Introduction
Osteosarcoma (OS) may be fatal, particularly in
adolescents (1). Greater than 50%
of bone cancers are OS (2).
Despite advances in chemotherapy and radiotherapy, the 5-year
survival rate for OS patients remains low (3). The long-term prognosis of OS patients
is poor, largely due to frequent metastasis, high recurrence rate
and lack of effective therapeutic intervention (4). Therefore, a more detailed
understanding of the underlying mechanisms of OS development and
effective therapeutics are required.
Long non-coding RNAs (lncRNAs) denote a class of
RNAs longer than 200 nucleotides in length (5,6), and
may serve a role in biological processes including development and
angiogenesis, differentiation and cell growth (7–9).
Recent studies have suggested that lncRNAs may act as either
oncogenes or tumor suppressors and contribute to the tumorigenesis
of multiple cancers including OS (10,11).
For example, the lnc-RNA, metastasis associated lung adenocarcinoma
transcript 1, promotes the development of OS (12). Overexpression of the lncRNA MFI2
also increases the proliferation and progression of OS cells
(11). Another report suggested an
oncogenic role of HULC which is another lncRNA and is highly
up-regulated in liver cancer in OS, when deregulated levels of it
predicted poor prognosis (13). A
well-characterized lncRNA, HOXA transcript at the distal tip, has
also been demonstrated to be upregulated in OS cells and in patient
samples and therefore contributes to the development of OS
(14). Another lncRNA, tumor
suppressor candidate 7, may instead inhibit OS progression via an
unknown mechanism (15).
Consistently, the lncRNA, long intergenic non-protein coding RNA
161, sensitizes OS cells to cisplatin-induced cell death and may be
a candidate tumor suppressor (16). Therefore, lncRNAs may be applied
for OS diagnosis and may act as novel therapeutic targets.
LncRNA, cancer susceptibility candidate 2 (CASC2) is
located at chromosome 10q26, and is a novel cancer-associated
lncRNA. Previously, decreased CASC2 expression and its tumor
suppressive function has been demonstrated in various tumor types
(17–21). However, the expression and
potential role of CASC2 in OS remains unclear.
In the current study, the function of CASC2 in OS
was investigated. CASC2 expression was significantly decreased in
OS tissues compared with normal adjacent tissues. The association
of CASC2 levels with different clinicopathological features was
also studied. Patients with greater CASC2 expression displayed
improved survival. Increased CASC2 expression impeded the
proliferation, invasion and migration of OS cells. Consistently,
increasing CASC2 expression also promoted apoptosis, whereas CASC2
knockdown exerted the opposite effect. In vivo implantation
studies suggested that CASC2 overexpression decreased the oncogenic
potential of OS cells. The data collectively suggested a tumor
suppressive role of CASC2 and may provide an insight into the
diagnosis of OS.
Materials and methods
Cell culture and human samples
OS cell lines used in the current study, 143B, U-2
OS, KHOS-240S, D22, Saos-2, HOS and MG-63, and a normal cell line,
hFOB, were purchased from the American Type Culture Collection
(Manassas, VA, USA). The OS cells were maintained in RPMI-1640
medium (Tiangen Biotech Co., Ltd., Beijing, China) supplemented
with 5% fetal calf serum (FCS; Tiangen Biotech Co., Ltd.),
streptomycin (50 µg/ml; Tiangen Biotech Co., Ltd.) and penicillin
(200 U/ml, Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 20°C
and 5% CO2. Matched fresh OS specimens and normal
adjacent tissues were collected from 97 patients who had undergone
resection at the Second Xiangya Hospital, Central South University
(Hunan, China) between March 2010 and October 2014. Immediately
after surgical resection, these tissues were stored at −80°C until
usage. None of the patients had received preoperative chemotherapy
or radiotherapy. All patients signed formal consent forms. Overall
survival was calculated from the day of primary surgery to death or
last follow-up. The research performed using human specimens was
reviewed and approved by the Ethics Committee of the Second Xiangya
Hospital, Central South University (Hunan, China).
CASC2 overexpression and
knockdown
A sequence of genomic DNA encoding the cDNA for
CASC2 and flanking sequences was amplified by PCR and cloned
into the EcoRI and XhoI (both purchased from Roche Diagnostics,
Shanghai, China) sites of pCDNA3.1 vector (50 nM, Tiangen Biotech
Co., Ltd.). For transfection, the empty pcDNA3.1 vector was used as
a control. The sequence for CASC2 insert was: 5-ACA ACA AGA AAC TTC
CCC AAG GTA TCA TTA TAG TCT TTA GAC TTC AGACA C ACA CCA CAC CTC AAA
TAT ATA CAC AAC TGA AAG GAA AAT TAA GGA AGT TTT TCA AAG AAC CCT ATT
CCG AGT AAG AAG TGT GTT GCA TGA ATT TCT AAG AGC CAG AAA ATG CAT GAC
ACA GGA GAA GAT GTA CCC TCA TCT GTT CAG TGA GAG ATG TGC AAA TCA ACA
TCA ACA CAG AAC TGC TGA AGA AAA AAA ATA TGT CTC TGA AAA GCA ACT TAT
TCA CTG GAG ATG TGA GGA GCC ATC CGC ACA TCA CAA TTC TAT AGACAT CAA
ACG CAT GAA GCA TTT CGG ATC TGC TTT AAG ACT GAG GCA GAC TTT CCA TCT
GGA CAC AGC CGA CCA TCC ATG TGT CAT TAC AAT GAA TCC AGC ACT TCC-3.
The negative control siRNA was also introduced (Shanghai
GenePharma, Co., Ltd., Shanghai, China). CASC2 small interfering
RNAs (si-CASC2, 50 nM) were synthesized by Shanghai GenePharma,
Co., Ltd. The si-CASC2 sequence was: 5-AAG GCT GTA TGC TGT ATC ATA
CCC TGT TCT CCC GGG TTT TGG CCA CTG ACT GAC CCG GGA GAA G-3.
Transfections were performed using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol for 24 h at 20°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNAs were isolated from 143B, U-2 OS,
KHOS-240S, D22, Saos-2, HOS, MG-63 and hFOB cell lines and human
samples with TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). A total of 5 ng RNA in a final volume of 10 µl containing 5
mM dNTP mix (Tiangen Biotech Co., Ltd.) was used to generate
complementary DNA using the SYBR Premix Taq™ toolkit
(Takara Biotechnology Co., Ltd., Dalian, China). The mixture was
maintained at 70°C for 5 min and then a mixture composed of 5xRT
buffer, 50 U/µl reverse transcriptase (M-MLV), 100 U/µl RNase
inhibitor was added (Tiangen Biotech Co., Ltd.). GAPDH was
used as the control. Reactions were performed using the ABI
PRISM® 7000 Sequence Detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The expression of CASC2 was
calculated using the 2−ΔΔCq method (22). The experiments were performed ≥3
times. The primer sequences were as follows: Sense,
5′-GAATGCTAGCTTACG-3′ and anti-sense, 5′-CTGAGTGCTTGACATGT-3′ for
CASC2; sense, 5′-GATTAGCTCTGCACGTT-3′ and anti-sense,
5′-ATGAGCATTACAGTGTT-3′ for GAPDH. The cycling conditions
were: 55°C for 2 min, 95°C for 15 min followed by 32 cycles of 95°C
for 15 sec and 60°C for 1 min. Experiments were performed in
triplicates.
Proliferation assay
In the proliferation study, a Cell Counting kit-8
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was used.
After treatment for 24 h, MG-63 and HOS cells were re-suspended and
seeded into a 6-well plate (1×105 cells/well) for 5
days. A total of 20 µl CCK-8 solution was added into the culture to
a final concentration of 10 mg/ml at 20°C. The crystalline formazan
product was resolved in 100 µl 10% SDS solution for one day at
20°C. The optical density was detected at a wavelength of 490 nm
and each assay was repeated three times.
Transwell invasion and migration
assays
Cell invasion and migration assays were performed
using 24-well Transwell chambers (8 µm pore size; BD Biosciences,
Franklin Lakes, NJ, USA). For the migration assay, 1×106
MG-63 and HOS cells were suspended in 100 µl RPMI-1640 serum-free
medium (Tiangen Biotech Co., Ltd., Beijing, China) and placed in
the top chambers. Dulbecco's modified Eagle's medium (500 µl,
Sigma-Aldrich; Merck KGaA, Drmstadt, Germany) containing 10% FCS
was added to the bottom chambers. Following 24 h of incubation at
37°C, the cells that did not migrate into the pores were removed
using a cotton swab, and cells on the lower surfaces of the
membrane were stained with crystal violet (0.40%) for 1 h at 20°C
and evaluated. The invasion assay was similar to that of the
migration assay except that the cell culture surface was firstly
coated with Matrigel (BD Biosciences, Franklin Lakes, NJ, CA, USA).
A fluorescence microscope (DM-IRB; Leica Microsystems GmbH,
Wetzlar, Germany) was used to visualize the results and CELLCOUNTER
(https://bitbucket.org/linora/cellcounter/downloads)
was used for quantification. A total of five fields were assessed
and experiments were performed in triplicates.
Cell cycle and apoptosis analysis
After a 48 h transfection, MG-63 and HOS cells were
harvested and washed with cold PBS. Subsequently, cells were fixed
with 70% ethanol at 4°C overnight. The fixed cells were then
stained with propidium iodide (PI; Sigma-Aldrich; Merck KGaA) at
4°C for 30 min in the dark. The fraction of cells in
G0/G1, S and G2/M phases were
measured using a fluorescence-activated cell sorting (FACS)
instrument (BD Biosciences). The experiments were performed in
triplicate.
For the apoptosis assay, transfected cells were
examined using a fluorescein isothiocyanate-labeled Annexin V/PI
Apoptosis Detection kit (Beyotime Institute of Biotechnology,
Haimen, China) following the manufacturer's protocol. Flow
cytometric analysis was performed immediately after staining. Data
acquisition and analysis were performed using a FACS instrument (BD
Biosciences).
In vivo implantation and
immunohistochemistry
Transduction of CASC2 into ~5×106 MG-63
cells was performed using pcDNA3.1 vector as described above. The
system was initially maintained for 24 h at 20°C. Following this,
cells were re-suspended and 1×106 cells were injected
subcutaneously into the rear flank of nude mice (n=32, female, 4~5
weeks old, average weight 16.7 g). Mice were housed at 20°C, with
55–60% humidity and a light-dark cycle of 12/12 h. Ad
libitum access to food and water was supplied. After 30 days,
mice were sacrificed by sodium amobarbital (Sigma-Aldrich; Merck
KGaA) overdose and solid tumors were resected and weighted. The
tumor sections were examined by two experienced pathologists who
were double blinded to the data. Nude mice were obtained from the
Model Animal Research Center of Nanjing University (Nanjing,
China). Tumor samples were fixed in 10% buffered formalin for 12 h
at 20°C, embedded in paraffin, and cut into 5 µm sections. After
deparaffinization and rehydration, antigens were retrieved with 1×
Cytomation target retrieval solution (Dako; Agilent Technologies,
Inc., Santa Clara, CA, USA) in a decloaker chamber at 95°C for 20
min and then at room temperature for 20 min followed by sequential
rinsing with distilled water and PBS at room temperature. Slides
were incubated with hydrogen peroxide for 5 min to quench
endogenous hydrogen peroxidase activity. After rinsing twice with
TBS with 0.1% Tween 20, slides were incubated with Ki-67 antibodies
(1:1,000, cat. no. P6834, Sigma-Aldrich; Merck KGaA). Slides were
counterstained with hematoxylin for 5 min and rinsed with distilled
water. 3,3-Diaminobenzidine (DAB) was used as a chromogen (cat. no.
D8001, Sigma-Aldrich; Merck KGaA). Slides were evaluated under an
optical light microscope (Olympus Corporation, Tokyo, Japan).
Images were captured using ×100 magnification. All the slides were
manually scored as previously described (23). Briefly, three random sections from
each animal of all groups were scored for the intensity of
staining. H-score were calculated as H-score=Σ(1+i)
pi, where i is the intensity score (0: no staining,
1: weak staining, 2: moderate staining; 3: Strong staining) and
pi is the percentage of cells exhibiting that intensity
(24). The median value was used
as the cutoff, as previously suggested (25).
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical significance was determined by Student's t-test or
one-way analysis of variance followed by the least significant
difference post hoc test, using SPSS software version 15.0 (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference. The Kaplan-Meier survival
curve was tested using log-rank test. Fisher exact test was used to
evaluate the association between CASC2 and clinicopathological
characteristics.
Results
lncRNA CASC2 is downregulated in
specimens and OS cell lines
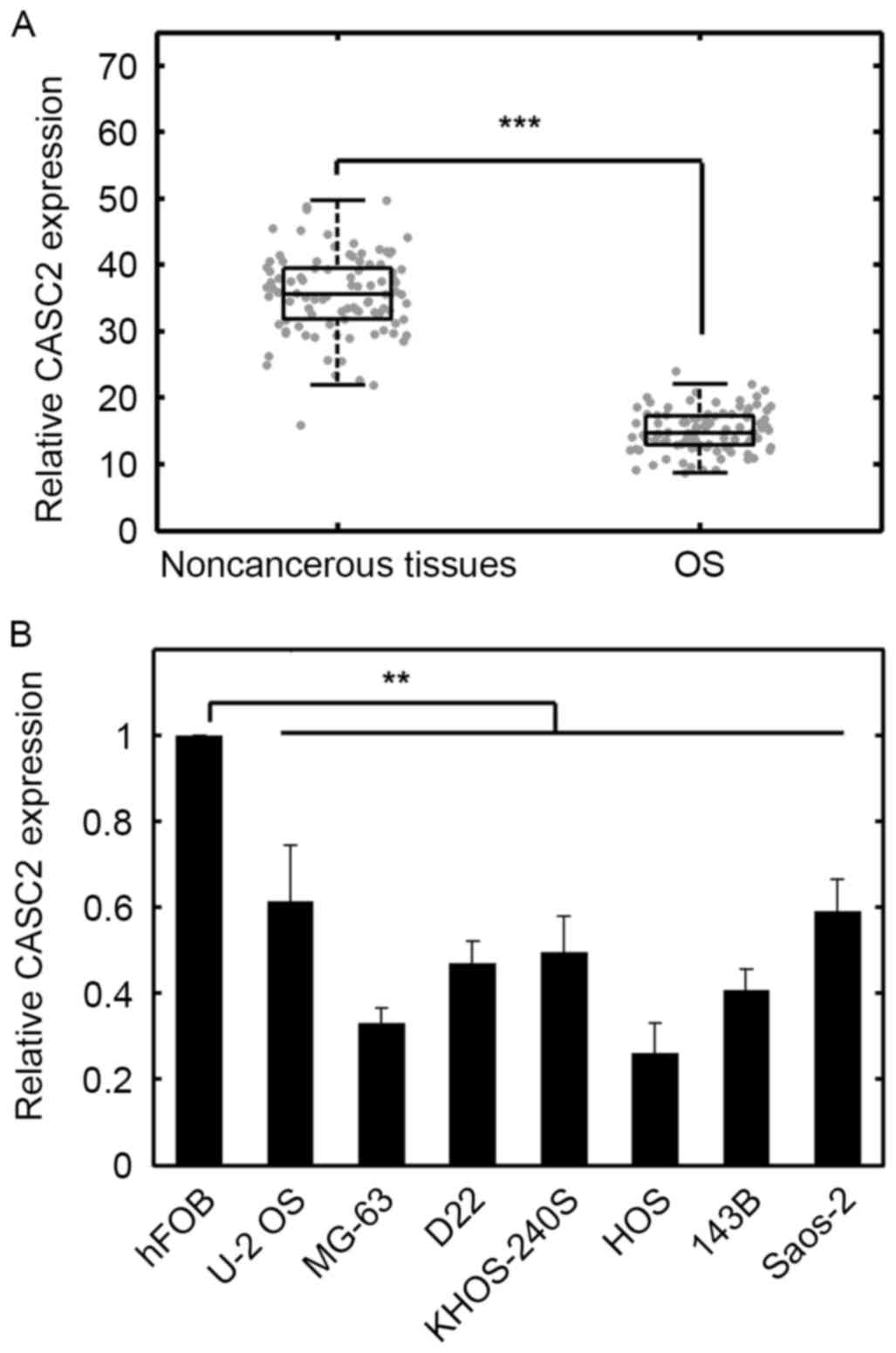
To quantify the expression levels of CASC2,
RT-qPCR was conducted. The results indicated that the expression
levels of CASC2 were significantly downregulated in OS
samples compared with normal adjacent tissues (Fig. 1A; P<0.001). In addition, in a
number of OS cell lines, CASC2 was decreased compared with
that of normal bone cells (Fig.
1B; P<0.01). The median value was used as the cutoff, as
previously suggested (25).
CASC2 protein expression in OS patient tissue was
not significantly associated with age and gender of the patients
(Table I); however, CASC2 levels
were significantly associated with differentiation, tumor, nodes,
metastasis (TNM) stages and metastasis (Table I). These results suggested that
CASC2 expression was lower in OS cell lines and in human OS tissue
samples, compared with normal controls. As downregulated CASC2 in
OS tissues implies a tumor suppressive role for CASC2, the tumor
cell lines with relatively low CASC2 expression may possibly
exhibit greater tumorigenic potential compared with the cell lines
with higher CASC2 expression levels. MG-63 and HOS cells
exhibited the lowest CASC2 expression (Fig. 1B). To evaluate the potential tumor
suppressive role of CASC2, we selected these two cell lines for
further analysis.
 | Table I.Association between CASC2 expression
and clinicopathological features. |
Table I.
Association between CASC2 expression
and clinicopathological features.
|
|
| CASC2 expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | No. | High [n (%)] | Low [n (%)] | P-value |
|---|
| Age |
|
|
|
|
|
<60 | 47 | 26 (55.3) | 21 (44.7) | 0.238 |
| ≥60 | 50 | 23 (46.0) | 27 (54.0) |
|
| Gender |
|
|
|
|
| Male | 74 | 36 (48.6) | 38 (51.4) | 0.337 |
|
Female | 23 | 13 (56.5) | 10 (43.5) |
|
| Differentiation |
|
|
|
|
|
Well/moderate | 60 | 38 (63.3) | 22 (36.7) | 0.001 |
|
Poor | 37 | 11 (29.7) | 26 (70.3) |
|
| Metastasis |
|
|
|
|
|
Absent | 23 | 8 (24.2) | 25 (75.8) | <0.0001 |
|
Present | 64 | 41 (64.1) | 23 (35.9) |
|
| TNM stage |
|
|
|
|
|
0/I | 45 | 30 (66.7) | 15 (33.3) | 0.003 |
|
II/III/IV | 52 | 19 (36.5) | 33 (63.5) |
|
CASC2 inhibits proliferation,
migration and invasion of OS cells
It was then determined whether CASC2 affects the
malignant phenotypes of OS cells. MG-63 and HOS cells were either
untreated or transfected with pcDNA-CASC2 or si-CASC2. The mRNA
expression levels of CASC2 in the cancer cells were then
quantified. Transfection with pcDNA-CASC2 significantly upregulated
the expression levels of CASC2 in MG-63 and HOS cells
compared with the control (Fig. 2A and
B). CASC2 overexpression significantly inhibited the
proliferation of MG-63 and HOS cells compared with the empty vector
control (Fig. 2C and D;
P<0.01). Knockdown of CASC2 using a specific siRNA
significantly promoted the proliferation of MG-63 and HOS cells
compared with the control (Fig. 2C and
D). pcDNA-CASC2 transfection attenuated the migration of MG-63
and HOS cells, compared with the control (Fig. 2E). Knockdown of CASC2 led to
elevated migration of MG-63 and HOS cells compared with the control
(Fig. 2E). The effect of CASC2 on
OS cell invasion was then investigated. The results demonstrated
that CASC2 knockdown resulted in increased invasion whereas
overexpression of CASC2 inhibited invasion compared with the
control (Fig. 2F). In addition,
overexpression of CASC2 significantly increased the
percentage of cells in G0/G1 phase of the cell cycle in both MG-63
and HOS cells compared with the control group (Fig. 2G). CASC2 knockdown cells
displayed the opposite effect (Fig.
2G). In addition, CASC2 overexpression increased apoptosis of
MG-63 and HOS cells (Fig. 2H),
whereas CASC2 knockdown led to a decrease in the percentage
of apoptotic cells, compared with the control (Fig. 2H). These results suggested that
CASC2 inhibits the malignancy of OS cells via the inhibition
proliferation, migration, invasion and induction of apoptosis.
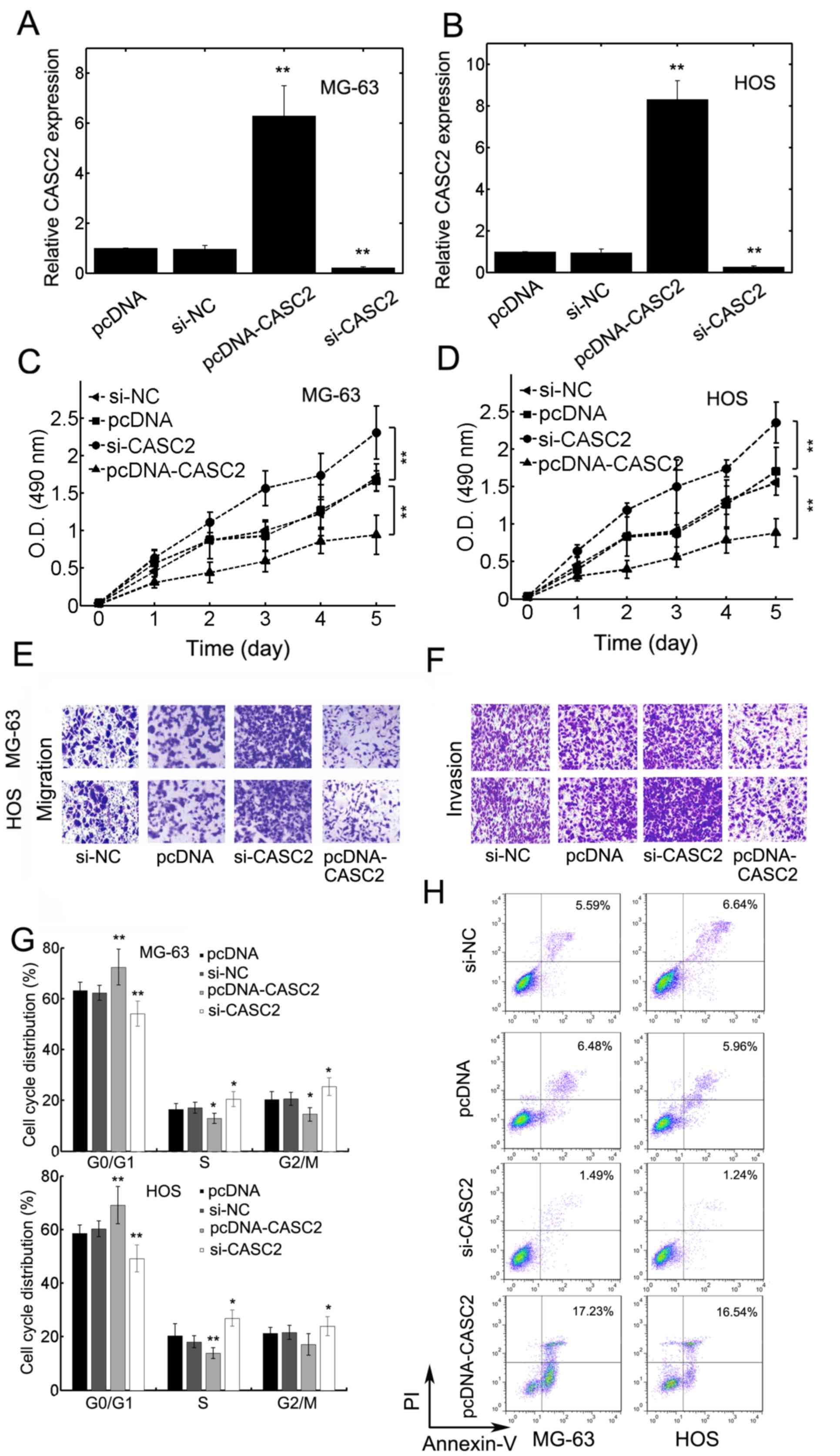 | Figure 2.CASC2 overexpression inhibited
osteosarcoma cell proliferation, migration and invasion. (A) MG-63
or (B) HOS cells were transfected with pcDNA, si-NC, pcDNA-CASC2 or
si-CASC2. The mRNA expression levels of CASC2 was quantified
using reverse transcription-quantitative polymerase chain reaction.
A five-day proliferation assay for (C) MG-63 and (D) HOS cells
transfected with pcDNA, si-NC, pcDNA-CASC2 or si-CASC2. (E)
Transwell migration assays and (F) transwell invasion assays for
MG-63 and HOS cells transfected with pcDNA, si-NC, pcDNA-CASC2 or
si-CASC2. Representative images are displayed. (G) Cell cycle was
measured for MG-63 (top graph) and HOS (bottom graph) cells
transfected with pcDNA, si-NC, si-CASC2 or pcDNA-CASC2 by flow
cytometry. (H) The percentage apoptotic cells were quantified using
flow cytometry in MG-63 (left panel) and HOS (right panel) cells
transfected with pcDNA, si-NC, si-CASC2 or pcDNA-CASC2. Data are
presented as the mean ± standard deviation. *P<0.05 and
**P<0.01. CASC2, cancer susceptibility candidate 2 gene;
CASC2, cancer susceptibility candidate 2; si-CASC2, CASC2 short
interfering RNA. |
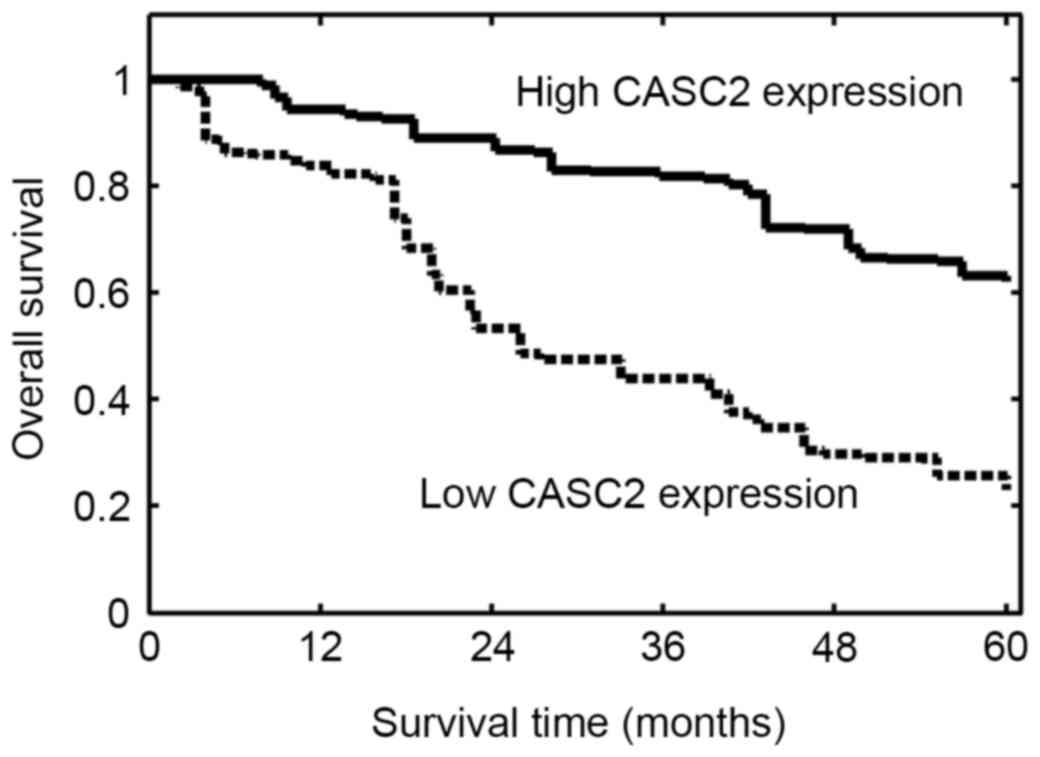
Association between CASC2 expression
and survival of OS patients
The prognostic significance of CASC2 expression in
OS was investigated. The follow-up data was complete for all
patients. The overall survival was evaluated from the day of
primary surgery to mortality or to the last follow-up study. Based
on these data, Kaplan-Meier survival curves were plotted. The
results demonstrated that OS patients with lower CASC2 expression
had a significantly reduced overall survival compared with those
with higher CASC2 expression (P=0.002; Fig. 3). These results suggested low
expression of CASC2 in OS may be associated with poor survival.
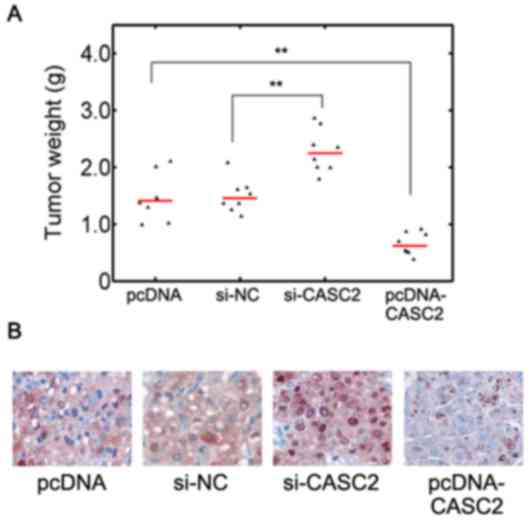
CASC2 inhibits OS progression in
vivo
In addition, in vivo implantation studies
were conducted in order to further verify the effect of CASC2. It
was observed that compared with the empty vector control,
CASC2 overexpression significantly decreased the tumor
weight (Fig. 4A; P<0.01).
CASC2 knockdown resulted in increased tumor weight compared
with the control (Fig. 4A).
Immunostaining for Ki-67 also identified that si-CASC2 treatment
led to elevated proliferation in resected mice tumors whereas
CASC2 overexpression reduced the proliferation (Fig. 4B). These results suggested that
lncRNA CASC2 may serve a tumor suppressive role in vivo.
Discussion
Advances in biological technology have improved
understanding of lncRNAs. A large number of lncRNAs have been
identified to serve important roles in different malignant tumor
types. Despite progress in cancer-associated research, the exact
underlying molecular mechanisms of OS occurrence and development
remain poorly understood.
Numerous efforts have been made to establishing an
association between lncRNA expression and tumor development
(11,26–28).
Owing to the complexity of the tumor microenvironment, lncRNA may
serve diverse roles in carcinogenesis based on tumor type.
Furthermore, as lncRNAs may also function as diagnostic or
prognostic markers, even at the early stages of the tumorigenesis,
determining the potential association between lncRNAs and various
tumors is a challenge (10,12–14).
In the present study, the CASC2 expression in OS tissues was
investigated, and results indicated that CASC2 was
substantially downregulated in cancerous tissues compared with
normal adjacent ones. CASC2 overexpression decreased the
malignant potential of OS MG-63 and HOS cells. In addition,
siRNA-mediated CASC2 knockdown increased the oncogenic
properties of OS cells, including proliferation, migration and
invasion. Higher CASC2 expression was associated with improved
prognosis of OS patients. The effect of CASC2 in an in vivo
model was investigated, which suggested that CASC2 serves a tumor
suppressive role.
The lncRNA CASC2 was originally identified as a
candidate tumor suppressor transcript in endometrial cancer in 2004
(21). Wang et al (18) observed that CASC2 was markedly
downregulated in human glioma cell lines. Overexpression of
CASC2 inhibited the malignancy of glioma cells including
proliferation and invasion (18).
Low CASC2 expression levels may additionally serve as an
unfavorable predictor for overall survival of patients with
non-small cell lung cancer (NSCLC). Overexpression of CASC2
has been previously reported to inhibit NSCLC progression in
vitro and in vivo (20). In colorectal cancer, decreased
CASC2 expression was significantly associated with advanced TNM
stage (19). As a result, CASC2
may behave as a potential tumor suppressive lncRNA in various types
of cancer. In the present study, a role of lncRNA CASC2 in OS was
reported and this further emphasized its potential function in
controlling cancer progression.
In conclusion, the function of lncRNA CASC2 in OS
was investigated in the present study and results suggested that it
may serve a tumor suppressive role. Low CASC2 expression was
significantly associated with OS progression. Overexpression of
CASC2 may attenuate the malignant phenotypes of OS in OS cell lines
in addition to in tumor xenograft tissues. The data suggested that
CASC2 may serve as a novel prognostic biomarker and a promising
therapeutic target for further intervention. With more advance
strategies such as high-throughput technology, more detailed
mechanisms regarding how CASC2 inhibits tumor progression may be
defined.
Acknowledgements
The present study was supported by the Science and
Technology Plan of Xiamen (grant no. 3502Z20154067).
References
|
1
|
Heare T, Hensley MA and Dell'Orfano S:
Bone tumors: Osteosarcoma and Ewing's sarcoma. Curr Opin Pediatr.
21:365–372. 2009. View Article : Google Scholar
|
|
2
|
Shackleton M, Quintana E, Fearon ER and
Morrison SJ: Heterogeneity in cancer: Cancer stem cells versus
clonal evolution. Cell. 138:822–829. 2009. View Article : Google Scholar
|
|
3
|
Levings PP, McGarry SV, Currie TP,
Nickerson DM, McClellan S, Ghivizzani SC, Steindler DA and Gibbs
CP: Expression of an exogenous human Oct-4 promoter identifies
tumor-initiating cells in osteosarcoma. Cancer Res. 69:5648–5655.
2009. View Article : Google Scholar :
|
|
4
|
Hattinger CM, Fanelli M, Tavanti E, Vella
S, Ferrari S, Picci P and Serra M: Advances in emerging drugs for
osteosarcoma. Expert Opin Emerg Drugs. 20:495–514. 2015. View Article : Google Scholar
|
|
5
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar :
|
|
6
|
Ernst C and Morton CC: Identification and
function of long non-coding RNA. Front Cell Neurosci. 7:1682013.
View Article : Google Scholar :
|
|
7
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar
|
|
8
|
Maass PG, Luft FC and Bähring S: Long
non-coding RNA in health and disease. J Mol Med (Berl). 92:337–346.
2014. View Article : Google Scholar
|
|
9
|
Carpenter S, Aiello D, Atianand MK, Ricci
EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB,
et al: A long noncoding RNA mediates both activation and repression
of immune response genes. Science. 341:789–792. 2013. View Article : Google Scholar :
|
|
10
|
Zhou Q, Chen F, Fei Z, Zhao J, Liang Y,
Pan W, Liu X and Zheng D: Genetic variants of lncRNA HOTAIR
contribute to the risk of osteosarcoma. Oncotarget. 7:19928–19934.
2016. View Article : Google Scholar :
|
|
11
|
Yin Z, Ding H, He E, Chen J and Li M:
Overexpression of long non-coding RNA MFI2 promotes cell
proliferation and suppresses apoptosis in human osteosarcoma. Oncol
Rep. 36:2033–2040. 2016. View Article : Google Scholar
|
|
12
|
Luo W, He H, Xiao W, Liu Q, Deng Z, Lu Y,
Wang Q, Zheng Q and Li Y: MALAT1 promotes osteosarcoma development
by targeting TGFA via MIR376A. Oncotarget. 7:54733–54743. 2016.
View Article : Google Scholar :
|
|
13
|
Uzan VR, Lengert A, Boldrini É, Penna V,
Scapulatempo-Neto C, Scrideli CA, Filho AP, Cavalcante CE, de
Oliveira CZ, Lopes LF and Vidal DO: High expression of HULC is
associated with poor prognosis in osteosarcoma patients. PLoS One.
11:e01567742016. View Article : Google Scholar :
|
|
14
|
Li F, Cao L, Hang D, Wang F and Wang Q:
Long non-coding RNA HOTTIP is up-regulated and associated with poor
prognosis in patients with osteosarcoma. Int J Clin Exp Pathol.
8:11414–11420. 2015.
|
|
15
|
Cong M, Li J, Jing R and Li Z: Long
non-coding RNA tumor suppressor candidate 7 functions as a tumor
suppressor and inhibits proliferation in osteosarcoma. Tumour Biol.
37:9441–9450. 2016. View Article : Google Scholar
|
|
16
|
Wang Y, Zhang L, Zheng X, Zhong W, Tian X,
Yin B, Tian K and Zhang W: Long non-coding RNA LINC00161 sensitises
osteosarcoma cells to cisplatin-induced apoptosis by regulating the
miR-645-IFIT2 axis. Cancer Lett. 382:137–146. 2016. View Article : Google Scholar
|
|
17
|
Cao Y, Xu R, Xu X, Zhou Y, Cui L and He X:
Downregulation of lncRNA CASC2 by microRNA-21 increases the
proliferation and migration of renal cell carcinoma cells. Mol Med
Rep. 14:1019–1025. 2016. View Article : Google Scholar
|
|
18
|
Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J
and Xue YX: Long non-coding RNA CASC2 suppresses malignancy in
human gliomas by miR-21. Cell Signal. 27:275–282. 2015. View Article : Google Scholar
|
|
19
|
Huang G, Wu X, Li S, Xu X, Zhu H and Chen
X: The long noncoding RNA CASC2 functions as a competing endogenous
RNA by sponging miR-18a in colorectal cancer. Sci Rep. 6:265242016.
View Article : Google Scholar :
|
|
20
|
He X, Liu Z, Su J, Yang J, Yin D, Han L,
De W and Guo R: Low expression of long noncoding RNA CASC2
indicates a poor prognosis and regulates cell proliferation in
non-small cell lung cancer. Tumour Biol. 37:9503–9510. 2016.
View Article : Google Scholar
|
|
21
|
Baldinu P, Cossu A, Manca A, Satta MP,
Sini MC, Rozzo C, Dessole S, Cherchi P, Gianfrancesco F, Pintus A,
et al: Identification of a novel candidate gene, CASC2, in a region
of common allelic loss at chromosome 10q26 in human endometrial
cancer. Hum Mutat. 23:318–326. 2004. View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Godbole GB, Modi DN and Puri CP:
Regulation of homeobox A10 expression in the primate endometrium by
progesterone and embryonic stimuli. Reproduction. 134:513–523.
2007. View Article : Google Scholar
|
|
24
|
Cheon KW, Lee HS, Parhar IS and Kang IS:
Expression of the second isoform of gonadotrophin-releasing hormone
(GnRH-II) in human endometrium throughout the menstrual cycle. Mol
Hum Reprod. 7:447–452. 2001. View Article : Google Scholar
|
|
25
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar :
|
|
26
|
Fan H, Liu G, Zhao C, Li X and Yang X:
Transcription factor Oct4 promotes osteosarcoma by regulating
lncRNA AK055347. Oncol Lett. 13:396–402. 2017.
|
|
27
|
Li Z, Shen J, Chan MT and Wu WK: TUG1: A
pivotal oncogenic long non-coding RNA of human cancers. Cell
Prolif. 49:471–475. 2016. View Article : Google Scholar
|
|
28
|
Ye K, Wang S, Zhang H, Han H, Ma B and Nan
W: Long noncoding RNA GAS5 suppresses cell growth and
epithelial-mesenchymal transition in osteosarcoma by regulating the
miR-221/ARHI pathway. J Cell Biochem. May 18–2017.(Epub ahead of
print). View Article : Google Scholar
|