Introduction
Colorectal cancer has become one of the malignant
tumor diseases with the highest incidence rates worldwide. The
liver is the main target organ for the hematogenous metastasis of
colorectal cancer, and liver metastasis is the leading cause of
colorectal cancer-associated mortality in patients (1). Clinical trials have shown that liver
metastasis is an independent predictor of poor prognosis and
increased risk of mortality in patients with colorectal cancer
(2). There remains no effective
therapy for liver metastasis despite promising developments
achieved in the treatment of colorectal cancer. Early diagnosis and
positive therapy may improve the survival rates of patients with
colorectal cancer at an early stage of liver metastasis. Therefore,
the present study aimed to examine the molecular mechanisms of
liver metastasis in colorectal cancer, and screen for biomarkers
associated with liver metastasis. These investigations are likely
to assist in improving the accuracy of early diagnosis, efficacy of
novel therapy and prognosis of patients with colorectal cancer
(3).
MicroRNAs are a type of non-coding RNA with a length
of 21–23 bp (4). Previous studies
have shown that microRNAs affect multiple cellular events via mRNA
interference (5). The microRNA
expression profile of colorectal cancer shows differential
expression of microRNAs in colorectal cancer, compared with normal
tissues, suggesting that the aberrant expression of microRNA is
associated with the metabolism and cell behavior of cancer
(6). Several microRNAs have been
confirmed to be associated with the clinicopathological features of
colorectal cancer, including microRNA (miR)-31, miR-143 and miR-145
(7,8). Zhang et al (9) found that miR-200 was important in the
regulatory mechanisms of liver cancer, and a previous study showed
that serum levels of miR-200 and miR-141 were increased in patients
with colorectal cancer (10).
These findings suggest that miR-200 and miR-141 may be involved in
the pathogenesis of colorectal cancer. However, whether the
aberrant expression of miR-200 and miR-141 is associated with liver
metastasis remains to be elucidated. The present study aimed to
examine the association between liver metastasis and levels of
serum miR-200 and miR-141 in patients with colorectal cancer.
Patients and methods
Study subjects
A total of 380 patients with colorectal cancer were
randomly enrolled into the experimental group at China-Japan Union
Hospital of Jilin University (Changchun, China) between June 2008
and June 2014 (male/female ratio, 218/162; age range, 42–73;
53.6±6.2 years). The patients with colorectal cancer were diagnosed
by an alimentary tract barium meal, B-ultrasonic examination and
histopathological analyses. Disease grading was performed for all
enrolled patients according to the criteria of the World Health
Organization grading standard for colorectal cancer (11). The grading comprised 142 grade I/II
cases and 238 III/IV cases. None of the patients had received
radiotherapy, chemotherapy or surgery prior to hospitalization. A
control group included healthy adults, who were age- and
sex-matched to patients in the experimental group. This clinical
trial was performed following approval and under supervision of the
Ethics Committee of the China-Japan Union Hospital of Jilin
University (Changchun, China). All participants signed informed
consent prior to enrollment in the study.
Reagents and instruments
RNA extraction kits (cat. no 74104) were purchased
from Qiagen, Inc. (Valencia, CA, USA). Routine kits for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis (cat. no. KR103-03) were purchased from TianGen Biotech
Co., Ltd. (Beijing, China). MicroRNA qRT-PCR kits (cat. no. AM1558)
were purchased from Ambion; Thermo Fisher Scientific, Inc.
(Waltham, MA, USA). Lipidosome INTERFERin™ transfection kits (cat.
no. 410-015) were purchased from Polyplus Transfection (Illkirch,
France). DMEM (cat. no. SH30081.01) was purchased from HyClone; GE
Healthcare Life Sciences (Logan, UT, USA). TUNEL apoptosis
detection kits were purchased from Roche Diagnostics (Basel,
Switzerland). Transwell kits were purchased from BD Biosciences
(Franklin Lakes, NJ, USA). The Mx3000P fluorescence ration PCR
instrument was purchased from Agilent Technologies, Inc. (Santa
Clara, CA, USA). The cell incubator was purchased from Thermo
Fisher Scientific, Inc.
Extraction of microRNA and RT-qPCR
analysis
Fasting blood samples (10 ml) were collected into
vacuum tubes with EDTA as an anticoagulant. Plasma was obtained via
two-step centrifugation (2,000 g for 10 min; 8,000 g for 10 min at
25°C). The supernatant was recycled for the extraction of total RNA
and microRNA, respectively. Primers were synthesized for miR-200
and miR-141 (Table I). RT-qPCR
analysis was performed in a total volume of 20 µl, including 10 µl
SYBR-Green qPCR Super mix, 0.5 µl forward primer (10 µM), 0.5 µl
reverse primer (10 µM), 5 µl cDNA and 4 µl sterile water using
routine protocols. The reaction conditions were as follows: 95°C
for 3 min, 95°C for 15 sec and 60°C for 30 sec (40 cycles). Applied
Biosystems 7500 software V2.02 (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used with the U6-RNA sequence as an internal
control. The 2−ΔΔCq method was used to analyze data
(12).
 | Table I.Primer sequences of miR-200 and
miR-141. |
Table I.
Primer sequences of miR-200 and
miR-141.
| Primer | Sequence |
|---|
| miR-200-F |
5′-GGTTCTTCCCTGGGCTTC-3′ |
| miR-200-R |
5′-GAGTAGGAGCTCCGGATGTG-3′ |
| miR-141-F |
5′-CAAACAAAGCCTGGGAGAG-3′ |
| miR-141-R |
5′-TTAAGGCCCCAGATTCCAC-3′ |
Transfection and overexpression of
microRNA
The miR-200 mimic and miR-141 mimic were synthesized
(Shanghai GenePharma Co., Ltd., Shanghai, China) for transfection
and overexpression. The medium comprised DMEM with 10% fetal bovine
serum, 5,000 units penicillin and 5,000 units streptomycin. The
HCT116 and SW480 colon carcinoma cells (American Type Culture
Collection, Manassas, VA, USA) were incubated separately in 96-well
plates with cell culture conditions of 37°C and 5% CO2.
Following culture for 24 h, lipidosome transfection was performed
to establish HCT116 and SW480 cells overexpressing miR-200 or
miR-141.
In situ detection of apoptosis using
TUNEL staining
The transfected HCT116 and SW480 cells were
transferred into 24-well plates following culture for 24 h.
Untransfected HCT116 cells were used as a control group, and six
parallel experiments were performed for each group. Staining for
was performed for the detection of apoptosis at different time
points (1, 2, 3 and 4 days) according to the manufacturer's
protocol of the apoptosis detection kit (Roche Diagnostics).
Subsequently, five clear visual fields were randomly selected under
a light microscope to analyze the total cell count and apoptotic
cell count. The apoptotic index was calculated as the ratio of
apoptotic cells to total cells (13).
Examination of cell migration using a
Transwell assay
Matrigel (60 µl; 5 mg/ml) was added into the upper
Transwell chamber and air-dried at 4°C. The HCT116 and SW480 cells
were cultured, respectively, until the logarithmic growth phase.
The cells were digested and resuspended with a diluted cell density
of 1×106, and 200 µl of the cell suspension was added
into the upper Transwell chamber. In the lower Transwell chamber,
600 µl fresh nutrient medium was added. After 24 h, the Matrigel
and HCT116 cells or SW480 cells were cleaned. Crystal violet
staining (30 min) was performed for the cells in the lower
Transwell chamber. The optical density at 570 nm was assessed via a
microplate assay following rinsing in 10% acetic acid (14). The above process was repeated three
times to produce steady results.
Statistical analysis
SPSS 21.0 software (IBM SPSS, Armonk, NY, USA) was
used for data processing. Measurement data are presented as the
mean ± standard deviation. Student's t-test was performed for
comparison of difference between two groups. Statistical
significance was assessed using one-way analysis of variance among
multiple treatment groups. A χ2 test was performed for
statistical analysis of enumeration data. P<0.05 was considered
to indicate a statistically significant difference.
Results
miR-200 and miR-141 are increased in
colorectal cancer
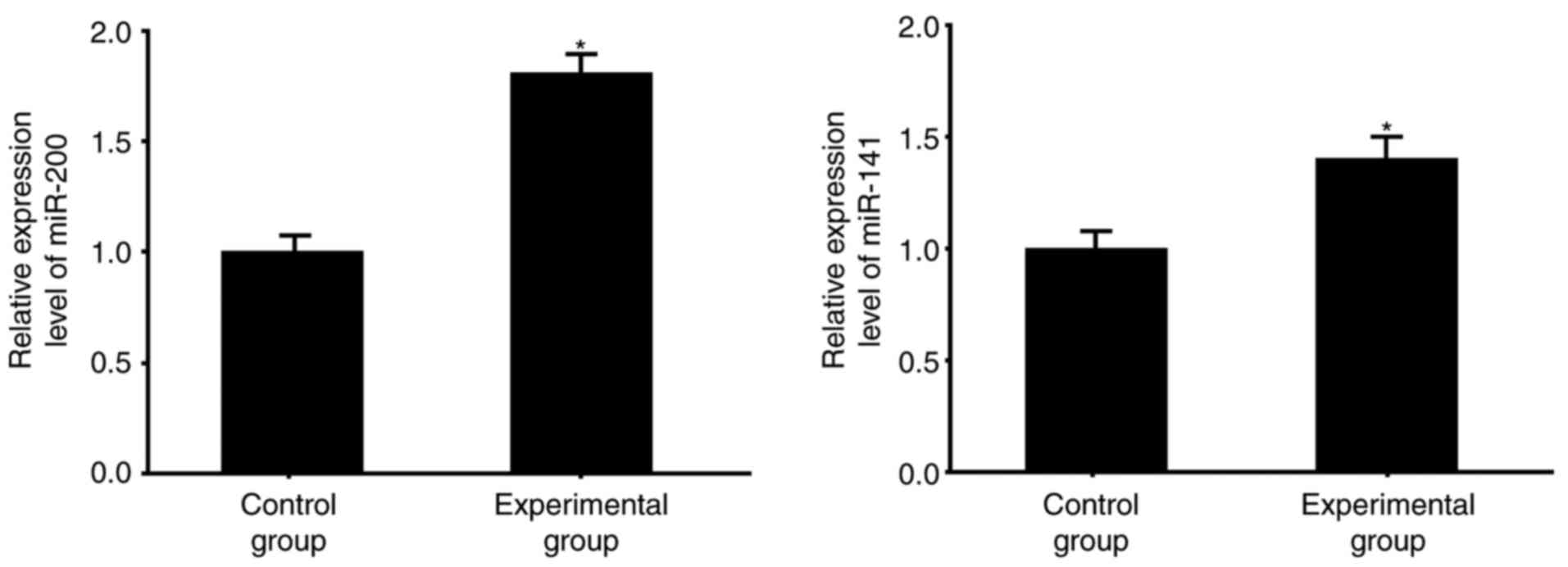
Compared with the normal tissues from the control
group, the colorectal cancer tissues had significantly higher mRNA
levels of miR-200 and miR-141 (P<0.05; Fig. 1), which were measured using RT-qPCR
analysis.
Increased expression of miR-200 and
miR-141 are associated with liver metastasis
The follow-up data showed that 110 patients (28.9%)
suffered from liver metastasis in the experimental group, whereas
32 patients (8.4%) suffered from metastasis in other organs
(Table II). Serum levels of
miR-200 and miR-141 were positively correlated with clinical
grading. In the patients with liver metastasis, 78.2% had increased
expression of miR-200, which was only observed in 20.4% of the
patients without liver metastasis. The same was observed for the
expression of miR-141, in that patients with liver metastasis had a
higher level, compared with those without liver metastasis (81.8,
vs. 22.0%, respectively).
 | Table II.Analysis of the association between
clinicopathological parameters and expression levels of miR-200 and
miR-141. |
Table II.
Analysis of the association between
clinicopathological parameters and expression levels of miR-200 and
miR-141.
|
| Expression of
miR-200 | Expression of
miR-141 |
|---|
|
|
|
|
|---|
| Parameter | High (n) | Low (n) | P-value | High (n) | Low (n) | P-value |
|---|
| Age (years) |
|
| >0.05 |
|
| >0.05 |
| ≥60 | 118 | 103 |
| 121 | 100 |
|
|
<60 | 87 | 72 |
| 78 | 81 |
|
| Sex |
|
| >0.05 |
|
| >0.05 |
| Male | 122 | 96 |
| 115 | 103 |
|
|
Female | 91 | 71 |
| 89 | 73 |
|
| Clinical grade |
|
| <0.05 |
|
| <0.05 |
| I+II | 46 | 96 |
| 37 | 105 |
|
|
III+IV | 186 | 52 |
| 177 | 61 |
|
| Liver metastasis |
|
| <0.05 |
|
| <0.05 |
| Yes | 86 | 24 |
| 90 | 20 |
|
| No | 55 | 215 |
| 62 | 208 |
|
| Metastasis in other
organs |
|
| <0.05 |
|
| <0.05 |
| Yes | 23 | 9 |
| 21 | 11 |
|
| No | 66 | 282 |
| 60 | 288 |
|
Transfection with miR-200 or
miR-141
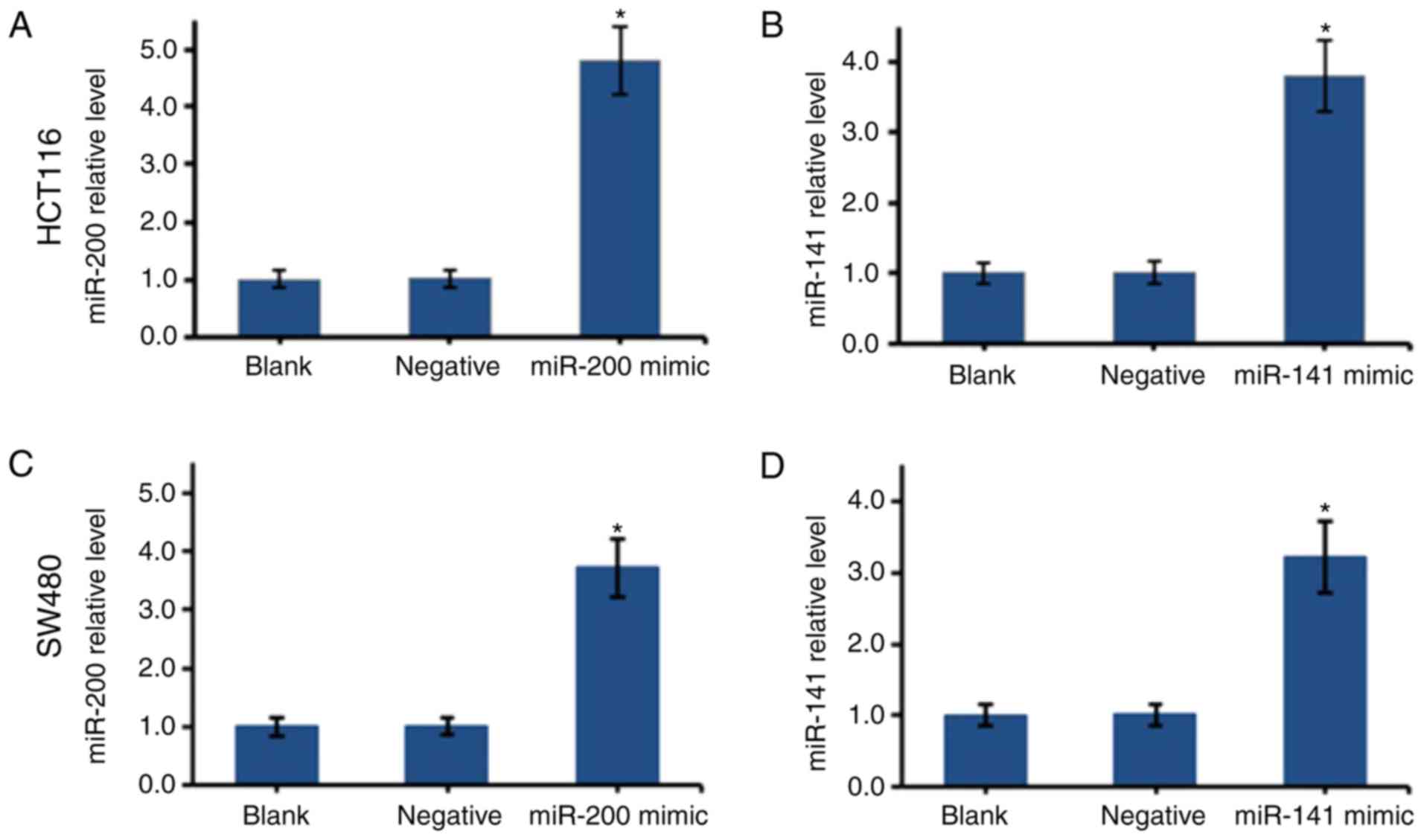
The overexpression of miR-200 or miR-141 in the
HCT116 and SW480 cells was established using mimics. The
transfected cells were cultured to the log phase, and total RNA was
extracted for RT-qPCR analysis (Fig.
2). Compared with the negative groups, the transfected HCT116
and SW480 cells had significantly higher expression levels of
miR-200 and miR-141 (P<0.05), suggesting transfection was
successful.
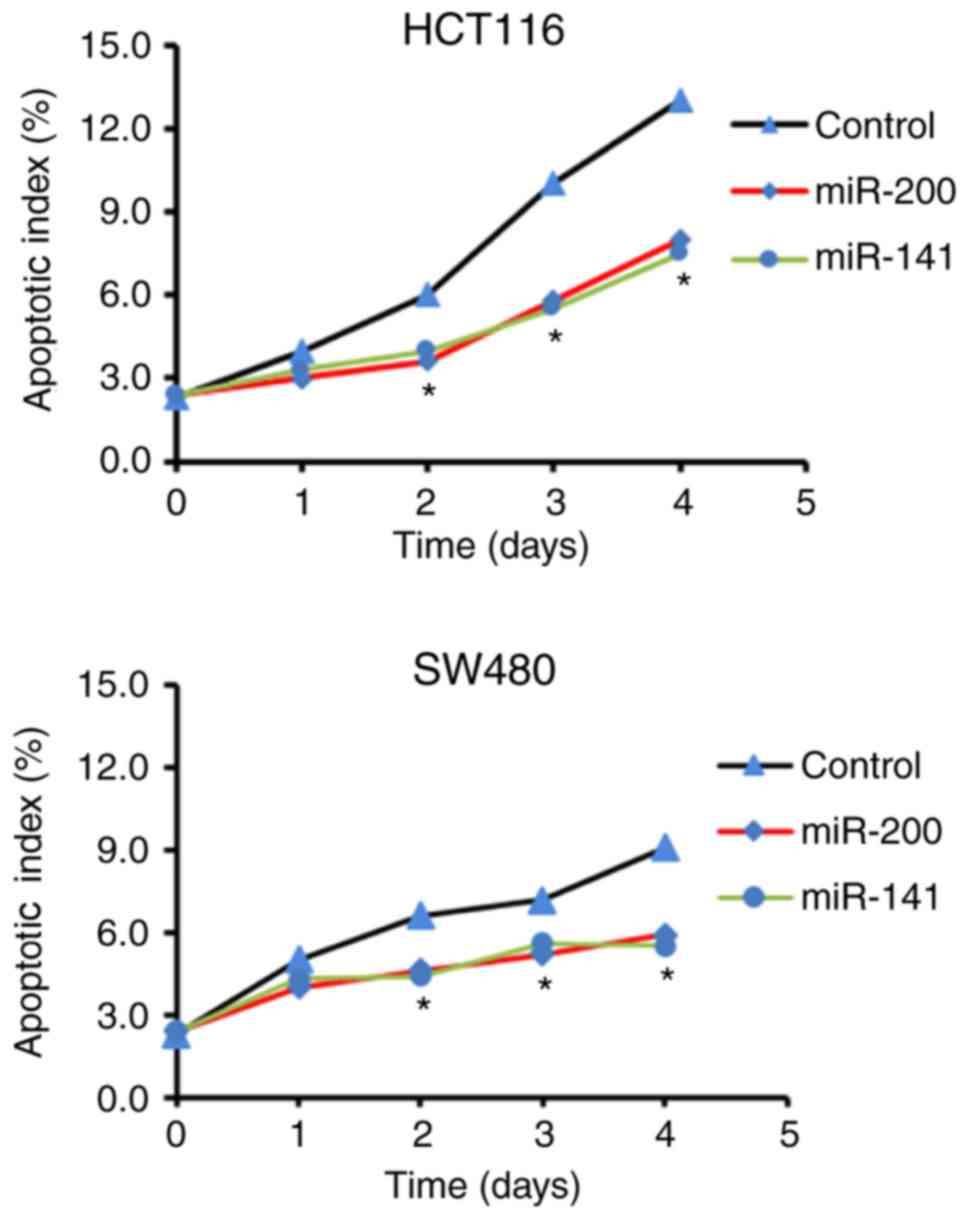
miR-200 and miR-141 inhibit the
apoptosis of HCT116 and SW480 cells
An apoptosis index-time curve is shown in Fig. 3. Compared with the control groups,
the transfected HCT116 and SW480 cells had significantly lower
apoptotic indices, suggesting that miR-200 and miR-141 inhibited
apoptosis.
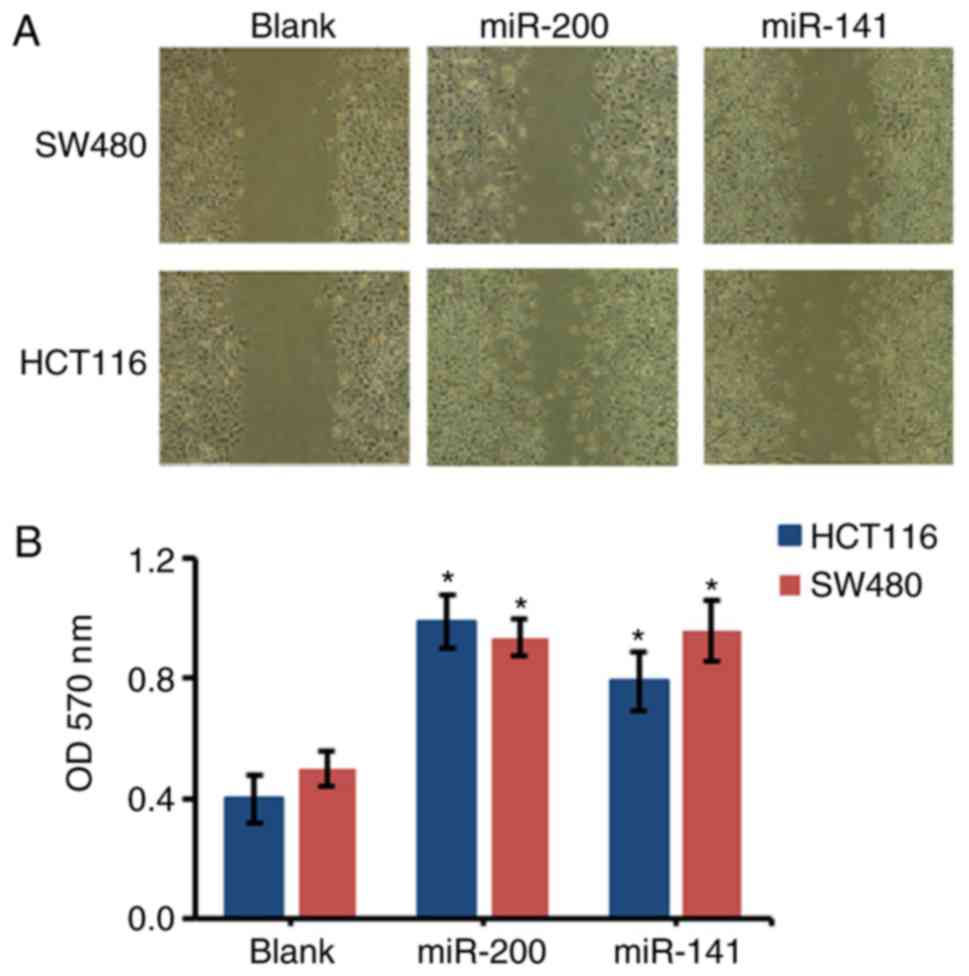
miR-200 and miR-141 enhance the
migration of HCT116 and SW480 cells
Compared with the control groups, the migration of
the miR-200-transfected HCT116 and SW480 cells significantly
increased. Consistently, the migration of the miR-141-transfected
cells was also significantly increased when compared with the blank
control group (P<0.05; Fig.
4).
Discussion
Colorectal cancer is the most common type of
malignant tumor of the digestive tract in clinical settings, and
liver metastasis in colorectal cancer leads to a poorer prognosis
(2). Several studies have shown
that microRNA is tightly correlated with the development of tumors
and survival rates of patients (6). The present study demonstrated that,
compared with normal tissues, colorectal cancer tissues expressed
significantly higher levels of miR-200 and miR-141. It was also
found that miR-200 and miR-141 inhibited apoptosis and enhanced the
migration of colorectal cancer cells, suggesting that miR-200 and
miR-141 may accelerate liver metastasis in colorectal cancer.
The way in which the profile of microRNAs, including
miR-200 and miR-141, is altered in colorectal cancer remains to be
fully elucidated (14). Tamagawa
et al (15) reported marked
demethylation in CpG regions of the miR-200 and miR-141 promoters
in cancer cells; this was confirmed by DNA methylation assays and
suggested that cancer cells regulated the profile of microRNAs,
particularly miR-200 and miR-141, via DNA methylation. In addition,
proteins encoded by microRNA target genes can reversely regulate
the profile of microRNAs (16,17).
However, further investigations are required to explain how
colorectal cancer affects the expression profile of microRNAs.
The migration and invasion of cancer cells are
complex biological processes. Studies have shown that
epithelial-mesenchymal transition (EMT) is pivotal in the migration
and invasion of cancer (18,19).
Studies have confirmed that miR-200 and miR-141 regulate EMT via
affecting the expression of E-cadherin, which is involved in EMT
(20,21). When miR-200 and miR-141 are
increased, transcription factors of E-cadherin are inhibited,
including zinc finger E-box binding homeobox (ZEB)1 and ZEB2,
leading to the downregulation of E-cadherin (20). These results are consistent with
the findings of the present study.
Early diagnosis and active treatment can improve the
survival rates of patients with colorectal cancer. Therefore, it is
important to identify effective biomarkers for liver metastasis in
colorectal cancer. The present study showed that increased serum
levels of miR-200 and miR-141 were associated with liver metastasis
in colorectal cancer, and this may improve the accuracy of
colorectal cancer and provide a basis for active treatment. In
addition, miR-200 and miR-141 enhanced the growth of colorectal
cancer, therefore, miR-200 and miR-141 maybe novel therapeutic
targets for the treatment of colorectal cancer.
In conclusion, the present study showed that
upregulation in the serum levels of miR-200 and miR-141 were
associated with liver metastasis in patients with colorectal
cancer. The overexpression of miR-200 and miR-141 exacerbated liver
metastasis of colorectal cancer via inhibiting apoptosis and
inducing migration.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abdalla EK, Bauer TW, Chun YS, D'Angelica
M, Kooby DA and Jarnagin WR: Locoregional surgical and
interventional therapies for advanced colorectal cancer liver
metastases: Expert consensus statements. HPB (Oxford). 15:119–130.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grothey A, Van Cutsem E, Sobrero A, Siena
S, Falcone A, Ychou M, Humblet Y, Bouché O, Mineur L, Barone C, et
al: Regorafenib monotherapy for previously treated metastatic
colorectal cancer (CORRECT): An international, multicentre,
randomised, placebo-controlled, phase 3 trial. Lancet. 381:303–312.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pritchard CC, Cheng HH and Tewari M:
MicroRNA profiling: Approaches and considerations. Nat Rev Genet.
13:358–369. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedman RC and Burge CB: MicroRNA target
finding by comparative genomics. Methods Mol Biol. 1097:457–476.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 9:8522017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schee K, Boye K, Abrahamsen TW, Fodstad Ø
and Flatmark K: Clinical relevance of microRNA miR-21, miR-31,
miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC
Cancer. 12:5052012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Faltejskova P, Svoboda M, Srutova K,
Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K,
Kiss I, et al: Identification and functional screening of microRNAs
highly deregulated in colorectal cancer. J Cell Mol Med.
16:2655–2666. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou
WP, Huo XS, Xu D, Bi HS, Wang F and Sun SH: Epigenetic activation
of the MiR-200 family contributes to H19-mediated metastasis
suppression in hepatocellular carcinoma. Carcinogenesis.
34:577–586. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toiyama Y, Hur K, Tanaka K, Inoue Y,
Kusunoki M, Boland CR and Goel A: Serum miR-200c is a novel
prognostic and metastasis-predictive biomarker in patients with
colorectal cancer. Ann Surg. 259:735–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hamilton SR, Bosman FT and Boffetta P:
Carcinoma of the colon and rectumWHO Classification of Tumours of
the Digestive System. Bosman FT, Carneiro F, Hruban RH and Theise
ND: Lyon: IARC Press; pp. 134–146. 2010
|
|
12
|
Kyrylkova K, Kyryachenko S, Leid M and
Kioussi C: Detection of apoptosis by TUNEL assay. Methods Mol Biol.
887:41–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stevens P, An Wen Y, Li X and Gao T:
Erbin-mediated regulation of colon cancer. FASEB J. 29:724–726.
2015.PubMed/NCBI
|
|
14
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tamagawa S, Beder LB, Hotomi M, Gunduz M,
Yata K, Grenman R and Yamanaka N: Role of miR-200c/miR-141 in the
regulation of epithelial-mesenchymal transition and migration in
head and neck squamous cell carcinoma. Int J Mol Med. 33:879–886.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lynch SM, O'Neill KM, McKenna MM, Walsh CP
and McKenna DJ: Regulation of miR-200c and miR-141 by methylation
in prostate cancer. Prostate. 76:1146–1159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gibbons DL, Lin W, Creighton CJ, Rizvi ZH,
Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y,
Pertsemlidis A and Kurie JM: Contextual extracellular cues promote
tumor cell EMT and metastasis by regulating miR-200 family
expression. Genes Dev. 23:2140–2151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Brabletz T: EMT and MET in metastasis:
Where are the cancer stem cells? Cancer Cell. 22:699–701. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu M, Marsters S, Ye X, Luis E, Gonzalez L
and Ashkenazi A: E-cadherin couples death receptors to the
cytoskeleton to regulate apoptosis. Mol Cell. 54:987–998. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu H, Shen Y, Hong J, Xia Q, Zhou F and
Liu X: The contribution of TGF-β in epithelial-mesenchymal
transition (EMT): down-regulation of E-cadherin via snail.
Neoplasma. 62:1–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|