Introduction
Hepatocellular carcinoma (HCC) is a commonly
diagnosed cancer worldwide, with ~782,500 newly diagnosed cases and
>700,000 HCC-related deaths in 2012 (1). Therapeutic strategies have been
improved greatly including liver surgical resection, liver
transplantation, chemotherapy and other modalities for HCC
(2). However, the prognosis
remains extremely dismal following surgical resection or liver
transplantation in HCC patients, of which a long-term 5-year
survival was reported ranging from 25–39%, and a 5-year recurrence
rate for whom had surgical treatment was reported ranging from
65–80% (3–5). Identification of a reliable oncogene
is essential to predict the propensity of metastasis, prognosis and
suggest further therapeutic decisions. Besides, integrated
assessment for the underlying mechanisms has potential to identify
novel therapeutic targets.
Long noncoding RNAs (lncRNAs) with >200
nucleotides are transcripts non-coding for proteins. LncRNAs have
recently been found to involve in the development and progression
of various kinds of cancer as key regulators of multiple biological
processes (6). Typically, the
lncRNA HOX transcript antisense intergenic RNA (HOTAIR) has been
reported as a potential prognostic biomarker for cancers since it
was discovered in 2007 (7). HOTAIR
as a long and polyadenylated RNA that does not code for protein,
and is expressed from the development HOXC locus located on
chromosome 12q13.13 (7). HOTAIR
overexpression has been extensively observed in multiple cancers
and is reported to predict poor prognosis in esophageal carcinoma,
gastric cancer, colorectal cancer, HCC, breast cancer and other
types of cancer (8,9). There have been plenty of
investigations carried out to study the potential molecular
mechanisms of HOTAIR in cancer progression. Likewise, efforts were
made to evaluate the expression pattern of HOTAIR in HCC and
research its clinical implications (10,11).
However, the sample size ranged from 60 to 64 HCC patients and
clinicopathological parameters of each individual previous study
were relatively limited. The absence of bioinformatics analysis of
the previous studies might have missed essential information for
functional evaluation.
Therefore, the current study incorporating a larger
HCC sample size, comprehensive clinicopathological parameters, and
combined analyses of in vitro assays and functional
assessments was designed to: 1) Evaluate the expression of HOTAIR
in HCC tissues and adjacent non-tumor tissues, 2) evaluate the
association between HOTAIR expression and clinical, histological,
pathological and other biological features in HCC patients, 3) test
the effects of HOTAIR on HCC using liver cancer cell lines, 4) find
out genes associated or coordinated with HOTAIR and enrich in
pathways.
Materials and methods
Patients
Eligible HCC participants were enrolled from the
First Affiliated Hospital of Guangxi Medical University (Nanning,
China) from March 2010 to December 2011. All participated samples
were pathologically diagnosed with HCC by two independent
pathologists. In brief, the clinicopathological parameters were
collected and summarized in Table
I, including age, gender, differentiation, tumor size, tumor
nodes, clinical TNM stages, portal vein tumor embolus, metastasis,
capsular infiltration and cirrhosis, vasoinvasion as well as other
biomarkers, such as serum alpha fetal protein level detected by
ELISA, nm23, P53, P21, vascular endothelial growth factor,
microvessel density stained by CD34 through immunohistochemistry.
The Ethical Committee of First Affiliated Hospital, Guangxi Medical
University (Nanning, China) approved the current study. Informed
consents were obtained from all the enrolled patients. Related
research procedure was conducted based on the Helsinki
Declaration.
 | Table I.Relationship between the expression
of HOTAIR and clinicopathological features in HCC. |
Table I.
Relationship between the expression
of HOTAIR and clinicopathological features in HCC.
|
|
| Relative HOTAIR
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
features | n (patients) | Mean ± standard
deviation | t | P-value |
|---|
| Tissue |
|
Adjacent liver | 95 |
4.7600±1.78941 | 5.823a | <0.001 |
|
HCC | 95 |
6.3618±1.99692 |
|
|
| Age |
|
<50 | 46 |
6.1169±1.99293 | 1.237 | 0.219 |
|
≥50 | 49 |
6.6226±1.98960 |
|
|
| Gender |
|
Male | 75 |
6.3641±1.96163 | 0.022 | 0.982 |
|
Female | 20 |
6.3530±2.17753 |
|
|
|
Differentiation |
|
High | 6 |
7.1333±1.58072 |
F=1.259b | 0.289 |
|
Moderate | 60 |
6.4928±2.19732 |
|
|
|
Low | 29 |
5.9310±1.55267 |
|
|
| Size |
| <5
cm | 18 |
6.3317±2.17744 | 0.071 | 0.944 |
| ≥5
cm | 77 |
6.3688±1.96758 |
|
|
| Tumor nodes |
|
Single | 52 |
6.0300±1.84271 | −1.805 | 0.075 |
|
Multiple | 43 |
6.7630±2.12144 |
|
|
| Metastasis |
| No | 46 |
5.8957±1.88997 | −2.252 | 0.027 |
|
Yes | 49 |
6.7994±2.01415 |
|
|
| Clinical TNM
stage |
|
I~II | 22 |
5.4091±1.16452 | −3.541 | 0.001 |
|
III~IV | 73 |
6.6489±2.10944 |
|
|
| Portal vein tumor
embolus |
| No | 63 |
6.0397±1.72295 | −2.039 | 0.047 |
|
Yes | 32 |
6.9959±2.35131 |
|
|
| Vasoinvasion |
| No | 59 |
6.0231±1.69528 | −2.000 | 0.050 |
|
Yes | 36 |
6.9169±2.33170 |
|
|
| Tumor capsular
infiltration |
| No | 45 |
5.8644±1.73548 | −2.358 | 0.020 |
|
Yes | 50 |
6.8094±2.12455 |
|
|
| HBV |
| − | 17 |
7.0106±1.83662 | 1.488 | 0.140 |
| + | 78 |
6.2204±2.01345 |
|
|
| AFP |
| − | 41 |
6.2466±2.16253 | 0.491 | 0.625 |
| + | 38 |
6.4779±2.01384 |
|
|
| Cirrhosis |
| No | 50 |
6.6014±1.97817 | −1.236 | 0.219 |
|
Yes | 45 |
6.0956±2.00590 |
|
|
| NM23 |
| − | 20 |
5.5600±1.10043 | −2.920 | 0.005 |
| + | 75 |
6.5756±2.12943 |
|
|
| P53 |
| − | 40 |
6.3548±1.91948 | −0.029 | 0.977 |
| + | 55 |
6.3669±2.06894 |
|
|
| P21 |
| − | 62 |
6.3227±1.92007 | −0.260 | 0.795 |
| + | 33 |
6.4352±2.16279 |
|
|
| VEGF |
| − | 25 |
5.8720±1.74464 | −1.437 | 0.154 |
| + | 70 |
6.5367±2.06297 |
|
|
| Ki-67 |
|
Low | 47 |
6.2809±1.94552 | −0.389 | 0.689 |
|
High | 48 |
6.4410±2.06347 |
|
|
| MVD |
|
Low | 47 |
6.2894±2.21763 | −0.348 | 0.728 |
|
High | 48 |
6.4327±1.77532 |
|
|
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of HOTAIR expression
levels
RNA isolation and RNA normalization were performed
by the method descried in our previous reports (12,13).
Extracted RNA was analyzed by RT-qPCR using Applied Biosystems
PCR7900 (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The expression of HOTAIR was assessed by reverse
transcription RT-qPCR kits (Qiagen GmbH, Hilden, Germany) based on
the instructions described previously (14,15).
Human GAPDH was selected as the housekeeping reference for HOTAIR
expression analysis. Primer sequences used in the PCR were as
follows: HOTAIR 5′-GAGGGAGCCCAGAGTTACAGA-3′ (sense),
5′-TCCTCCATTTCAGCCTTTCT-3′ (antisense) and GAPDH:
5′-TGACTTCAACAGCGACACCCA-3′ (sense), 5′-CACCCTGTTGCTGTAGCCAAA-3′
(antisense). The HOTAIR expression was calculated with the formula
2−∆∆Cq (16).
Cell line and culture
The liver cancer cell line SMMC-7221 was purchased
from Shanghai Institute of Cell Biology (Shanghai, China). The cell
line was cultured at 37°C in 5% carbon dioxide in a humidified
incubator following the recommended culture conditions (14,15).
Lentiviral infection and gene
transfection
Lentivirus containing HOTAIR shRNA segments (HOTAIR
shRNA sequence was 5′-GAACGGGAGUACAGAGAGAUU-3′) was purchased from
Shanghai GenePharma Co., Ltd. (Shanghai, China). SMMC-7721 cells
were infected with the viral suspension. HOTAIR 3′ domain
(nucleotides 1 to 300 of HOTAIR) and 5′ domain (nucleotides 1500 to
2146 of HOTAIR) were inserted into pcDNA3.1 (+) plasmid. pcDNA3.1
(+)-3′ domain and pcDNA3.1 (+)-5′ domain plasmids were transfected
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. The authors
collected the transfected cell samples at 24 and 48 h points
following transfection.
Analysis of cell line
After the SMMC-7721 cells were transfected with
HOTAIR shRNA stably, the following steps to investigate the role
and molecular mechanisms of HOTAIR in HCC progression were
conducted at different time points (24 and 48 h).
i) Cell viability was evaluated using fluorimetric
detection of resorufin according to the manufacturer's protocols
(CellTiter-Blue Cell Viability Assay, G8080, Promega Corporation,
Madison, WI, USA). FL600 fluorescence plate reader was used for
fluorimetry (ex: 560 nm/em: 590 nm; Bio-Tek Instruments, Inc.,
Winooski, VT, USA).
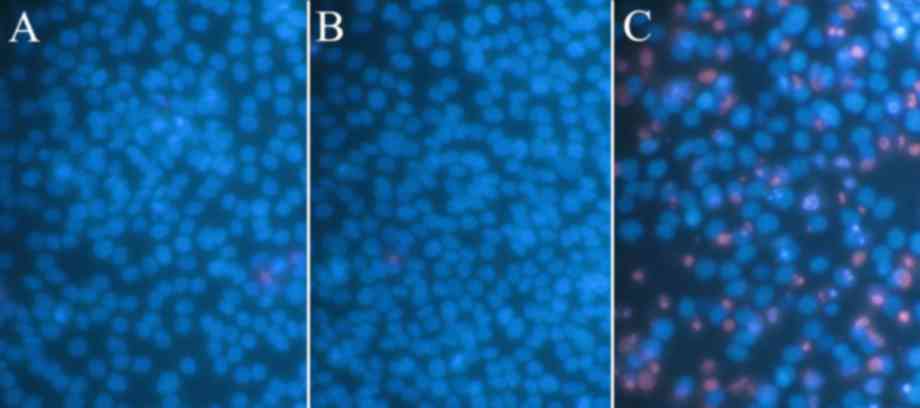
ii) Cell growth and apoptosis were evaluated with
Hoechst 33342 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and
propidium iodide (Sigma-Aldrich; Merck KGaA) double fluorescent
chromatin staining. The number of cells in different states
(necrotic, apoptotic and viable) were counted under ×200
magnification, and each type of cell was calculated in 10 diverse
fields in each well. The mean values compared with the mock control
group were shown as the final results.
iii) A microplate reader was used to measure the
cell proliferation ability (Scientific Multiskan MK3, Thermo Fisher
Scientific, Inc.) at 490 nm after the liver cancer cells were
processed by colorimetric tetrazolium (MTS) assay based on
instruction (CellTiter96 Aqueous One Solution Cell Proliferation
Assay G3580, Promega Corporation).
iv) The SMMC-7221 cells were plated into 96-well
plate and treated as indicated. The cell lysates were harvested for
caspase activity assays with caspase-Glo3/7 reagents (Promega
Corporation). The luminescence of each sample was measured with a
FL600 fluorescence plate-reading luminometer.
Construction of HOTAIR co-expression
network and biological function analysis
HOTAIR coexpression genes were calculated with
EBcoexpress package of R (17).
The gene network map was drawn in Cytoscape Web (18), thereby providing figures to
visually understand the relationship among coexpression genes. To
clarify the functions of the HOTAIR and coexpression genes, the
authors used the Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway (http://www.genome.ad.jp/kegg/) and Gene Ontology (GO)
(http://www.geneontology.org) analysis.
GO can assign genes into hierarchical categories based on the
associated aspects: Biological process, cellular component and
molecular function (19).
P<0.05 was considered to indicate a statistically significant
difference and was set as the threshold in GO and KEGG
analyses.
Statistical analysis
SPSS software (version, 20.0; IBM SPSS, Armonk, NY,
USA) was used for statistical analyses. For the analysis of the
significance of two groups, the authors performed Student's test.
Accordingly, they also applied one-way analysis of variance
followed by Dunnett's post hoc test to analysis data from
experiments in vitro. The associations between HOTAIR
expression and clinicopathological parameters were tested using the
Spearman rank correlation. The value of HOTAIR for differentiating
the HCC from noncancerous tissues was tested by receiver operating
characteristic (ROC) curve. All tests were two-sided, and P<0.05
was considered to indicate a statistically significant
difference.
Results
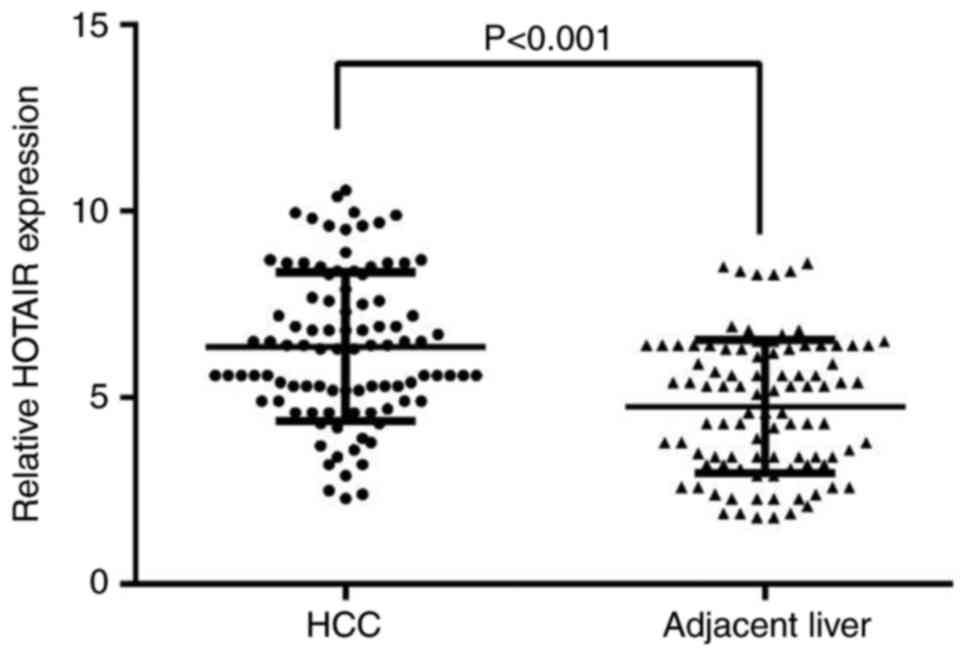
HOTAIR expression in HCC
A total of 95 eligible patients with diagnosed HCC,
available cancer and adjacent non-cancer tissues were finally
enrolled in the study. The age of the included individuals ranged
from 29 to 82 years old, and the mean age was 52 years. HOTAIR
expression was evaluated in 95 paired FFPE HCC and adjacent
non-tumor tissues using RT-qPCR. The HOTAIR expression was
significantly higher in HCC tissues, compared with the adjacent
non-tumor tissue (at least 2 cm away from the tumor node; Fig. 1). All the clinicopathological
parameters are presented in Table
I.
HOTAIR expression and
clinicopathological features of HCC
The relative expression of HOTAIR was 6.7994±2.01415
in tissues with distant metastasis, significantly higher than those
without distant metastasis (5.8957±1.88997, P=0.027). In patients
with advanced (III–IV) stage, higher HOTAIR expression
(6.6489±2.10944) was observed than patients in early (I~II) stage
(5.4091±1.16452, P=0.001). Compared with those without portal vein
tumor embolus (6.0397±1.72295), the expression level of HOTAIR was
obviously higher in patients with portal vein tumor embolus
(6.9959±2.35131, P=0.047). Similarly, HOTAIR expression was
remarkably higher in patients with vasoinvasion (6.9169±2.33170)
than those without vasoinvasion (6.0231±1.69528, P=0.005). In
addition, the level of HOTAIR distinctly upregulated in patients
with tumor capsular infiltration (6.8094±2.12455) than those with
complete liver capsule (5.8644±1.73548, P=0.020). In addition,
HOTAIR expression was visibly higher in positive nm23 expression
group of patients (6.5756±2.12943) than negative nm23 expression
group (5.5600±1.10043, P=0.005). In Spearman analysis, the current
result indicated that the relative expression of HOTAIR was
significantly positive correlated with distant metastasis (r=0.210,
P=0.041), clinical TNM stage (r=0.295, P=0.004), tumor capsular
infiltration (r=0.235, P=0.022) and nm23 (r=0.228, P=0.027).
Nevertheless, no association was identified between HOTAIR
expression and others clinicopathological features (P>0.05),
including age, gender, differentiation, size and other parameters
(Table I). Collectively, the
results above demonstrated that the expression level of HOTAIR was
associated with specific clinicopathological features that related
to tumor deterioration and HOTAIR may play a vital role in
promoting HCC progression.
Diagnostic significance of HOTAIR in
HCC
The authors took advantage of ROC analysis to
calculate the potential value of HOTAIR for diagnosing HCC. ROC
analysis demonstrated that the Area Under The Curve (AUC) of HOTAIR
was 0.715 [95% confidence interval (CI): 0.643–0.787] with a
sensitivity of 45.3% and a specificity of 86.3% in distinguishing
HCC and the cut off was 6.45. The ROC analysis of HOTAIR and
clinicopathological features revealed that HOTAIR levels remarkably
discriminated HCC patients with distant metastasis, advanced TNM
stage, tumor capsular infiltration and positive nm23 expression and
the AUC was 0.621 (95%CI: 0.509–0.734, sensitivity=24.5%,
specificity=97.8%), 0.701 (95%CI: 0.595–0.808, sensitivity=58.9%,
specificity=81.8%), 0.636 (95%CI: 0.524–0.747, sensitivity=74.0%,
specificity=51.1%) and 0.661 (95%CI:0.552–0.770, sensitivity=49.3%,
specificity=90.0%), respectively.
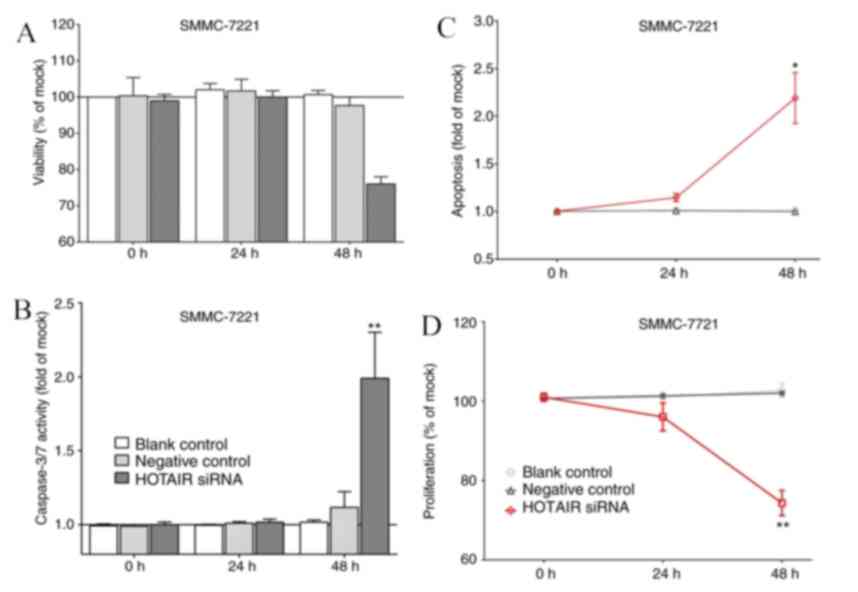
Expression of HOTAIR in the cell line
and transfected cells
The behavior alterations of SMMC-7221cells on
viability, proliferation, caspase-3/7 and apoptosis are presented
in Fig. 2. The viability and
proliferation of HOTAIR siRNA cells were significantly inhibited
compared to that in control cells in 48 h. Both levels of the
caspase3/7 activity and apoptosis in the transfected cells
exhibited higher than those in the control cells. The efficacy of
the apoptosis measurement with SMMC-7221 cell line is presented in
Fig. 3.
GO and KEGG pathway annotation
A total of 150 genes were identified in the
coexpression analysis. The coexpression genes were then annotated
through GO and KEGG pathway analyses. GO can organize genes into
hierarchical categories and show the gene functions based on
regulatory network of biological process, cellular component and
molecular functions (Table II)
(19). The genes showed dominant
enrichments in the KEGG pathways of ECM-receptor interaction, focal
adhesion and pathways in cancer (Table III).
 | Table II.Gene ontology terms enriched in
HOTAIR and coexpression genes (P<0.05, FDR<0.1). |
Table II.
Gene ontology terms enriched in
HOTAIR and coexpression genes (P<0.05, FDR<0.1).
| GO analysis | Count | P-value | FDR |
|---|
| Biological
process |
|
|
|
|
GO:0001501~skeletal system
development | 26 |
2.78×1018 |
4.37×1015 |
|
GO:0048598~embryonic
morphogenesis | 20 |
3.54×1012 |
5.57×109 |
|
GO:0048705~skeletal system
morphogenesis | 13 |
8.17×1011 |
1.29×107 |
|
GO:0048706~embryonic skeletal
system development | 11 |
4.84×1010 |
7.61×107 |
|
GO:0003002~regionalization | 14 |
5.47×109 |
8.60×106 |
|
GO:0009952~anterior/posterior
pattern formation | 12 |
1.44×108 |
2.27×105 |
|
GO:0007389~pattern
specification process | 15 |
2.58×108 |
4.07×105 |
|
GO:0030326~embryonic limb
morphogenesis | 10 |
2.87×108 |
4.52×105 |
|
GO:0035113~embryonic appendage
morphogenesis | 10 |
2.87×108 |
4.52×105 |
|
GO:0035108~limb
morphogenesis | 10 |
8.95×108 |
1.41×104 |
|
GO:0035107~appendage
morphogenesis | 10 |
8.95×108 |
1.41×104 |
|
GO:0060173~limb
development | 10 |
1.26×107 |
1.99×104 |
|
GO:0048736~appendage
development | 10 |
1.26×107 |
1.99×104 |
|
GO:0048704~embryonic skeletal
system morphogenesis | 8 |
3.23×107 |
5.09×104 |
|
GO:0006355~regulation of
transcription, DNA-dependent | 35 |
5.25×107 |
8.26×104 |
|
GO:0051252~regulation of RNA
metabolic process | 35 |
8.87×107 |
1.40×103 |
|
GO:0048562~embryonic organ
morphogenesis | 10 |
1.12×106 |
1.77×103 |
|
GO:0048568~embryonic organ
development | 11 |
1.12×106 |
1.77×103 |
|
GO:0043009~chordate embryonic
development | 14 |
2.27×106 |
3.57×103 |
|
GO:0009792~embryonic
development ending in birth or egg hatching | 14 |
2.50×106 |
3.94×103 |
|
GO:0006357~regulation of
transcription from RNA polymerase II promoter | 20 |
4.27×106 |
6.73×103 |
|
GO:0060348~bone
development | 9 |
6.12×1006 |
9.64×103 |
|
GO:0045893~positive regulation
of transcription, DNA-dependent | 15 |
2.55×105 |
4.01×102 |
|
GO:0051216~cartilage
development | 7 |
2.68×105 |
4.23×102 |
|
GO:0051254~positive regulation
of RNA metabolic process | 15 |
2.79×105 |
4.40×102 |
|
GO:0031328~positive regulation
of cellular biosynthetic process | 18 |
2.84×105 |
4.46×102 |
|
GO:0009954~proximal/distal
pattern formation | 5 |
3.03×105 |
4.76×102 |
|
GO:0009891~positive regulation
of biosynthetic process | 18 |
3.41×105 |
5.36×102 |
|
GO:0001503~ossification | 8 |
3.67×105 |
5.77×102 |
|
GO:0007155~cell adhesion | 18 |
3.73×105 |
5.87×102 |
|
GO:0022610~biological
adhesion | 18 |
3.80×105 |
5.98×102 |
|
GO:0045944~positive regulation
of transcription from RNA polymerase II promoter | 13 |
3.95×105 |
6.22×102 |
|
GO:0045941~positive regulation
of transcription | 16 |
4.02×105 |
6.33×102 |
|
GO:0021515~cell
differentiation in spinal cord | 5 |
4.27×105 |
6.72×102 |
|
GO:0051173~positive regulation
of nitrogen compound metabolic process | 17 |
4.94×105 |
7.78×102 |
|
GO:0010628~positive regulation
of gene expression | 16 |
5.64×105 |
8.88×102 |
|
GO:0010557~positive regulation
of macromolecule biosynthetic process | 17 |
5.94×105 |
9.34×102 |
| Cellular
component |
|
|
|
|
GO:0031012~extracellular
matrix | 18 |
4.61×1010 |
5.51×107 |
|
GO:0005578~proteinaceous
extracellular matrix | 16 |
1.07×108 |
1.28×105 |
|
GO:0044420~extracellular
matrix part | 10 |
1.83×107 |
2.20×104 |
|
GO:0044421~extracellular
region part | 22 |
4.77×106 |
5.713×103 |
| Molecular
function |
|
|
|
|
GO:0043565~sequence-specific
DNA binding | 25 |
5.21×1011 |
6.68×108 |
|
GO:0003700~transcription
factor activity | 29 |
1.59×109 |
2.04×106 |
|
GO:0030528~transcription
regulator activity | 33 |
1.36×107 |
1.74×104 |
|
GO:0005201~extracellular
matrix structural constituent | 7 |
6.40×105 |
8.21×102 |
 | Table III.KEGG pathways enriched in HOTAIR and
coexpression genes (P<0.05). |
Table III.
KEGG pathways enriched in HOTAIR and
coexpression genes (P<0.05).
| Pathway | Count | P-value | FDR | Genes |
|---|
| KEGG |
|
|
|
|
|
hsa04512:ECM-receptor
interaction | 5 |
7.85×104 |
7.28×101 | TNC, COL1A2,
COL6A1, LAMB1, COL5A2 |
|
hsa04510:Focal adhesion | 5 |
1.79×102 |
1.55×101 | TNC, COL1A2,
COL6A1, LAMB1, COL5A2 |
|
hsa05200:Pathways in
cancer | 6 |
2.32×102 |
1.96×101 | FGFR1, HDAC2,
TGFB3, LAMB1, GLI3, MMP2 |
Discussion
HCC is a critical health problem worldwide and is an
aggressive disease. The high morbidity and mortality rates of this
malignance are not arbitrary but raise the researcher's attention.
In the present study, the HOTAIR expression level was detected
significantly higher in the HCC tissues than the adjacent non-tumor
tissues. The HOTAIR expression levels were observed significantly
higher in tumor samples from patients with distant metastasis,
advanced stage, portal vein tumor embolus, vasoinvasion, tumor
capsular infiltration or positive nm23 than those from patients
without distant metastasis, advanced stage, portal vein tumor
embolus, vasoinvasion, tumor capsular infiltration or positive nm23
correspondingly. In vitro assays, the silencing of HOTAIR in
liver cancer cells induced the promotion of apoptosis and
inhibition of cell proliferation. ECM-receptor interaction, focal
adhesion and pathways in cancer were annotated with the HOTAIR and
coexpression genes.
HOTAIR has been evaluated extensively within the
context of cancer prognosis (20).
A meta-analysis for patients with solid tumors reported that higher
HOTAIR expression could significantly predict worse overall
survival (21). In gastric cancer,
high expression of HOTAIR was significantly associated with the
depth of tumor invasion, lymph node metastasis, vessel invasion,
lymphatic vessel involvement and TNM stage (22). The incidence of lymph node
metastasis was illustrated to be higher in cancer patients with
high HOTAIR expression compared to patients with low HOTAIR
expression (23). Likewise, HOTAIR
was indicated to be a potential predictor of poor relapse-free
survival, metastasis-free survival and disease-free survival in
cervical, ovarian, breast and endometrial cancer patients (24).
To the best of the authors' knowledge, the current
study contained a larger HCC sample than the previous
investigations for HCC and HOTAIR. Compared with adjacent non-HCC
tissue, the analysis yielded that HOTAIR expressed higher in the
HCC tissue, which was observed in several previous reports,
implying the diagnosis value of HOTAIR in HCC patients (11,25–27).
Ishibashi et al (11) found
that patients with HOTAIR expression had significantly larger
primary tumor size than those without HOTAIR expression. A vast
majority of samples from study of Geng et al (26) showed higher levels of HOTAIR in
tumor tissues than in adjacent non-tumor tissues, and HOTAIR
expression were significantly higher in tumor tissues from patients
with lymph node metastasis. The HOTAIR expression was reported
increased at least two-fold in HCC tumor tissues relative to that
in non-tumor tissues (27). These
evidences were robust to illustrate that HOTAIR might be an
oncogene for HCC.
In the current study, a Spearman's rank correlation
coefficient analysis showed that the higher HOTAIR expression level
was significantly correlated with metastasis, advanced TNM stage
and portal vein tumor embolus. Besides, HOTAIR expression was
remarkably higher in patients with vasoinvasion than those without
vasoinvasion, as well as in capsular infiltration than those with
complete liver capsule. The clinicopathological characteristics of
vasoinvasion, capsular infiltration always correlated to invasion
and progression, suggesting HOTAIR might be involved in HCC
invasion and serve as poor prognosis predictor. These findings
allowed HOTAIR to be a potential molecular for risk classification
of HCC patients, and that molecular-based tumor risk stratification
was important for the decision of individual diagnosis and therapy
(25,28). ROC analysis demonstrated that the
AUC of HOTAIR was 0.715 (95%CI: 0.643–0.787) with a sensitivity of
45.3% and a specificity of 86.3% in distinguishing HCC. The authors
speculated that an individual biomarker was insufficient to
diagnosis malignant disease, and a combined analysis of the
well-studied diagnosis biomarkers might provide novel insight into
the diagnosis method for HCC.
Given the clinical implications of HOTAIR upon HCC
and the currently poor interpretation of its mechanisms, a series
of in vitro assays were conducted to uncover the effects of
HOTAIR on HCC. After silencing HOTAIR expression, the proliferation
and viability of SMMC-7221 cells were depressed, while the
significant promotion of apoptosis and caspase ability was
observed. Furthermore, the level of the depressed and promoted
effects was associated with the transfection processing time,
indicating the high HOTAIR expression level was related to the
increased HCC progression risk. Taken the in vitro results
together, it demonstrated that HOTAIR involved in promoting HCC
cell growth and inhibiting apoptosis, explaining the phenomenon
that the increased HOTAIR expression was found in HCC tissues and
the significant associations with the advanced TNM stage (26). Moreover, the ROC analysis of HOTAIR
and distant metastasis, advanced TNM stage and tumor capsular
infiltration also indicated expression of HOTAIR might be used for
molecular based tumor staging or progressive risk stratification.
Since increased HOTAIR expression was not exclusively specific in
one particular cancer, and observed promoting cell proliferation,
invasion and migration in variant types of tumor cells, the authors
speculated high HOTAIR expression could be a potential marker for
predicting poor prognosis in multiple types of cancers (20,29,30).
It was well recognized that HOTAIR interacted with
multiple genes in a manner dependent on polycomb repressive complex
2 (PRC2), which could induce histone H3 lysine 27 (H3K27)
methylation and thus silence transcription of the HOXD locus, and
eventually cause epigenetic silencing of metastasis suppressor
genes (31–33). Knockdown of HOTAIR was associated
with reductions in levels of vascular endothelial growth factor
protein and matrix metalloproteinase-9, which were vital for cell
motility and metastasis (26).
Besides, HOTAIR was found to promote cell migration and invasion
via inhibiting RNA binding motif protein 38 (RBM38) in HCC cells,
since knockdown of HOTAIR raised the expression RBM38 both on mRNA
levels and protein levels, and the increased expression of RBM38
could restrain cell motility (10). A previous study reported HOTAIR
enhanced cell viability and escaped G1-phase arrest through
suppressing miRNA-218 expression and inhibiting P14 and P16
signaling. Suppressing oncogene Bmi-1 was shown to be a functional
target of miR-218, and the main downstream targets signaling,
P16(lnk4a) and P14(ARF), were inactivated in HOTAIR tumorigenesis
(34). In HepG2 cells, the
phenomenon that microRNA miR-125a-5p decreased and released caspase
2 to promote HCC cell apoptosis was reported after HOTAIR knockdown
(35). Therefore, HOTAIR might
control HCC cell proliferation through interacting with microRNAs.
The identification of microRNAs and target genes of HOTAIR in HCC
is of great importance to understand HCC pathogenesis.
Although accumulating evidence has revealed
substantial biological functions of HOTAIR, the precise mechanisms
remained largely to be explored. Coexpression network analysis,
which was used to find genes having similar or coordinated
expression patterns, was carried out to identify HOTAIR
coexpression genes (36,37). GO enrichment analysis revealed that
a majority of the HOTAIR and coexpression genes were involved in
pathways related to cell adhesion, biological adhesion,
transcription regulator activity, transcription factor activity and
sequence-specific DNA binding. These data supported that most
lncRNAs worked with DNA-binding proteins, and epigenetically
regulated the expression of multiple genes (7,31).
It is noteworthy that in HCC patients with positive nm23, which has
also been reported to contribute to metastasis (38), HOTAIR expression level was higher
than that in nm23 negative patients. To some extent, the
significant correlation between HOTAIR expression and nm23 status
suggested that they might function in a manner of cooperation.
However, a previous study reports that the absence of nm23 in HCC
is significantly correlated with extrahepatic metastasis (39). In mice models, nm23 has been also
observed overexpressed in the HCC, whereas lung metastasis was
found to be promoted in the transgenic nm23-null mice (40). The oncogenic and metastasis role of
the nm23 is still debated. The contradiction between the results
suggests future studies with larger samples are needed to
illustrate the function of nm23.
In addition, three pathways were screened out using
KEGG analysis for the HOTAIR and coexpression genes, such as
ECM-receptor interaction, focal adhesion and pathways in cancer.
Most of these pathways were reported involving in cancer
progression (41). Typically,
pathways linked to cell spread and migration was both annotated in
GO and KEGG, such as cell adhesion, biological adhesion and focal
adhesion (42,43). It strongly suggests that HOTAIR
might be a therapeutic target to decrease metastasis risk, which
remained a field to be investigated (44).
Concerning with the limitations, the absence of
survival data hindered this study to gain prognosis value of HOTAIR
in HCC. As for control tissue origin, only the adjacent
noncancerous tissues including cirrhotic and noncirrhotic liver
tissues were available. Given the multi-stages hepatocarcinogenesis
process of HCC (45), the authors
suggested that the combined analysis of normal tissues might reveal
more in-depth mechanisms of HOTAIR in HCC progression. A future
study to validate HOTAIR mechanisms in HCC patients is
necessary.
The current study suggested that HOTAIR was an
oncogene in HCC. It functioned though promoting tumor cell growth
and inhibiting apoptosis. HOTAIR potentially regulated HCC
metastatic progression by several pathways correlated cell
adhesion, and might be a therapeutic target in future.
Acknowledgements
The present study was supported by grants from the
Fund of Future Academic Star of Guangxi Medical University (grant
no. WLXSZX16001) and the Scientific Research Project of the
Department of Education in Guangxi Zhuang Autonomous Region (grant
nos. LX2014064 and 201204LX044).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kanda T, Ogasawara S, Chiba T, Haga Y,
Omata M and Yokosuka O: Current management of patients with
hepatocellular carcinoma. World J Hepatol. 7:1913–1920. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamiyama T, Nakanishi K, Yokoo H, Kamachi
H, Tahara M, Suzuki T, Shimamura T, Furukawa H, Matsushita M and
Todo S: Recurrence patterns after hepatectomy of hepatocellular
carcinoma: Implication of Milan criteria utilization. Ann Surg
Oncol. 16:1560–1571. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sakon M, Umeshita K, Nagano H, Eguchi H,
Kishimoto S, Miyamoto A, Ohshima S, Dono K, Nakamori S, Gotoh M and
Monden M: Clinical significance of hepatic resection in
hepatocellular carcinoma: Analysis by disease-free survival curves.
Arch Surg. 135:1456–1459. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Portolani N, Coniglio A, Ghidoni S,
Giovanelli M, Benetti A, Tiberio GA and Giulini SM: Early and late
recurrence after liver resection for hepatocellular carcinoma:
Prognostic and therapeutic implications. Ann Surg. 243:229–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kugel JF and Goodrich JA: Non-coding RNAs:
Key regulators of mammalian transcription. Trends Biochem Sci.
37:144–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang S, Chen S, Yang G, Gu F, Li M, Zhong
B, Hu J, Hoffman A and Chen M: Long noncoding RNA HOTAIR as an
independent prognostic marker in cancer: A meta-analysis. PLoS One.
26:e1055382014. View Article : Google Scholar
|
|
9
|
Deng Q, Sun H, He B, Pan Y, Gao T, Chen J,
Ying H, Liu X, Wang F, Xu Y and Wang S: Prognostic value of long
non-coding RNA HOTAIR in various cancers. PLoS One. 9:e1100592014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding C, Cheng S, Yang Z, Lv Z, Xiao H, Du
C, Peng C, Xie H, Zhou L, Wu J and Zheng S: Long non-coding RNA
HOTAIR promotes cell migration and invasion via down-regulation of
RNA binding motif protein 38 in hepatocellular carcinoma cells. Int
J Mol Sci. 15:4060–4076. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen G, Umelo IA, Lv S, Teugels E, Fostier
K, Kronenberger P, Dewaele A, Sadones J, Geers C and De Grève J:
miR-146a inhibits cell growth, cell migration and induces apoptosis
in non-small cell lung cancer cells. PLoS One. 8:e603172013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rong M, He R, Dang Y and Chen G:
Expression and clinicopathological significance of miR-146a in
hepatocellular carcinoma tissues. Ups J Med Sci. 119:19–24. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang S, He R, Rong M, Dang Y and Chen G:
Synergistic effect of MiR-146a mimic and cetuximab on
hepatocellular carcinoma cells. Biomed Res Int.
2014:3841212014.PubMed/NCBI
|
|
15
|
Dang YW, Zeng J, He RQ, Rong MH, Luo DZ
and Chen G: Effects of miR-152 on cell growth inhibition, motility
suppression and apoptosis induction in hepatocellular carcinoma
cells. Asian Pac J Cancer Prev. 15:4969–4976. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dawson JA, Ye S and Kendziorski C:
R/EBcoexpress: An empirical Bayesian framework for discovering
differential co-expression. Bioinformatics. 28:1939–1940. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lopes CT, Franz M, Kazi F, Donaldson SL,
Morris Q and Bader GD: Cytoscape web: An interactive web-based
network browser. Bioinformatics. 26:2347–2348. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Serghiou S, Kyriakopoulou A and Ioannidis
JP: Long noncoding RNAs as novel predictors of survival in human
cancer: A systematic review and meta-analysis. Mol Cancer.
15:502016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Miao Z, Ding J, Chen B, Yang Y and Chen Y:
HOTAIR overexpression correlated with worse survival in patients
with solid tumors. Minerva Med. 107:392–400. 2016.PubMed/NCBI
|
|
22
|
Liu FT, Qiu C, Luo HL, Zhang Y, Xia GF,
Hao TF and Zhu PQ: The association of HOTAIR expression with
clinicopathological features and prognosis in gastric cancer
patients. Panminerva Med. 58:167–174. 2016.PubMed/NCBI
|
|
23
|
Cai B, Wu Z, Liao K and Zhang S: Long
noncoding RNA HOTAIR can serve as a common molecular marker for
lymph node metastasis: A meta-analysis. Tumour Biol. 35:8445–8450.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li J, Wen W, Zhao S, Wang J, Chen J, Wang
Y and Zhang Q: Prognostic role of HOTAIR in four estrogen-dependent
malignant tumors: A meta-analysis. Onco Targets Ther. 8:1471–1482.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Geng YJ, Xie SL, Li Q, Ma J and Wang GY:
Large intervening non-coding RNA HOTAIR is associated with
hepatocellular carcinoma progression. J Int Med Res. 39:2119–2128.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gao JZ, Li J, Du JL and Li XL: Long
non-coding RNA HOTAIR is a marker for hepatocellular carcinoma
progression and tumor recurrence. Oncol Lett. 11:1791–1798.
2016.PubMed/NCBI
|
|
28
|
Schmidt C and Marsh JW: Molecular
signature for HCC: Role in predicting outcomes after liver
transplant and selection for potential adjuvant treatment. Curr
Opin Organ Transplant. 15:277–282. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Wu Z, Mei Q, Li X, Guo M, Fu X and
Han W: Long non-coding RNA HOTAIR, a driver of malignancy, predicts
negative prognosis and exhibits oncogenic activity in oesophageal
squamous cell carcinoma. Br J Cancer. 109:2266–2278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jing L, Yuan W, Ruofan D, Jinjin Y and
Haifeng Q: HOTAIR enhanced aggressive biological behaviors and
induced radio-resistance via inhibiting p21 in cervical cancer.
Tumour Biol. 36:3611–3619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fu WM, Zhu X, Wang WM, Lu YF, Hu BG, Wang
H, Liang WC, Wang SS, Ko CH, Waye MM, et al: Hotair mediates
hepatocarcinogenesis through suppressing miRNA-218 expression and
activating P14 and P16 signaling. J Hepatol. 63:886–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tang L, Shen H, Li X, Li Z, Liu Z, Xu J,
Ma S, Zhao X, Bai X, Li M, et al: MiR-125a-5p decreases after long
non-coding RNA HOTAIR knockdown to promote cancer cell apoptosis by
releasing caspase 2. Cell Death Dis. 7:e21372016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Carlson MR, Zhang B, Fang Z, Mischel PS,
Horvath S and Nelson SF: Gene connectivity, function, and sequence
conservation: Predictions from modular yeast co-expression
networks. BMC Genomics. 7:402006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ma S, Shi M, Li Y, Yi D and Shia BC:
Incorporating gene co-expression network in identification of
cancer prognosis markers. BMC Bioinformatics. 11:2712010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Guo H, Nan K, Hu T, Meng J, Hui W, Zhang
X, Qin H and Sui C: Prognostic significance of co-expression of
nm23 and p57 protein in hepatocellular carcinoma. Hepatol Res.
40:1107–1116. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
An R, Meng J, Shi Q, Dai XX, Chen JH, Lei
YJ, Shan B, Gao C, Chu YL and Dong XP: Expressions of nucleoside
diphosphate kinase (nm23) in tumor tissues are related with
metastasis and length of survival of patients with hepatocellular
carcinoma. Biomed Environ Sci. 23:267–272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Boissan M, Wendum D, Arnaud-Dabernat S,
Munier A, Debray M, Lascu I, Daniel JY and Lacombe ML: Increased
lung metastasis in transgenic NM23-Null/SV40 mice with
hepatocellular carcinoma. J Natl Cancer Inst. 97:836–845. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu P, Jiang W, Ren H, Zhang H and Hao J:
Exploring the molecular mechanism and biomakers of liver cancer
based on gene expression microarray. Pathol Oncol Res.
21:1077–1083. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim DH and Wirtz D: Predicting how cells
spread and migrate: Focal adhesion size does matter. Cell Adh Migr.
7:293–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang CC, Chang SF, Chao JK, Lai YL, Chang
WE, Hsu WH and Kuo WH: Activation of AMP-activated protein kinase
attenuates hepatocellular carcinoma cell adhesion stimulated by
adipokine resistin. BMC Cancer. 14:1122014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bugianesi E, Leone N, Vanni E, Marchesini
G, Brunello F, Carucci P, Musso A, De Paolis P, Capussotti L,
Salizzoni M and Rizzetto M: Expanding the natural history of
nonalcoholic steatohepatitis: From cryptogenic cirrhosis to
hepatocellular carcinoma. Gastroenterology. 123:134–140. 2002.
View Article : Google Scholar : PubMed/NCBI
|