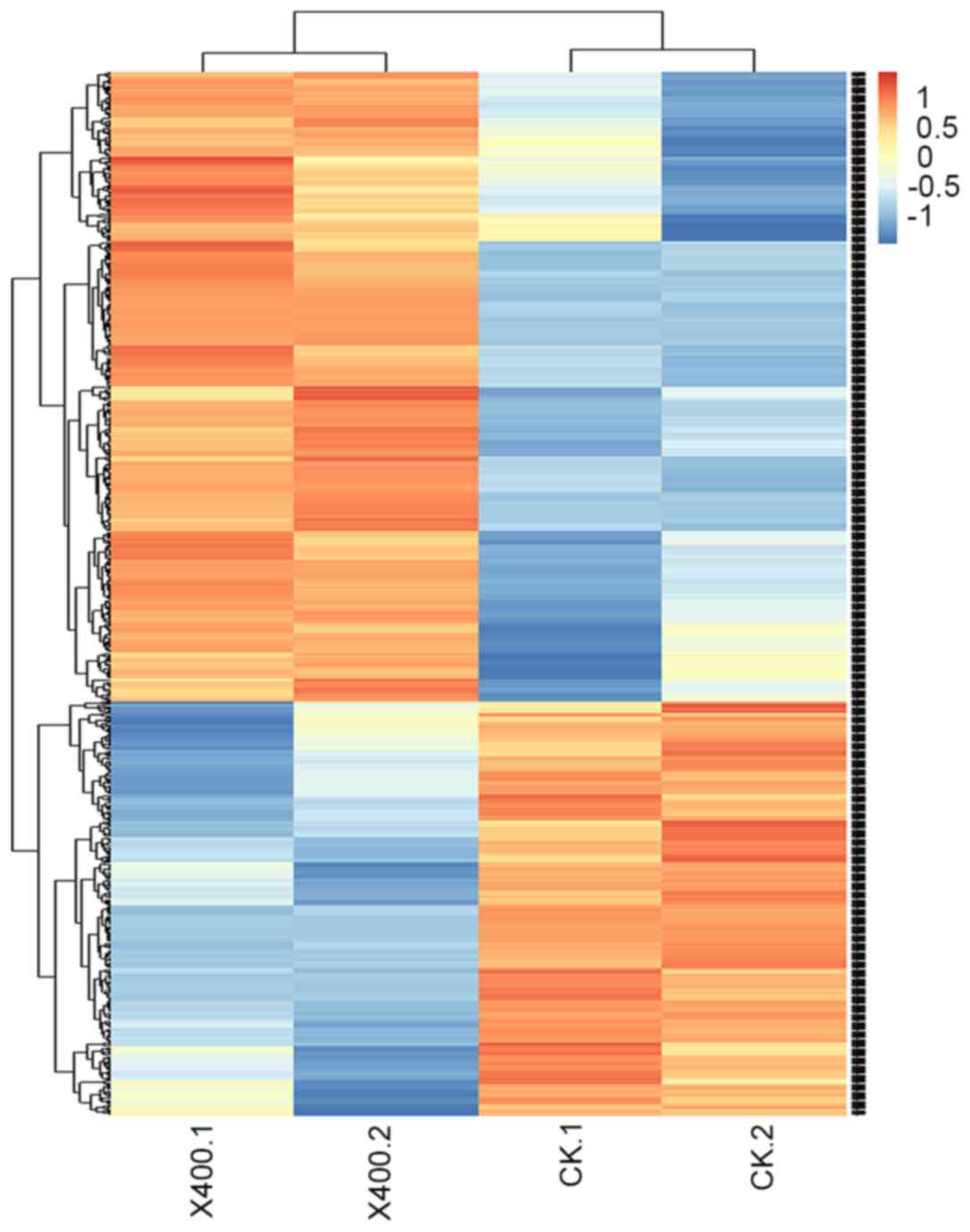
Hierarchical cluster analysis
Hierarchical cluster analysis was performed to allow
for the rapid detection of alterations in the levels of gene
expression in addition to identification of important genes, as
presented in Fig. 3. Pathway
analysis was conducted to identify the significant pathways of the
differentially-expressed gene sets, according to KEGG. Through a
selection process, the differentially-expressed genes were
determined to be primarily involved in metabolic pathways (the
metabolism of exogenous substances, sugars, amino acids,
glutathione and steroid hormones), apoptotic pathways, and those
relevant to reproductive function.
Confirmation of gene expression by
RT-qPCR analysis
In order to verify the data obtained by the
microarray analysis (data not shown), RT-qPCR analysis was
performed on 20 differentially-expressed genes. The results of the
RT-qPCR analysis were consistent with those from the microarray
analysis. As presented in Fig. 4,
treatment with AOB in MEF cells resulted in the downregulation of
dehydrogenase/reductase 9, phospholipase A2 group IVE and platelet
derived growth factor B, while the other 17 genes were
upregulated.
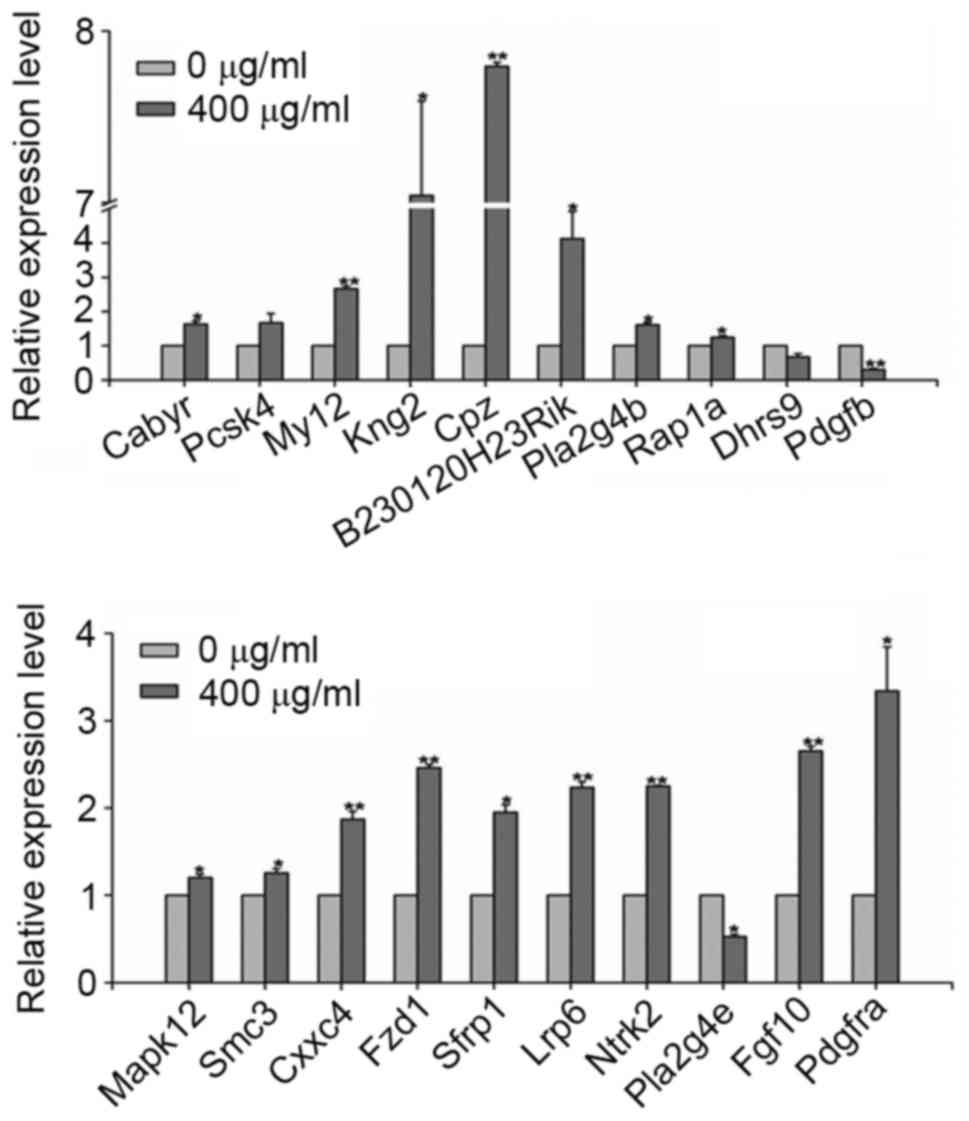 | Figure 4.Reverse transcription-quantitative
polymerase chain reaction analysis of differentially-expressed
genes. *P<0.05, **P<0.01 vs. 0 µg/ml AOB. AOB, antioxidant of
bamboo leaves. Cabyr, calcium-binding tyrosine
phosphorylation-regulated protein; Pcsk4, proprotein convertase
subtilisin/kexin type 4; Myl2, myosin, light polypeptide 2; Kng2,
kininogen v2; Cpz, carboxypeptidase Z; Pla2g4b, phospholipase A2
group IVB; Dhrs9, dehydrogenase/reductase 9; Pdgfb, platelet
derived growth factor B; Mapk12, mitogen activated protein kinase
12; Smc3, structural maintenance of chromosomes 3; Cxxc4, CXXC
finger protein 4; Fzd1, frizzled class receptor 1; Sfrp1, secreted
frizzled related protein 1; Lrp6, LDL receptor related protein 6;
Ntrk2, neutrotrophic tyrosine kinase receptor type 2; Pla2g4e,
phospholipase A2 group IVE; Fgf10, fibroblast growth factor 10;
Pdgfra, platelet derived growth factor receptor α. |
Effects of AOB on the
mitogen-activated protein kinase (MAPK)/ERK and the proto-oncogene
Wnt (Wnt) pathways
As presented in Fig.
5, treatment with AOB notably increased the expression of
p-ERK, and the ERK pathway inhibitor U0126 suppressed the
expression of p-ERK in AOB-treated MEF cells. Additionally, the
expression of β-catenin, SOX17, CABYR and P450scc was increased
following treatment with AOB, although it was decreased by
treatment with AOB+U0126 and AOB+Dickkopf-related protein 1 (DKK1;
a Wnt pathway inhibitor).
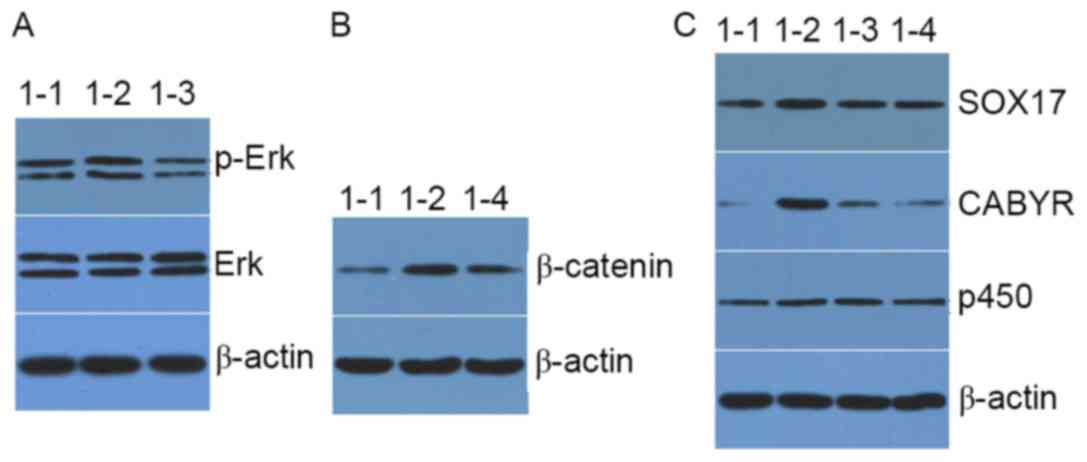 | Figure 5.Western blot analysis of the
expression level of (A) p-ERK and ERK, (B) β-catenin, and (C) P450,
CABYR and SOX17 in mouse embryonic fibroblast cells. 1–1, blank
control group; 1–2, AOB group; 1–3, AOB+U0216 group; 1–4, AOB+DKK1
group; AOB, antioxidant of bamboo leaves; p-, phosphorylated; ERK,
extracellular signal-regulated kinase; P450, cholesterol side-chain
cleavage enzyme mitochondrial; CABYR, calcium-binding tyrosine
phosphorylation-regulated protein; SOX17, transcription factor
SOX-17. |
Discussion
MAPK signaling pathways are involved in mediating
cell growth, survival and death processes. Three of the MAPK
members are c-Jun N-terminal kinase, p38 and ERK (10–12).
ERK is a widely-expressed protein kinase intracellular signaling
molecule which is involved in the regulation of meiosis, mitosis,
and postmitotic functions in differentiated cells (13). A number of different stimuli,
including growth factors, cytokines, viral infection, ligands for
heterotrimeric G protein-coupled receptors, transforming agents and
carcinogens, may activate the ERK pathway (14). ERK serves a prominent role in the
bone morphogenetic protein (BMP) and ERK/MAPK signaling pathways
(15). The phosphorylation of ERK
activates the BMP pathway by interacting with mothers against
decapentaplegic homolog 1, 5 and 8. In addition, p-ERK exerts a
function in the process of apoptosis by interacting with son of
sevenless homolog and ribosomal protein S6 kinase α-1 (16).
In the results of western blot analysis performed in
the present study, the expression of p-ERK significantly increased
in AOB-treated MEF cells compared with control MEF cells. No
notable changes in ERK expression occurred between the experimental
and control groups. The results indicated that AOB may promote the
phosphorylation of ERK and activate the ERK signaling pathway.
Additionally, adding ERK inhibitor U0126 to AOB-treated cells
downregulated the expression of p-ERK. The results of the present
study demonstrated that AOB may enhance the activation of the ERK
pathway, thereby influencing cellular processes.
The Wnt/β-catenin pathway facilitates an
accumulation of β-catenin in the cytoplasm and its eventual
translocation into the nucleus to act as a transcriptional
coactivator of transcription factors (17). The results of the present study
demonstrated that the expression of β-catenin increased markedly in
AOB-treated MEF cells. Dkk1 is an antagonistic inhibitor of the Wnt
signaling pathway that acts by isolating the low-density
lipoprotein receptor-related protein 6 co-receptor so that it is
unable to activate the Wnt signaling pathway (18). DKK1 has been demonstrated to
antagonize the Wnt/β-catenin pathway via a reduction in β-catenin
expression (18). It was
previously observed that treatment with DDK1 led to a decrease in
β-catenin expression in AOB-treated MEF cells (19). These previous results suggested
that AOB may activate the Wnt signaling pathway by enhancing
β-catenin expression, thus influencing cellular processes. CABYR
and SOX17 proteins are associated with the morphological and
molecular maturational processes of spermatozoa (20). P450scc, encoded by the CYP11A1
gene, is able to convert cholesterol to pregnenolone to initiate
steroidogenesis (7,21). In the results of the presents
study, the expression of CABYR, SOX17 and P450scc was upregulated
in AOB-treated MEF cells, demonstrating that AOB may affect the
regulation of reproductive function. In addition, U0126 and DDK1
inhibited the expression of CABYR, SOX17 and P450scc in AOB-treated
MEF cells. The results of the present study demonstrated that these
proteins were involved in the ERK and Wnt signaling pathways, and
that AOB markedly upregulated the expression of CABYR, SOX17 and
P450scc.
In conclusion, the present study established an MEF
cell model to investigate the effects of AOB on the expression of
genes associated with murine reproduction and embryonic
development. The results of the present study provided evidence
that AOB may impact the expression of proteins associated with
reproduction. Additionally, the present findings suggested that AOB
may enhance the activation of the ERK and Wnt signaling pathways,
thereby influencing cellular processes. Elucidating the AOB role on
signaling pathways in the MEF cell model may provide useful
information for clinical reproduction and embryonic
development.
Acknowledgements
The Research Fund for Giant Panda Breeding of
Chengdu (grant no. CPF2012-15) and the China Agriculture Research
System (grant no. CARS-05).
References
|
1
|
Lu B, Wu X, Shi J, Dong Y and Zhang Y:
Toxicology and safety of antioxidant of bamboo leaves. Part 2:
Developmental toxicity test in rats with antioxidant of bamboo
leaves. Food Chem Toxicol. 44:1739–1743. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma X, Wang E, Lu Y, Wang Y, Ou S and Yan
R: Acylation of antioxidant of bamboo leaves with fatty acids by
lipase and the acylated derivatives' efficiency in the inhibition
of acrylamide formation in fried potato crisps. PLoS One.
10:e01306802015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi XC, Liu XQ, Xie XL, Xu YC and Zhao ZX:
Gene chip array for differentiation of mycobacterial species and
detection of drug resistance. Chin Med J (Engl). 125:3292–3297.
2012.PubMed/NCBI
|
|
4
|
Lu L, Gao Y, Xu M, Ge RC and Lu L: Gene
expression profiles associated with osteoblasts differentiated from
bone marrow stromal cells. Asian Pac J Trop Med. 7:344–351. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rieske P, Krynska B and Azizi SA: Human
fibroblast-derived cell lines have characteristics of embryonic
stem cells and cells of neuro-ectodermal origin. Differentiation.
73:474–483. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ngo C, Nicco C, Leconte M, Chéreau C,
Arkwright S, Vacher-Lavenu MC, Weill B, Chapron C and Batteux F:
Protein kinase inhibitors can control the progression of
endometriosis in vitro and in vivo. J Pathol. 222:148–157. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Slominski A, Zbytek B, Nikolakis G, Manna
PR, Skobowiat C, Zmijewski M, Li W, Janjetovic Z, Postlethwaite A,
Zouboulis CC and Tuckey RC: Steroidogenesis in the skin:
Implications for local immune functions. J Steroid Biochem Mol
Biol. 137:107–123. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang N, Zhang W, Cui J, Zhang H, Chen X,
Li R, Wu N, Chen X, Wen S, Zhang J, et al: The piggyBac
transposon-mediated expression of SV40 T antigen efficiently
immortalizes mouse embryonic fibroblasts (MEFs). PLoS One.
9:e973162014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bandyopadhyay M, Bishop CP and Bidwai AP:
The Conserved MAPK Site in E(spl)-M8, an effector of Drosophila
notch signaling, controls repressor activity during eye
development. PLoS One. 11:e01595082016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li W, Ma Z, Ma J, Li X, Xu Q, Duan W, Chen
X, Lv Y, Zhou S, Wu E, et al: Hydrogen peroxide mediates
hyperglycemia-induced invasive activity via ERK and p38 MAPK in
human pancreatic cancer. Oncotarget. 6:31119–31133. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aye IL, Jansson T and Powell TL: TNF-α
stimulates System A amino acid transport in primary human
trophoblast cells mediated by p38 MAPK signaling. Physiol Rep.
3(pii): e125942015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Williams KA, Zhang M, Xiang S, Hu C, Wu
JY, Zhang S, Ryan M, Cox AD, Der CJ, Fang B, et al: Extracellular
signal-regulated kinase (ERK) phosphorylates histone deacetylase 6
(HDAC6) at serine 1035 to stimulate cell migration. J Biol Chem.
288:33156–33170. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ghanian MH, Farzaneh Z, Barzin J, Zandi M,
Kazemi-Ashtiani M, Alikhani M, Ehsani M and Baharvand H:
Nanotopographical control of human embryonic stem cell
differentiation into definitive endoderm. J Biomed Mater Res A.
103:3539–3553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim HK, Kim MG and Leem KH: Osteogenic
activity of collagen peptide via ERK/MAPK pathway mediated boosting
of collagen synthesis and its therapeutic efficacy in osteoporotic
bone by back-scattered electron imaging and microarchitecture
analysis. Molecules. 18:15474–15489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang YH, Cheng F, Du XT, Gao JL, Xiao XL,
Li N, Li SL and de L Dong: GDF11/BMP11 activates both smad1/5/8 and
smad2/3 signals but shows no significant effect on proliferation
and migration of human umbilical vein endothelial cells.
Oncotarget. 7:12063–12074. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bikorimana E, Lapid D, Choi H and Dahl R:
Retroviral infection of murine embryonic stem cell derived embryoid
body cells for analysis of hematopoietic differentiation. J Vis
Exp. 20:e520222014.
|
|
18
|
Liu L, Miao XP and Lin DX: The challenge
and opportunity in the post genome-wide association study era.
Zhonghua Yu Fang Yi Xue Za Zhi. 46:198–201. 2012.(In Chinese).
PubMed/NCBI
|
|
19
|
Atlasi Y, Mowla SJ, Ziaee SA and Bahrami
AR: OCT-4, an embryonic stem cell marker, is highly expressed in
bladder cancer. Int J Cancer. 120:1598–1602. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Naaby-Hansen S, Mandal A, Wolkowicz MJ,
Sen B, Westbrook VA, Shetty J, Coonrod SA, Klotz KL, Kim YH, Bush
LA, et al: CABYR, a novel calcium-binding tyrosine
phosphorylation-regulated fibrous sheath protein involved in
capacitation. Dev Biol. 242:236–254. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tee MK, Abramsohn M, Loewenthal N, Harris
M, Siwach S, Kaplinsky A, Markus B, Birk O, Sheffield VC, Parvari
R, et al: Varied clinical presentations of seven patients with
mutations in CYP11A1 encoding the cholesterol side-chain cleavage
enzyme, P450scc. J Clin Endocrinol Metab. 98:713–720. 2013.
View Article : Google Scholar : PubMed/NCBI
|