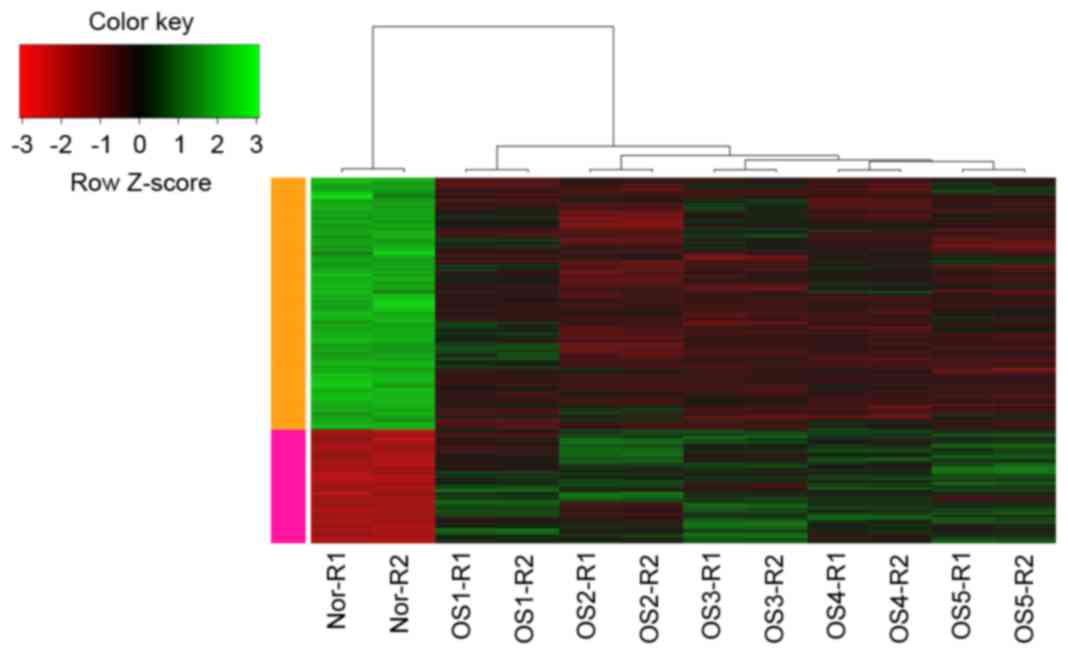
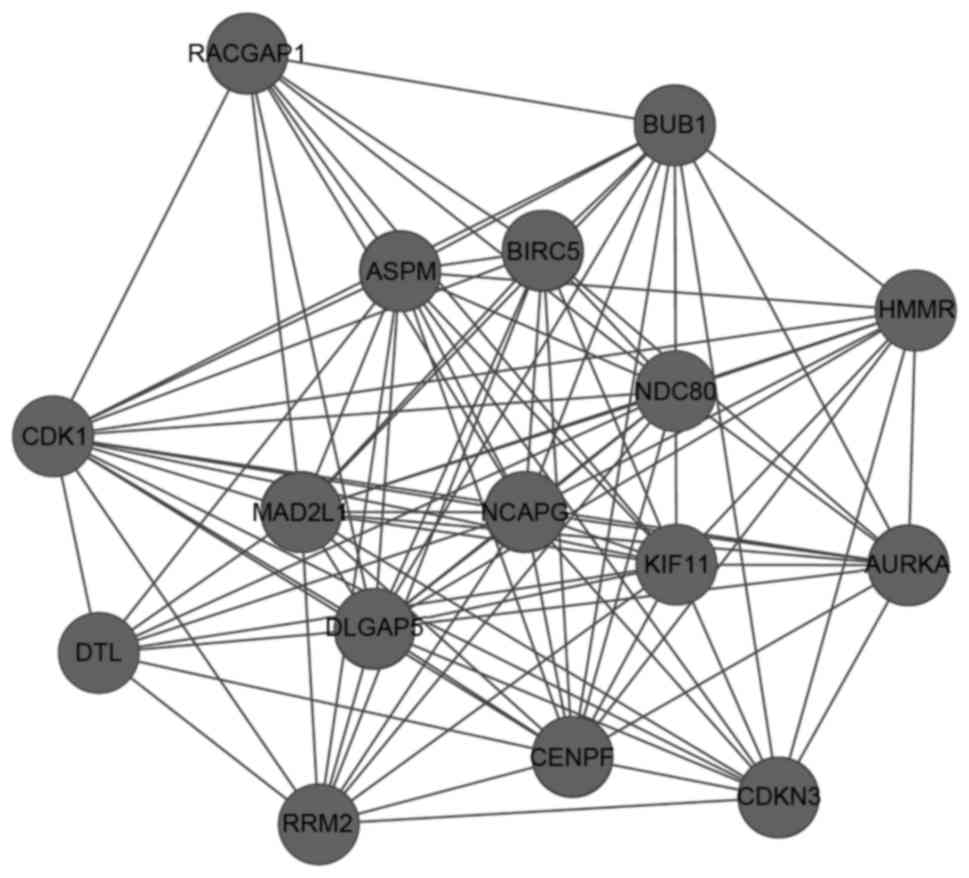
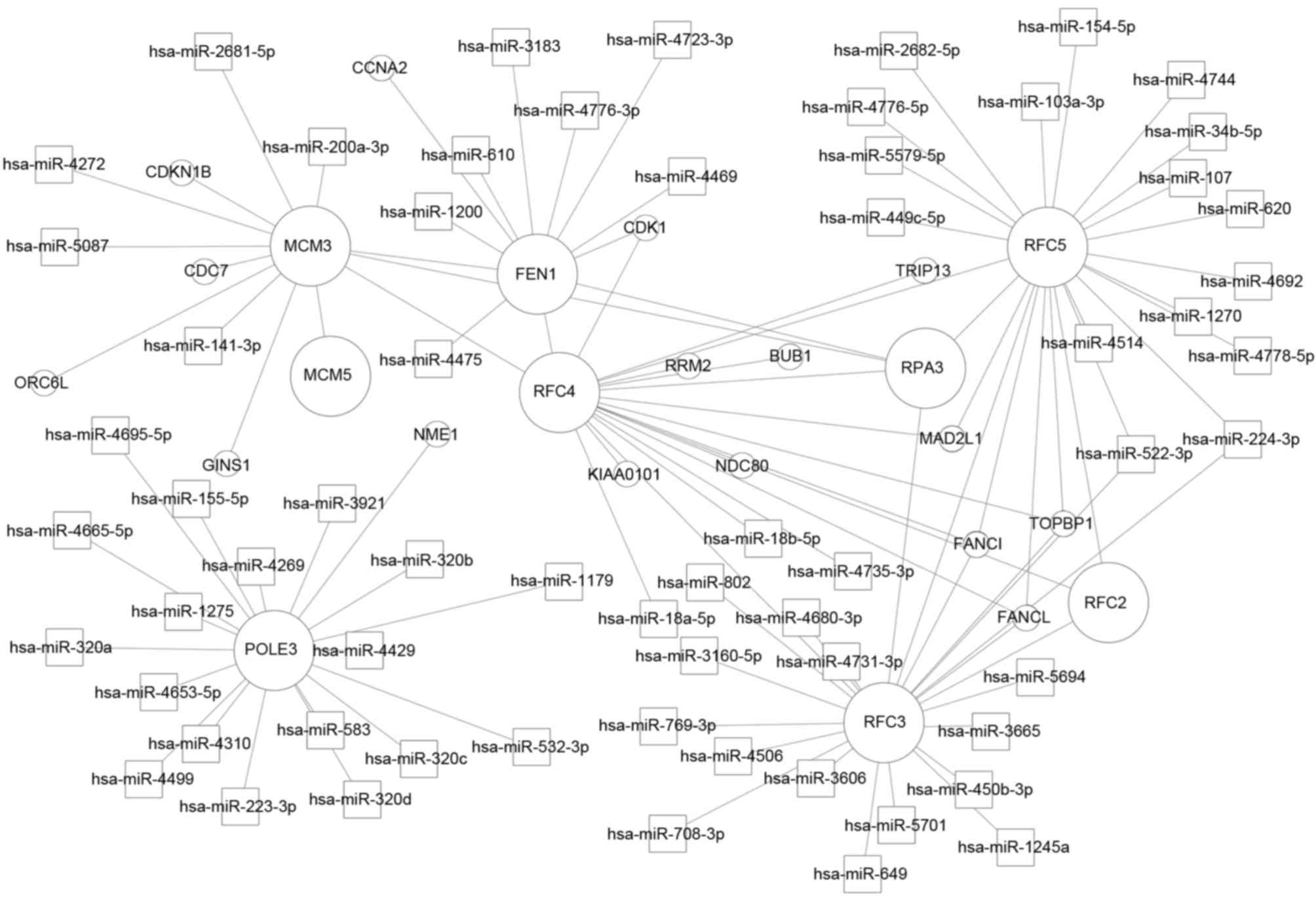
|
1
|
Messerschmitt PJ, Garcia RM, Abdul-Karim
FW, Greenfield EM and Getty PJ: Osteosarcoma. J Am Acad Orthop
Surg. 17:515–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mohseny AB, Tieken C, Van Der Velden PA,
Szuhai K, de Andrea C, Hogendoorn PC and Cleton-Jansen AM: Small
deletions but not methylation underlie CDKN2A/p16 loss of
expression in conventional osteosarcoma. Genes Chromosomes Cancer.
49:1095–1103. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bae Y, Yang T, Zeng HC, Campeau PM, Chen
Y, Bertin T, Dawson BC, Munivez E, Tao J and Lee BH: miRNA-34c
regulates Notch signaling during bone development. Hum Mol Genet.
21:2991–3000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Silva A, Yunes JA, Cardoso BA, Martins LR,
Jotta PY, Abecasis M, Nowill AE, Leslie NR, Cardoso AA and Barata
JT: PTEN posttranslational inactivation and hyperactivation of the
PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin
Invest. 118:3762–3774. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Freeman SS, Allen SW, Ganti R, Wu J, Ma J,
Su X, Neale G, Dome JS, Daw NC and Khoury JD: Copy number gains in
EGFR and copy number losses in PTEN are common events in
osteosarcoma tumors. Cancer. 113:1453–1461. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao G, Cai C, Yang T, Qiu X, Liao B, Li
W, Ji Z, Zhao J, Zhao H, Guo M, et al: MicroRNA-221 induces cell
survival and cisplatin resistance through PI3K/Akt pathway in human
osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Namløs HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fritsche-Guenther R, Noske A, Ungethüm U,
Kuban RJ, Schlag PM, Tunn PU, Karle J, Krenn V, Dietel M and Sers
C: De novo expression of EphA2 in osteosarcoma modulates activation
of the mitogenic signalling pathway. Histopathology. 57:836–850.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fritsche-Guenther R, Gruetzkau A, Noske A,
Melcher I, Schaser KD, Schlag PM, Kasper HU, Krenn V and Sers C:
Therapeutic potential of CAMPATH-1H in skeletal tumours.
Histopathology. 57:851–861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Luo Y, Deng Z and Chen J: Pivotal
regulatory network and genes in osteosarcoma. Arch Med Sci.
9:569–575. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Z, Chen Y, Fu Y, Yang Y, Zhang Y,
Chen Y and Li D: Meta-analysis of differentially expressed genes in
osteosarcoma based on gene expression data. BMC Med Genet.
15:802014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Z, Irizarry RA, Gentleman R,
Martinez-Murillo F and Spencer F: A model-based background
adjustment for oligonucleotide expression arrays. Journal of the
American Statistical Association. 99:909–917. 2004. View Article : Google Scholar
|
|
13
|
Seo J and Hoffman EP: Probe set
algorithms: Is there a rational best bet? BMC bioinformatics.
7:3952006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article32004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. Journal of the Royal Statistical Society Series B
(Methodological). 57:289–300. 1995.
|
|
16
|
Olson CF: Parallel algorithms for
hierarchical clustering. Parallel Computing. 21:1313–1325. 1995.
View Article : Google Scholar
|
|
17
|
Kolde R: pheatmap: Pretty Heatmaps. R
package. version 0.7. 7. 2012.
|
|
18
|
Huang DW, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X: miRDB: A microRNA target
prediction and functional annotation database with a wiki
interface. RNA. 14:1012–1017. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Reynolds N, Fantes PA and MacNeill SA: A
key role for replication factor C in DNA replication checkpoint
function in fission yeast. Nucleic Acids Res. 27:462–469. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Redondo-Muñoz J, Rodríguez MJ, Silió V,
Pérez-García V, Valpuesta JM and Carrera AC: Phosphoinositide
3-kinase beta controls replication factor C assembly and function.
Nucleic Acids Res. 41:855–868. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Loeb LA, Springgate CF and Battula N:
Errors in DNA replication as a basis of malignant changes. Cancer
Res. 34:2311–2321. 1974.PubMed/NCBI
|
|
26
|
Tsaniras S Champeris, Kanellakis N,
Symeonidou IE, Nikolopoulou P, Lygerou Z and Taraviras S: Licensing
of DNA replication, cancer, pluripotency and differentiation: An
interlinked world? Semin Cell Dev Biol. 30:174–180. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
PosthumaDeBoer J, van Royen B and Helder
M: Mechanisms of therapy resistance in osteosarcoma: A review.
Oncol Discov. 1:82013. View Article : Google Scholar
|
|
28
|
Kim MJ, Lee JY and Lee SJ: Transient
suppression of nuclear Cdc2 activity in response to ionizing
radiation. Oncol Rep. 19:1323–1329. 2008.PubMed/NCBI
|
|
29
|
Fu W, Ma L, Chu B, Wang X, Bui MM, Gemmer
J, Altiok S and Pledger WJ: The cyclin-dependent kinase inhibitor
SCH 727965 (dinacliclib) induces the apoptosis of osteosarcoma
cells. Mol Cancer Ther. 10:1018–1027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Doak SH, Jenkins GJ, Parry EM, Griffiths
AP, Baxter JN and Parry JM: Differential expression of the MAD2,
BUB1 and HSP27 genes in Barrett's oesophagus-their association with
aneuploidy and neoplastic progression. Mutat Res. 547:133–144.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Giantin M, Granato A, Baratto C, Marconato
L, Vascellari M, Morello EM, Vercelli A, Mutinelli F and Dacasto M:
Global gene expression analysis of canine cutaneous mast cell
tumor: Could molecular profiling be useful for subtype
classification and prognostication? PLoS One. 9:e954812014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu L, Guo WC, Zhao SH, Tang J and Chen JL:
Mitotic arrest defective protein 2 expression abnormality and its
clinicopathologic significance in human osteosarcoma. Apmis.
118:222–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Trougakos IP, Chondrogianni N, Amarantos
I, Blake J, Schwager C, Wirkner U, Ansorge W and Gonos ES:
Genome-wide transcriptome profile of the human osteosarcoma Sa OS
and U-2 OS cell lines. Cancer Genet Cytogenet. 196:109–118. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuijjer ML, Peterse EF, van den Akker BE,
Briaire-de Bruijn IH, Serra M, Meza-Zepeda LA, Myklebost O, Hassan
AB, Hogendoorn PC and Cleton-Jansen AM: IR/IGF1R signaling as
potential target for treatment of high-grade osteosarcoma. BMC
Cancer. 13:2452013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cao ZQ, Shen Z and Huang WY: MicroRNA-802
promotes osteosarcoma cell proliferation by targeting p27. Asian
Pac J Cancer Prev. 14:7081–7084. 2013. View Article : Google Scholar : PubMed/NCBI
|