Introduction
Gallbladder cancer (GBC) is the most prevalent
biliary malignancy, and is the sixth most common type of
gastrointestinal cancer in the United States of America, with
>10,000 novel cases of GBC diagnosed each year (1). GBC is a highly progressive and
aggressive malignancy, with survival in the majority of patients
being ≤1 year after diagnosis. Early diagnosis of GBC is difficult
due to a lack of specific clinical symptoms, and a large number of
patients with GBC are diagnosed at an advanced stage with
substantial metastasis and invasion to other organs (2,3).
Currently, cholecystectomy is the only effective treatment strategy
for ~10 % of early-stage patients with GBC (4,5).
Therefore, identification of clinical biomarkers that may be used
to accurately evaluate tumor metastasis and prognosis may allow the
most appropriate treatment regimen to be selected for individual
patients.
MicroRNAs (miRNAs/miRs) are a class of non-coding
RNAs that consist of 18–25 nucleotides with no protein-coding
function (6,7). Previous reports have demonstrated
that the aberrant expression miRNAs have important roles in the
regulation of carcinogenesis and progression in various tumor
types, including brain, lung, breast, liver, bladder and colorectal
cancers (8,9). For example, miRNAs modulate
epigenetic regulation, the immune system and the tumor
microenvironment in lung cancer (10). In addition, miRNA-21 was reported
to have diagnostic, prognostic and predictive value in patients
with breast cancer, their daughters and healthy individuals
(11), and miRNA-143 was
demonstrated to be a predictive factor for the response to
fluoropyrimidine-based chemotherapy in patients with colorectal
cancer metastasis (12).
Although the functions of aberrant miRNA expression
in malignancies have not been fully illustrated, miRNAs may become
novel diagnostic biomarkers or therapeutic targets for GBC
(13). hsa-miR-372 is transcribed
from the human genomic region on chromosome 19q13.42. Together with
hsa-miR-371a, hsa-miR-371b and hsa-miR-373, hsa-miR-372 forms part
of the miR-371-373 cluster, which is reported to exhibit a key role
in tumorigenesis and progression (14). However, to the best of our
knowledge, the clinical effects of aberrant hsa-miR-372 expression
have not been previously reported in GBC.
The present study investigated hsa-miR-372
expression levels in 80 pairs of GBC tissues and adjacent normal
gallbladder tissues, and assessed their association with the
prognosis of patients with GBC. Furthermore, a dual-luciferase
reporter assay and western blot analysis demonstrated that chloride
intracellular channel 1 (CLIC1) was a direct target gene of
hsa-miR-372.
Materials and methods
Patients
A total of 80 samples from patients with
pathologically confirmed GBC were collected from Hunan Provincial
People's Hospital, The First Affiliated Hospital of Hunan Normal
University (Changsha, China) between 2008 and 2012 following
cholecystectomy. None of the patients that participated in the
present study were treated with radiotherapy, chemotherapy or other
treatments prior to cholecystectomy. The patients included 30 males
and 50 females, with an average age of 48.2±2.6 years. The
pathological stage of GBC was classified according to 6th edition
of the tumor-node-metastasis (TNM) classification of the American
Joint Committee on Cancer (15).
Poorly differentiated tumor tissues exhibit poor maturity and high
degree of malignancy, while well-differentiated tumor tissues are
similar in appearance to normal cells and exhibit a low degree of
malignancy. The histological grade was evaluated by hematoxylin and
eosin staining. Tumor invasion and lymph metastasis were assessed
according to the standard criteria proposed by the Chinese Research
Society for GBC (16). The
clinicopathological features of all patients are presented in
Table I. The overall survival time
of each patient was measured from the date of initial surgical
operation to mortality. The follow-up time ranged from 1 to 60
months.
 | Table I.Association between hsa-miR-372
expression in patients with gallbladder cancer and
clinicopathological characteristics. |
Table I.
Association between hsa-miR-372
expression in patients with gallbladder cancer and
clinicopathological characteristics.
|
|
| hsa-miR-372
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | Number of cases | Low | High | P-value |
|---|
| Age, years |
|
|
| 0.105 |
|
<50 | 47 | 29 | 18 |
|
| ≥50 | 33 | 26 | 7 |
|
| Gender |
|
|
| 0.071 |
| Male | 30 | 17 | 13 |
|
|
Female | 50 | 38 | 12 |
|
| Tumor size, cm |
|
|
| 0.191 |
| ≤3 | 27 | 16 | 11 |
|
|
>3 | 53 | 39 | 14 |
|
| Histological
grade |
|
|
| <0.001 |
|
G1-G2 | 16 | 5 | 11 |
|
|
G3-G4 | 64 | 50 | 14 |
|
| TNM stage |
|
|
| <0.001 |
|
I–II | 23 | 8 | 15 |
|
|
III–IV | 57 | 47 | 10 |
|
| Lymph node
metastasis |
|
|
| 0.014 |
|
Negative | 35 | 19 | 16 |
|
|
Positive | 45 | 36 | 9 |
|
| Distant
metastasis |
|
|
| <0.001 |
|
Negative | 42 | 18 | 24 |
|
|
Positive | 38 | 37 | 1 |
|
| Gallbladder
stones |
|
|
| 0.228 |
|
Absent | 40 | 25 | 15 |
|
|
Present | 40 | 30 | 10 |
|
This medicinal study was approved by the Research
Ethics Committee of Hunan Provincial People's Hospital, The First
Affiliated Hospital of Hunan Normal University, and written
informed consent was also obtained from all patients or their
families. All patient specimens were made anonymous and handled
according to the legal and ethical standards for protecting human
rights.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from 80 samples of fresh GBC tissues and
adjacent normal gallbladder tissues or GBC cell lines were
extracted using a miRNeasy mini kit (Qiagen, Inc., Valencia, CA,
USA). Subsequently, for hsa-miR-372, the cDNA was synthesized by
using One Step Prime script miRNA cDNA Synthesis kit (Qiagen, Inc.)
according to the manufacturer's protocol. qPCR was performed on the
Roche Light Cycler 480 II real-time PCR System using a SYBR Premix
Ex Taq II kit (Takara Bio, Inc., Otsu, Japan). The qPCR conditions
were: 95°C for 5 min then 40 cycles of denaturation at 95°C for 10
sec and for the annealing/elongation step, 60°C for 30 sec.
Hsa-miR-372 was amplified by using forward primers with the DNA
sequence of the mature miRNA (5′-ACTATTCTGATGTCCAAGTGGA-3′) and
common reverse primers provided in the First Strand cDNA Synthesis
kit. U6 small nuclear RNA was used as an internal control with the
following primers: forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-ACGCTTCACGAATTTGCGT-3′. For CLIC1, the first-strand cDNA was
synthesized by using a RevertAid H Minus First Strand cDNA
Synthesis kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's instructions. qPCR was also
performed on the Roche Light Cycler 480 II real-time PCR System
using a SYBR Premix Ex Taq II kit. The qPCR conditions were 98°C
for 10 min and 40 cycles of denaturation at 98°C for 10 sec and for
the annealing/elongation step, 65°C for 30 sec. GAPDH was used as
an internal control. The primer sequences of CLIC1 and GAPDH were
as follows: CLIC1 forward, 5′-TCTGAACCCTGAGTCCAAC-3′ and reverse,
5′-GAGTCCCTTCTCCAGATTGT-3′; and GAPDH forward,
5′-GAAGGTGAAGGTCGGAG-3′ and reverse, 5′-GAAGATGGTGATGGGAT-3′. All
reactions were run in triplicate, the relative expression levels of
hsa-miR-372 and CLIC1 were normalized to the expression of U6 and
GAPDH, respectively using the 2−ΔΔCq method (17).
Target gene prediction and plasmid
construction
Target genes of hsa-miR-372 were predicted by using
TargetScanHuman (version 7.1; www.targetscan.org) and miRanda (2010 version;
www.microrna.org) algorithms.
To generate a reporter plasmid for the
dual-luciferase reporter assay, the full length 3′-untranslated
region (UTR) of the CLIC1 gene was synthesized by PCR. The
following primers were used: 5′-GCCCCTCCTGGGACTCCCTCAACCC-3′
(forward) and 5′-TTGCGTAAAAACACTTGATTTTTAT-3′ (reverse).
Subsequently, PCR products were cloned into a psiCHECK-2 vector
(Promega Corporation, Madison, WI, USA) to produce psiCHECK-2-CLIC1
wild type luciferase reporter plasmid. The correct clones were
confirmed by Sanger DNA sequencing. The mutant CLIC1 3′-UTR
sequence plasmid was synthesized by GeneCopoeia (GeneCopoeia, Inc.,
Rockville, MD, USA) and also inserted into the psiCHECK-2 vector to
construct psiCHECK-2-CLIC1 mutant (MUT) luciferase reporter
plasmid.
Cell culture and dual-luciferase
reporter assay
G-415, OCUG-1 and SGC-996 human GBC cell lines were
purchased from the Japan Health Science Research Resources Bank
(Osaka, Japan). All cells were cultured in Dulbecco's modified
Eagle's medium/F12 (Thermo Fisher Scientific, Inc.) with 10% fetal
bovine serum (Thermo Fisher Scientific, Inc.) in a humidified
atmosphere with 5% CO2 at 37°C.
GBC cells were seeded onto 24-well plates
(4×105/well) and cotransfected with 100 nM hsa-miR-372
mimics (5′-AAAGUGCUGCGACAUUUGAGCGU-3′) or corresponding mimics
negative control (5′-CAGCUCAAGUGUCGCCAUGUTCU-3′) and WT or MUT
luciferase reporter plasmids (100 ng) by using Lipofectamine 2000
reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. After transfection for 24 h, the
luciferase activities were analyzed using a dual luciferase
reporter gene assay kit (Promega Corporation) in a TD-20/20
luminometer (Turner Designs, San Jose, CA, USA). Renilla
luciferase activity was normalized to firefly luciferase
activity.
Western blot analysis
Total protein from G-415, OCUG-1 and SGC-996 cells
was extracted by radioimmunoprecipitation assay lysis buffer
(Thermo Fisher Scientific, Inc.) at 65°C and quantified using a
bicinchoninic assay kit (Thermo Fisher Scientific, Inc.).
Subsequently, the quantified proteins (50 µg) were denatured and
subjected to 15% SDS-PAGE, and blotted onto polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA). The
membranes were then blocked with 5% non-fat milk for 2 h at 37°C.
Following washing with TBS-Tween-20 (0.1%) three times, the
membranes were incubated with mouse anti-human CLIC1 (1:1,000; cat.
no. ab77214; Abcam, Cambridge, MA, USA) and anti-GAPDH (1:2,000;
cat. no. ab8245; Abcam) antibodies overnight at 4°C. Then, the
membranes were incubated with a horseradish peroxidase-conjugated
goat anti-mouse IgG (1:2,000; cat. no. 7076; Cell Signaling
Technology, Inc., Danvers, MA, USA) for 1 h at 37°C and visualized
using an enhanced chemiluminescence western blot detection kit (GE
Healthcare, Pittsburgh, PA, USA) according to the manufacturer's
protocols.
Statistical analysis
Statistical analysis was performed using SPSS 19.0
(IBM Corp., Armonk, NY, USA). Each experiment was repeated at least
three times. Data are presented as the mean ± standard deviation. A
paired Student's t-test was performed to compare hsa-miR-372
expression in GBC tissues with adjacent normal gallbladder tissues.
The χ2 test was used to detect the association between
hsa-miR-372 expression and clinicopathological features. The
expression of hsa-miR-372 between the three GBC cell lines was
compared using one-way analysis of variance and Dunnett's post-hoc
test. Overall survival curves were evaluated by the Kaplan-Meier
method and logrank test. Univariate Cox regression analysis was
performed for prognostic parameters, and the importance of multiple
parameters for survival was assessed by the Cox's proportional
hazards model for multivariate analysis. P<0.05 was considered
to indicate a statistically significant difference.
Results
hsa-miR-372 expression is reduced in
GBC tissues
The expression of hsa-miR-372 was determined in 80
GBC tissue samples and adjacent normal gallbladder tissues by
RT-qPCR. Following normalization to U6, the hsa-miR-372 expression
in GBC tissues was significantly lower compared with adjacent
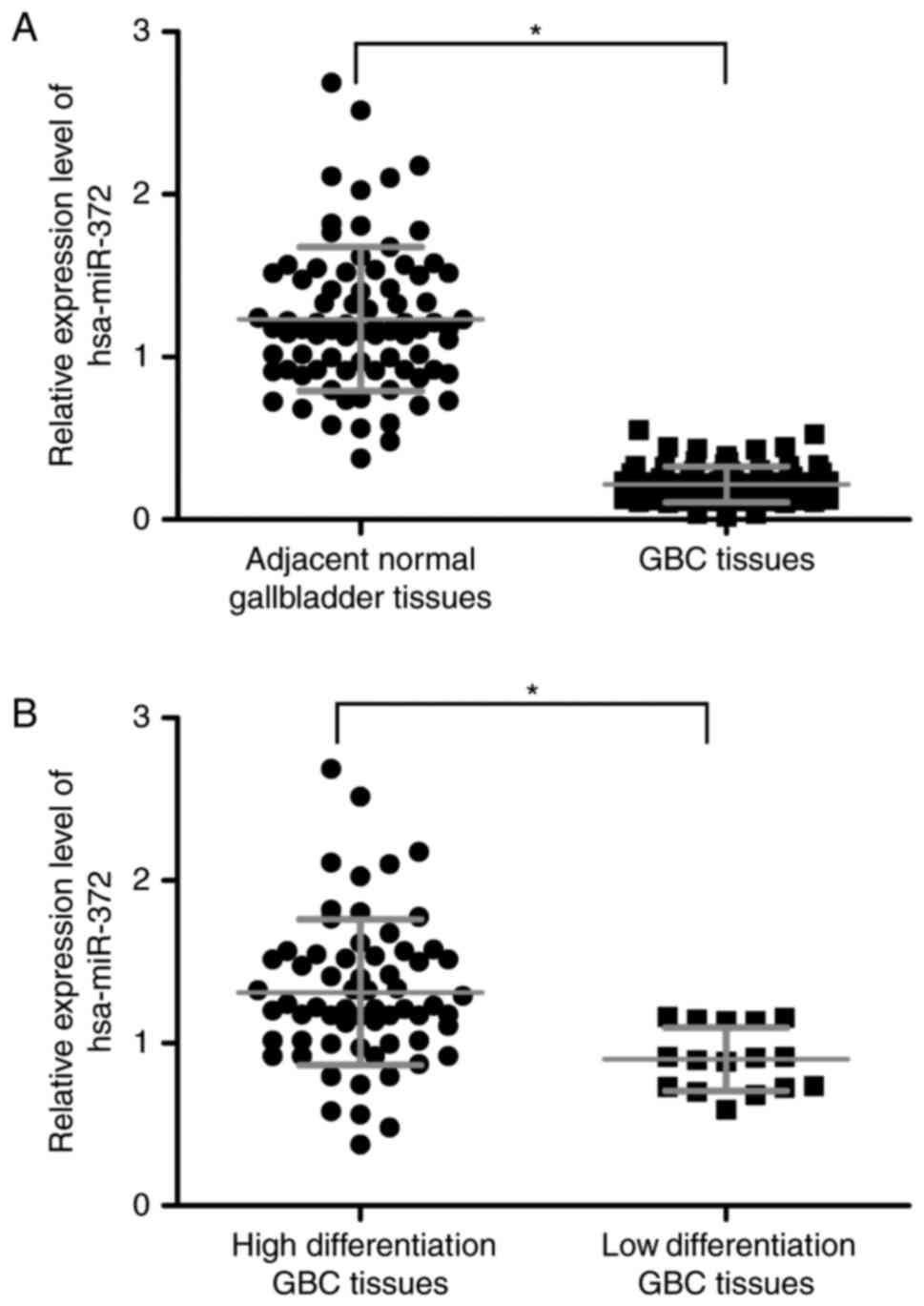
normal gallbladder tissues (P=0.002; Fig. 1A). Additionally, hsa-miR-372
expression was also downregulated in GBC tissues with low
differentiation compared with highly differentiated tissues
(P=0.042; Fig. 1B).
Association between hsa-miR-372
expression and clinicopathological parameters of patients with
GBC
RT-qPCR results indicated that hsa-miR-372
expression was significantly downregulated in patients with GBC. In
order to further determine the potential associations between
hsa-miR-372 expression and the clinicopathological features, 80
samples of GBC tissues were divided into low and high expression
groups based on the average expression of hsa-miR-372 in GBC
tissues. Patients with GBC that exhibited expression of hsa-miR-372
at levels lower than the average expression level (2.41) were
assigned to the low expression group (n=55), and those samples with
expression equal to or above the average value were assigned to the
high expression group (n=25).
As summarized in Table
I, low hsa-miR-372 expression was significantly associated with
the presence of GBC histological grade (P<0.001), TNM stage
(P<0.001), lymph node metastasis (P=0.014) and distant
metastasis (P<0.001). However, there were no significant
differences in the gender of patients with GBC (P=0.071), patient
age (P=0.105), tumor size (P=0.191) or the presence of gallbladder
stones (P=0.228) between the high and low hsa-miR-372 expression
groups. Collectively, these results indicate that decreased
expression of hsa-miR-372 may be relevant to tumor differentiation
and metastasis, which are involved in tumorigenesis and the
progression of GBC.
Reduced expression of hsa-miR-372 is
associated with poor prognosis in patients with GBC
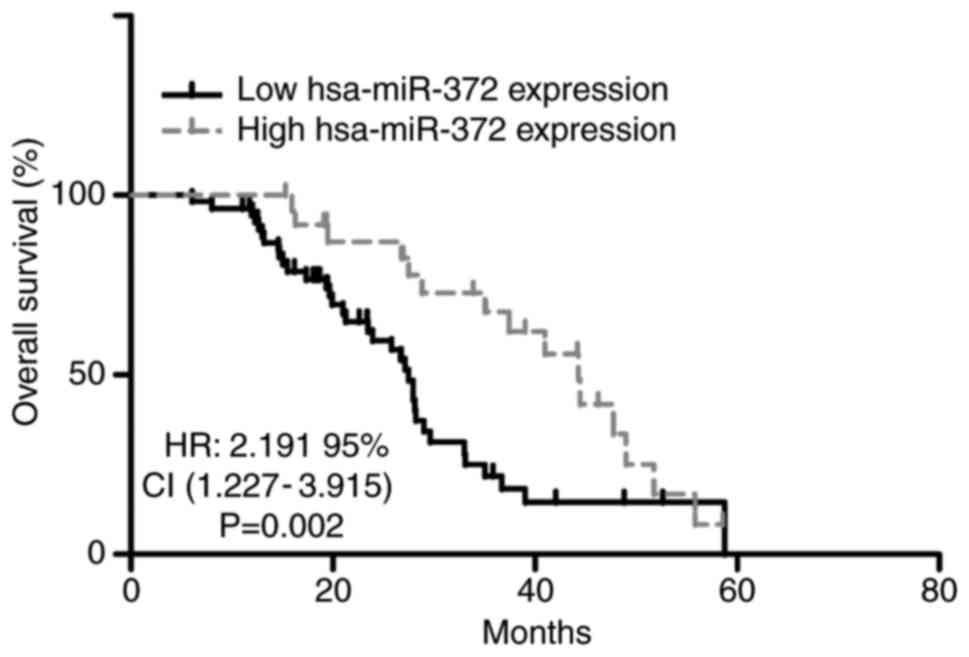
The association between hsa-miR-372 expression and
the overall survival of GBC patients was evaluated by Kaplan-Meier
analysis. As demonstrated in Fig.
2, low hsa-miR-372 expression was associated with a shorter
overall survival time compared with patients with high hsa-miR-372
expression (P=0.002). Furthermore, multivariate Cox regression
analyses indicated that hsa-miR-372 expression (P=0.004),
histological grade (P=0.024) and lymph node metastasis (P=0.001)
maintained an independent prognostic influence on the overall
survival of patients with GBC (Table
II).
 | Table II.Univariate and multivariate Cox
regression analyses of prognostic parameters patients with
gallbladder cancer. |
Table II.
Univariate and multivariate Cox
regression analyses of prognostic parameters patients with
gallbladder cancer.
|
| Univariate Cox
regression | Multivariate Cox
regression |
|---|
|
|
|
|
|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, ≥50 vs.
<50 | 0.991
(0.525–1.467) | 0.724 | – | – |
| Gender, male vs.
female | 1.275
(0.838–1.529) | 0.522 | – | – |
| Tumor size, ≤3 vs.
>3 cm | 1.643
(0.587–2.965) | 0.458 | – | – |
| Histological grade,
G1-G2 vs. G3-G4 | 2.047
(1.198–4.325) | 0.026a | 2.013
(1.112–4.214) | 0.024a |
| TNM stage, I–II vs.
III–IV | 1.028
(0.468–2.093) | 0.791 | – | – |
| Lymph node
metastasis, negative vs. positive | 2.431
(1.752–7.043) | 0.001a | 2.419
(1.682–6.931) | 0.001a |
| Distant metastasis,
negative vs. positive | 1.125
(0.369–2.467) | 0.652 | – | – |
| Gallbladder stones,
absent vs. present | 1.019
(0.728–1.646) | 0.435 | – | – |
| hsa-miR-372
expression, low vs. high | 2.559
(1.578–4.927) | 0.006a | 2.410
(1.467–4.532) | 0.004a |
Endogenous expression levels of
hsa-miR-372 in three GBC cell lines
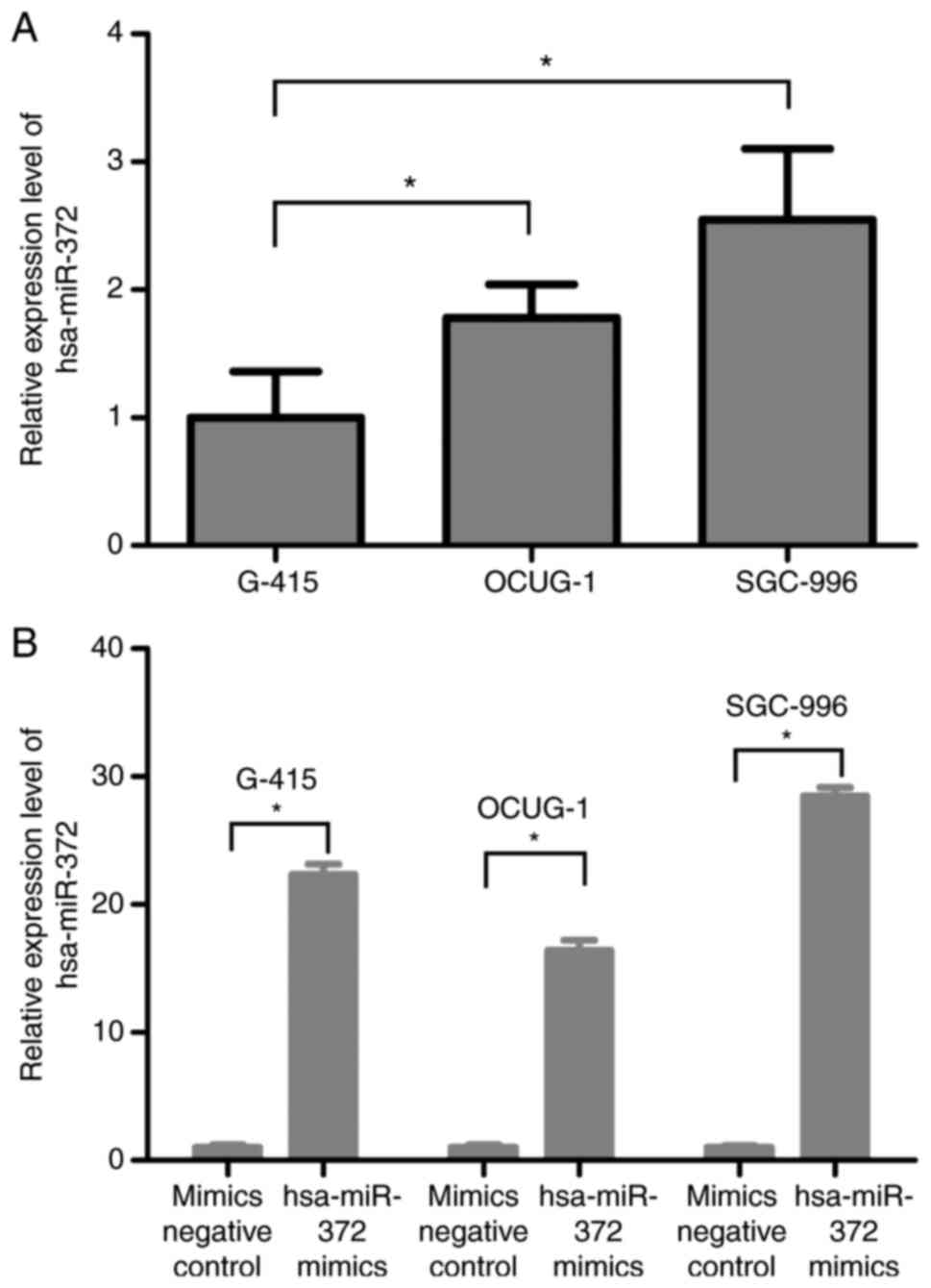
The endogenous expression level of hsa-miR-372 in
OCUG-1 and SGC-996 cells were relatively higher compared with G-415
cell, as determined by RT-qPCR (Fig.
3A). The expression levels of hsa-miR-372 in G-415, OCUG-1 and
SGC-996 cells transfected with hsa-miR-372 mimics were
significantly upregulated by 22.33-, 16.38- and 28.45-fold,
respectively, when compared with levels in the mimics negative
control transfection group (Fig.
3B), indicating that the transfection efficiency is suitable
for further cell experiments.
CLIC1 is a direct target of
hsa-miR-372 in GBC cells
As miRNAs primarily function by inhibiting their
target genes, the targets of hsa-miR-372 in GBC were predicted by
using bioinformatics analysis. Previous research demonstrated that
CLIC1 expression was closely associated with GBC progression and
may be regarded as an effective biomarker for predicting the
prognosis of GBC (18,19). Complementary sequences were
identified between hsa-miR-372 and the 3′UTR of CLIC1 (Fig. 4A), which indicated CLIC1 may be a
target gene of hsa-miR-372. To further investigate whether
hsa-miR-372 inhibits CLIC1 expression in GBC cells, G-415, OCUG-1
and SGC-996 cells were treated with hsa-miR-372 mimics or mimics
negative control. RT-qPCR and western blot analysis demonstrated
that transfection with hsa-miR-372 mimics markedly decreased the
expression of CLIC1 mRNA and protein in all three GBC cell lines
(Fig. 4B). In addition, a
dual-luciferase reporter assay was performed to confirm the direct
interaction between hsa-miR-372 and the 3′UTR of CLIC1. Notably,
the results demonstrated that the relative luciferase activity of
the wild-type CLIC1 3′UTR plasmid decreased in response to
treatment with hsa-miR-372 mimics compared with negative control
mimics (Fig. 4C). By contrast,
hsa-miR-372 mimics exhibited no inhibitory effect on luciferase
activity when transfected with the mutant CLIC1 3′UTR plasmid
(Fig. 4C).
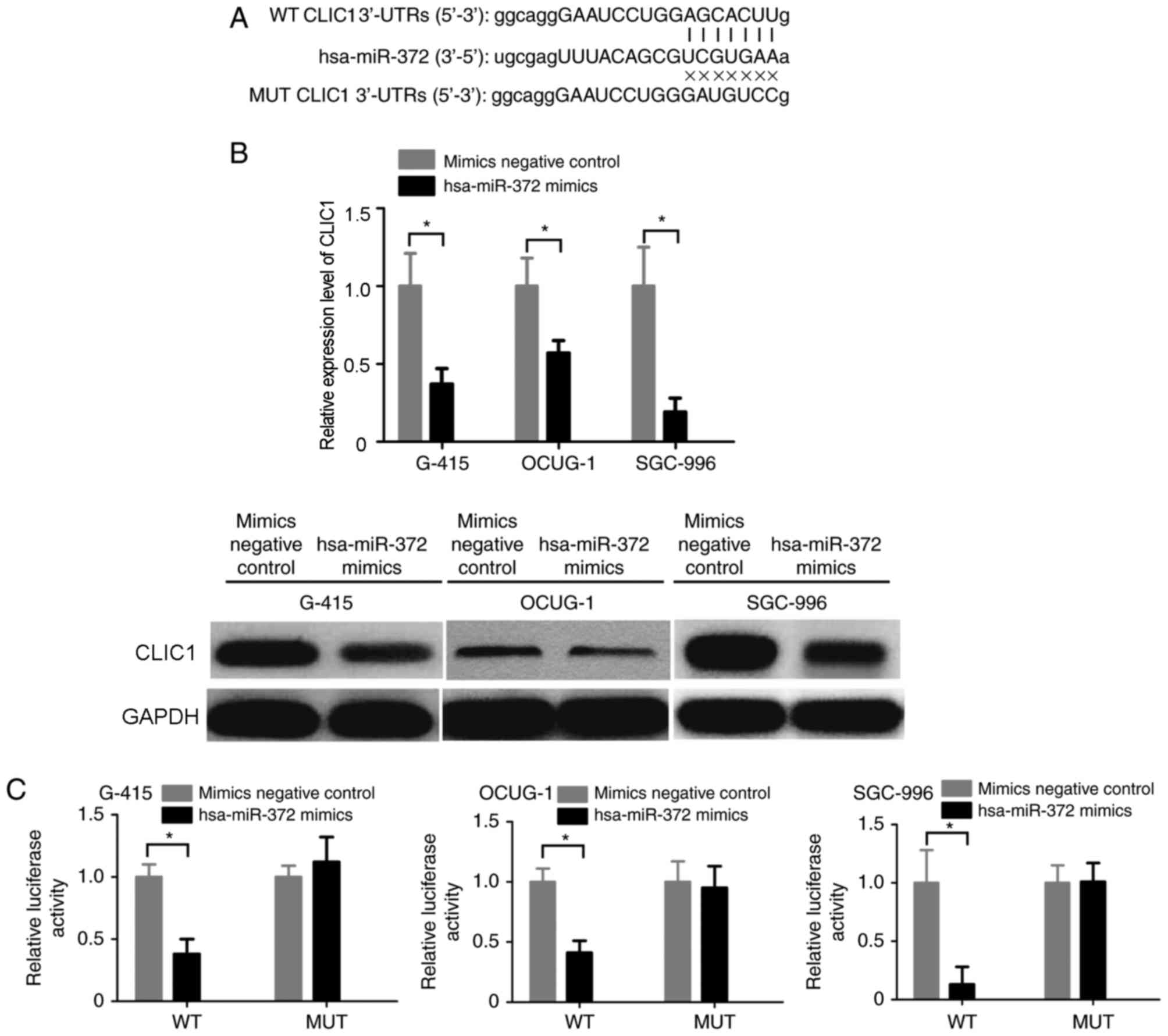 | Figure 4.CLIC1 is a direct target of
hsa-miR-372 in GBC cells. (A) Sequence alignment of hsa-miR-372 and
the CLIC1 3′-UTR using miRanda algorithm. (B) Reverse
transcription-quantitative polymerase chain reaction and western
blot analysis was performed to determine CLIC1 expression in G-415,
OCUG-1 and SGC-996 cells following transfection with hsa-miR-372
mimics or mimics negative control. GAPDH was used as an internal
control. (C) G-415, OCUG-1 and SGC-996 cells were cotransfected
with hsa-miR-372 mimics or mimics negative control, and the
luciferase reporter plasmid containing a fragment of the CLIC1
3′-UTR harboring either the WT hsa-miR-372 binding sites or MUT
binding sites. *P<0.05, as indicated. CLIC1, chloride
intracellular channel 1; miR, microRNA; GBC, gallbladder cancer;
UTR, untranslated region; WT, wild-type; MUT, mutant. |
Discussion
At present, GBC is associated with a high mortality
rate and poor prognosis (2,3).
Although certain clinicopathological parameters have been the
standard for predicting the clinical outcome of patients with GBC,
these classification schemes are not precise indexes of prognosis
for patients with GBC. Thus, it is essential to identify novel and
effective biomarkers that are associated with advanced GBC
progression to allow early diagnosis and treatment.
hsa-miR-372 has been reported to function as either
an oncogenic gene or a tumor suppressor gene in several types of
human cancer. For example, Cho et al (20) demonstrated that hsa-miR-372 has an
oncogenic function by directly targeting the tumor suppressor gene
large tumor suppressor kinase 2 in gastric cancer. In addition,
Yamashita et al (21)
indicated that the upregulation of hsa-miR-372 was an independent
prognostic factor in colon cancer, and was strongly associated with
synchronous liver metastasis. Furthermore, hsa-miR-372 was reported
to regulate glioma cell proliferation and invasion by targeting PH
domain and leucine rich repeat protein phosphatase 2 (22), and was downregulated and
potentially contributes to tumorigenesis in human cervical cancer
by targeting cyclin-dependent kinase 2 and cyclin A1 (23). hsa-miR-372 was also demonstrated to
downregulate the expression of the oncogene ATPase family AAA
domain containing 2 (ATAD2) to affect hepatocellular carcinoma cell
proliferation and metastasis (24). In addition, hsa-miR-372 was
reported to be downregulated in breast cancer (25), and may regulate protein folding by
suppressing two chaperones [DnaJ heat shock protein family (Hsp40)
members A2 and C9] and Sec61 translocon α 2 subunit, which is
considered to be an essential complex for correct protein folding
(25).
The present study demonstrated that hsa-miR-372
expression was decreased in human GBC tissues compared with
adjacent normal gallbladder tissues. hsa-miR-372 was already
established as a potential marker and regulated a member of the
ATPase family, ATAD2, in lung cancer (24). In addition, the results of the
current study indicated that the downregulated expression of
hsa-miR-372 in patients with GBC was associated with advanced tumor
progression. Kaplan-Meier analysis demonstrated that patients with
low hsa-miR-372 expression exhibited poorer overall survival
compared with those with higher expression. Furthermore,
multivariate Cox regression analysis demonstrated that low
hsa-miR-372 expression was a statistically significant risk index
influencing overall survival rates in patients with GBC, which
indicated that downregulation of hsa-miR-372 in GBC is a predictor
of overall survival as well as a grade-dependent factor. These
results were consistent with previous studies (24,25),
confirming the downregulation pattern of hsa-miR-372 in GBC and
indicating a potential important role for hsa-miR-372 in the
regulation of GBC progression.
Increasing evidence has demonstrated that miRNAs
regulate genes expression at transcriptional and/or
post-transcriptional levels via mRNA degradation and/or
translational repression (26,27).
At present, it is estimated that ~60% of human genes may be
controlled by >1,900 identified miRNAs (28). miRNAs mediate translational
repression and/or mRNA degradation by binding to the 3′UTRs of
their target genes (29,30). CLIC1 is a novel member of the p64
chloride channel protein family, which has been reported to have
important roles in various tumor types (31–34).
A recent study indicated that CLIC1 expression is associated with
total postoperative survival of patients with GBC, and CLIC1 was a
potential poor prognostic factor for GBC (35). The present study identified CLIC1
as a direct target of hsa-miR-372 in GBC, as hsa-miR-372 binds to
the 3′-UTR of CLIC1 mRNA in vitro, which indicates that the
association between the downregulation of hsa-miR-372 and poor
prognosis of patients with GBC may occur via CLIC1.
In conclusion, the present study provides evidence
that hsa-miR-372 expression was downregulated in GBC tissues, which
was associated with advanced tumor progression. Notably, the
present study, to the best of our knowledge, is the first to
demonstrated that hsa-miR-372 may regulate GBC progression and
development by directly targeting CLIC1. As a result, the
manipulation of hsa-miR-372 may be an effective and promising
therapeutic method for patients with GBC in the future.
References
|
1
|
Randi G, Franceschi S and La Vecchia C:
Gallbladder cancer worldwide: Geographical distribution and risk
factors. Int J Cancer. 118:1591–1602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wistuba II and Gazdar AF: Gallbladder
cancer: Lessons from a rare tumour. Nat Rev Cancer. 4:695–706.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gourgiotis S, Kocher HM, Solaini L,
Yarollahi A, Tsiambas E and Salemis NS: Gallbladder cancer. Am J
Surg. 196:252–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Balachandran P, Agarwal S, Krishnani N,
Pandey CM, Kumar A, Sikora SS, Saxena R and Kapoor VK: Predictors
of long-term survival in patients with gallbladder cancer. J
Gastrointest Surg. 10:848–854. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim WS, Choi DW, You DD, Ho CY, Heo JS and
Choi SH: Risk factors influencing recurrence, patterns of
recurrence, and the efficacy of adjuvant therapy after radical
resection for gallbladder carcinoma. J Gastrointest Surg.
14:679–687. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Farh KK, Grimson A, Jan C, Lewis BP,
Johnston WK, Lim LP, Burge CB and Bartel DP: The widespread impact
of mammalian MicroRNAs on mRNA repression and evolution. Science.
310:1817–1821. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bagga S and Pasquinelli AE: Identification
and analysis of microRNAs. Genet Eng (N Y). 27:1–20. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nikitina EG, Urazova LN and Stegny VN:
MicroRNAs and human cancer. Exp Oncol. 34:2–8. 2012.PubMed/NCBI
|
|
10
|
Rusek AM, Abba M, Eljaszewicz A, Moniuszko
M, Niklinski J and Allgayer H: MicroRNA modulators of epigenetic
regulation, the tumor microenvironment and the immune system in
lung cancer. Mol Cancer. 14:342015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Usmani A, Shoro AA, Memon Z, Hussain M and
Rehman R: Diagnostic, prognostic and predictive value of
MicroRNA-21 in breast cancer patients, their daughters and healthy
individuals. Am J Cancer Res. 5:2484–2490. 2015.PubMed/NCBI
|
|
12
|
Simmer F, Venderbosch S, Dijkstra JR,
Vink-Börger EM, Faber C, Mekenkamp LJ, Koopman M, De Haan AF, Punt
CJ and Nagtegaal ID: MicroRNA-143 is a putative predictive factor
for the response to fluoropyrimidine-based chemotherapy in patients
with metastatic colorectal cancer. Oncotarget. 6:22996–23007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kono H, Nakamura M, Ohtsuka T, Nagayoshi
Y, Mori Y, Takahata S, Aishima S and Tanaka M: High expression of
microRNA-155 is associated with the aggressive malignant behavior
of gallbladder carcinoma. Oncol Rep. 30:17–24. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nikaki A, Piperi C and Papavassiliou AG:
Role of microRNAs in gliomagenesis: Targeting miRNAs in
glioblastoma multiforme therapy. Expert Opin Investig Drugs.
21:1475–1488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang CS, Yu W, Cui H, Wang YJ, Zhang L,
Han F and Huang T: Increased expression of miR-21 predicts poor
prognosis in patients with hepatocellular carcinoma. Int J Clin Exp
Pathol. 8:7234–7238. 2015.PubMed/NCBI
|
|
16
|
Cai J, Xu L, Cai Z, Wang J, Zhou B and Hu
H: MicroRNA-146b-5p inhibits the growth of gallbladder carcinoma by
targeting epidermal growth factor receptor. Mol Med Rep.
12:1549–1555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang JW, Peng SY, Li JT, Wang Y, Zhang ZP,
Cheng Y, Cheng DQ, Weng WH, Wu XS, Fei XZ, et al: Identification of
metastasis-associated proteins involved in gallbladder carcinoma
metastasis by proteomic analysis and functional exploration of
chloride intracellular channel 1. Cancer Lett. 281:71–81. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Wang Z, Li M, Ye Y, Xu Y, Zhang Y,
Yuan R, Jin Y, Hao Y, Jiang L, et al: Chloride intracellular
channel 1 regulates the antineoplastic effects of metformin in
gallbladder cancer cells. Cancer Sci. 108:1240–1252. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cho WJ, Shin JM, Kim JS, Lee MR, Hong KS,
Lee JH, Koo KH, Park JW and Kim KS: miR-372 regulates cell cycle
and apoptosis of ags human gastric cancer cell line through direct
regulation of LATS2. Mol Cells. 28:521–527. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamashita S, Yamamoto H, Mimori K, Nishida
N, Takahashi H, Haraguchi N, Tanaka F, Shibata K, Sekimoto M, Ishii
H, et al: MicroRNA-372 is associated with poor prognosis in
colorectal cancer. Oncology. 82:205–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen X, Hao B, Han G, Liu Y, Dai D, Li Y,
Wu X, Zhou X, Yue Z, Wang L, et al: miR-372 regulates glioma cell
proliferation and invasion by directly targeting PHLPP2. J Cell
Biochem. 116:225–232. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian RQ, Wang XH, Hou LJ, Jia WH, Yang Q,
Li YX, Liu M, Li X and Tang H: MicroRNA-372 is down-regulated and
targets cyclin-dependent kinase 2 (CDK2) and cyclin A1 in human
cervical cancer, which may contribute to tumorigenesis. J Biol
Chem. 286:25556–25563. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu G, Liu H, He H, Wang Y, Lu X, Yu Y, Xia
S, Meng X and Liu Y: miR-372 down-regulates the oncogene ATAD2 to
influence hepatocellular carcinoma proliferation and metastasis.
BMC Cancer. 14:1072014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cava C, Bertoli G, Ripamonti M, Mauri G,
Zoppis I, Rosa PA Della, Gilardi MC and Castiglioni I: Integration
of mRNA expression profile, copy number alterations, and microRNA
expression levels in breast cancer to improve grade definition.
PLoS One. 9:e976812014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
de Moor CH, Meijer H and Lissenden S:
Mechanisms of translational control by the 3′ UTR in development
and differentiation. Semin Cell Dev Biol. 16:49–58. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng HH, Zhang YD, Gong LS, Liu WD and
Zhang Y: Increased expression of microRNA-335 predicts a favorable
prognosis in primary gallbladder carcinoma. Onco Targets Ther.
6:1625–1630. 2013.PubMed/NCBI
|
|
29
|
Chang Y, Liu C, Yang J, Liu G, Feng F,
Tang J, Hu L, Li L, Jiang F, Chen C, et al: MiR-20a triggers
metastasis of gallbladder carcinoma. J Hepatol. 59:518–527. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jin K, Xiang Y, Tang J, Wu G, Li J, Xiao
H, Li C, Chen Y and Zhao J: miR-34 is associated with poor
prognosis of patients with gallbladder cancer through regulating
telomere length in tumor stem cells. Tumour Biol. 35:1503–1510.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berry KL and Hobert O: Mapping functional
domains of chloride intracellular channel (CLIC) proteins in vivo.
J Mol Biol. 359:1316–1333. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qiu MR, Jiang L, Matthaei KI,
Schoenwaelder SM, Kuffner T, Mangin P, Joseph JE, Low J, Connor D,
Valenzuela SM, et al: Generation and characterization of mice with
null mutation of the chloride intracellular channel 1 gene.
Genesis. 48:127–136. 2010.PubMed/NCBI
|
|
33
|
Tulk BM, Kapadia S and Edwards JC: CLIC1
inserts from the aqueous phase into phospholipid membranes, where
it functions as an anion channel. Am J Physiol Cell Physiol.
282:C1103–C1112. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ulmasov B, Bruno J, Woost PG and Edwards
JC: Tissue and subcellular distribution of CLIC1. BMC Cell Biol.
8:82007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ding Q, Li M, Wu X, Zhang L, Wu W, Ding Q,
Weng H, Wang X and Liu Y: CLIC1 overexpression is associated with
poor prognosis in gallbladder cancer. Tumour Biol. 36:193–198.
2015. View Article : Google Scholar : PubMed/NCBI
|