Introduction
Cardiac remodeling exerts an important role in the
development of various cardiovascular diseases, including
myocardial infarction and dilated cardiomyopathy. At the initial
stage, cardiac remodeling is an adaptive response to external
stimuli which maintains normal cardiac function. However, it may
become maladaptive and progress into severe decompensation
(1). A number of regulators have
been reported to be associated with cardiac remodeling, including
cytokines and growth factors. For example, transforming growth
factor (TGF)-β signaling inhibits the expression of inflammatory
cytokines and chemokines by disabling macrophages and promoting the
differentiation of myofibroblasts in myocardial infarction and
cardiac remodeling (2).
Granulocyte colony-stimulating factor has a preventative effect on
cardiac dysfunction and remodeling (3).
It has been established that the renin-angiotensin
system (RAS) is an important factor that promotes the progression
of cardiac remodeling. The effector hormone of RAS, angiotensin II
(AngII), has been observed to induce the expression of fibroblast
genes and to promote fibroblast proliferation via interactions with
the AngII type 1 receptor (4).
Therefore, AngII may contribute to cardiac remodeling and is
frequently used to induce cardiac remodeling for in vitro
experiments or in rat model systems (5,6).
Although the role of AngII in cardiac remodeling has been
established, the downstream factors that affect its function remain
to be elucidated. It has been reported that xin actin binding
repeat containing 2, a target gene of myocyte-specific enhancer
factor 2A, has been demonstrated to be an important regulator in
the AngII signaling pathway (7).
In addition, platelet-derived growth factor is considered to serve
as a downstream factor of AngII and has been proposed as an
important regulator of atrial fibrosis (8). However, additional downstream
regulators are yet to be identified.
In the present study, an RNA-sequencing (RNA-seq)
method was applied to identify differentially-expressed genes
(DEGs) in AngII-treated rat cardiac fibroblasts (CFs), compared
with those without treatment with AngII. Enrichment analysis was
performed to reveal the potential functions and pathways of these
important DEGs, and their potential interactions were examined via
protein-protein interaction (PPI) network analysis. The expression
of genes of interest was validated by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis. Via these comprehensive bioinformatics analyses and gene
expression validation experiments, the present study aimed to
elucidate the underlying mechanism of AngII-induced cardiac
remodeling and to identify potential therapeutic targets for its
prevention.
Materials and methods
Cell culture
Rat CFs were purchased from the Type Culture
Collection of the Chinese Academy of Sciences (Shanghai, China).
Primary rat CFs (n=2) at 50% confluence were cultured in a 12-well
plate supplemented with 10 nM AngII for 12 h (AngII group), while
those without AngII treatment were used as controls (control group;
n=2). Following 12 h of culture, samples in the two groups were
harvested for RNA-seq.
RNA isolation and sequencing
Total RNA from each sample was isolated using an
RNAiso Plus kit (Takara Biotechnology Co., Ltd., Dalian, China)
using the phenol/chloroform extraction method (9). Following dilution, the RNA purity was
determined using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA). The total RNA (3 µg/sample) was
reverse-transcribed to construct a cDNA library, using a
NEBNext® Ultra™ RNA Library Prep kit for
Illumina® (cat no. E7530L; New England BioLabs, Inc.,
Ipswich, MA, USA), according to the manufacturer's protocol. The
mRNAs were enriched on a magnetic bead, and subsequently sheared
into fragments. The random primers, buffer, dNTPs RNase H and DNA
polymerase I were added to generate the cDNA. The cDNA was
blunt-ended and a single 3′ adenosine moiety and Illumina adapters
were added to the repaired ends. The cDNAs were amplified via 20
cycles of repair chain reaction, and the thermocycling conditions
was as follows: 94°C for 2 min, 94°C for 30 sec, then subjected to
5 cycles at 94°C for 5 sec, 70°C for 4 min, and subjected to
another 5 cycles of 94°C for 5 sec, 72°C for 4 min, then 20 cycles
of 94°C for 5 sec, 68°C for 4 min and a final extension at 70°C for
10 min. The cDNA clusters were sequenced using the Illumina Hiseq
4000 platform (Illumina, Inc., San Diego, CA, USA) using the 150
paired end method (10). The
RNA-seq data was uploaded to the Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) with the accession no.
SRP082129.
Pretreatment of RNA-seq data
Quality control of the raw reads was implemented
using the prinseq-lite (sourceforge.net/projects/prinseq/files/standalone/)
and fastx_toolkit (hannonlab.cshl.edu/fastx_toolkit/index.html)
tools as follows: i) Removal of barcode sequences and adaptors of
the reads; ii) filtering out of reads with an N content >5%;
iii) elimination of the base at each end with a quality <10; iv)
exclusion of low-quality reads in which bases with Q <20
accounted for >20% of the length; and v) exclusion of reads with
a length <30. Clean reads from the four samples were obtained
and aligned with the rat reference genome using TopHat software
(version 2.0.8; ccb.jhu.edu/software/tophat/index.shtml) with the
default parameters. StringTie (version 1.2.2; ccb.jhu.edu/software/stringtie), a
computational method which is able to improve the reconstruction of
a transcriptome from RNA-seq reads (11), was utilized to obtain gene
annotation information for the aligned raw reads, based on Ensembl
(www.ensembl.org).
Identification of DEGs and enrichment
analysis
Following pretreatment of the raw RNA-seq data, gene
expression levels were obtained by calculating the fragments per
kilobase of exon per million fragments mapped (FPKM), using the
Cufflinks software (2.2.1 released, cufflinks.cbcb.umd.edu/index.html). Subsequently, the
linear models for microarray analysis (limma; www.bioconductor.org/packages/release/bioc/html/limma.html)
package in R (www.r-project.org) was used to select DEGs between the
treatment and control groups. The thresholds for DEG selection were
|log2 fold change (FC)|>1 and P<0.05.
The gene ontology database (www.geneontology.org) was utilized to conduct
functional enrichment analysis to identify potential biological
processes of the DEGs, while the Kyoto Encyclopedia of Genes and
Genomes database (www.genome.jp/kegg/pathway.html) was used to perform
pathway enrichment analysis to reveal the pathways that may involve
the DEGs. The function and pathway enrichment analyses were
performed using the Database for Annotation, Visualization and
Integrated Discovery online tool (david.abcc.Ncifcrf.gov) (12). The cut-off value for significant
function and pathway selection was P<0.05.
Construction of PPI network
In order to examine the potential interactions of
the DEGs at the protein level, the STRING database online tool
(string-db.org) was used to establish the PPI
network, under the condition of a combined score >0.4 (13). Cytoscape software (version 3.2.0;
cytoscape.org) was used to visualize the network.
A node in the PPI network represented the protein product of a DEG,
and the degree of a node was deemed to be the number of proteins
interacting with this specific node.
RT-qPCR analysis
RNA isolation was performed as described above for
RNA-seq. RNAs were transcribed to cDNA using the PrimeScript™ RT
Master Mix (Takara Biotechnology Co., Ltd.). qPCR was subsequently
performed on an ABI fluorescence quantitative PCR machine (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) within a 20 µl reaction
system: 10 µl SYBR Green PCR Master Mix (Thermo Fisher Scientific,
Inc.), 0.4 µl each primer, 2 µl cDNA templates and 7.2 µl
RNase-Free dH2O. PCR conditions were as follows:
Denaturation at 95°C for 10 min; 40 cycles of 95°C for 15 sec and
60°C for 60 sec; and melting at 95°C for 15 sec, 60°C for 60 sec
and 95°C for 15 sec. The primer sequences are listed in Table I. β-actin was used as an internal
reference gene. The relative expression of each gene was measured
using the 2−ΔΔCq method (14).
 | Table I.Primers of validated genes. |
Table I.
Primers of validated genes.
| Gene | Sequences |
|---|
| β-actin | F:
3′-CCCATCTATGAGGGTTACGC-5′ |
|
| R:
3′-TTTAATGTCACGCGATTTC-5′ |
| IL1B | F:
3′-AGCAGCTTTCGACAGTGAGG-5′ |
|
| R:
3′-CACACACTAGCAGGTCGTCA-5′ |
| NOS2 | F:
3′-TGCTATTCCCAGCCCAACAA-5′ |
|
| R:
3′-GAATTCAATGGCTTGAGGCA-5′ |
| DYRK3 | F:
3′-CAAGTGCCCACCCTATACAA-5′ |
|
| R:
3′-CAGTTGCTCCACCTTCATCAC-5′ |
| MMP3 | F:
3′-TTCCTTGGGCTGAAGATGAC-5′ |
|
| R:
3′-TCCTGGAGAATGTGAGTGGG-5′ |
| BOLL | F:
3′-CCTGCTTCTTCTGCTCCATTC-5′ |
|
| R:
3′-GGCACTTGAAGCATAAACCTG-5′ |
| TNNC2 | F: 3′-
AGGATGCTAGGGCAGACAC-5′ |
|
| R:
3′-TCAAAGATGCGGAAACACTCA-5′ |
| ACACB | F:
3′-CACAAGAAACTGGACCTGC-5′ |
|
| R:
3′-CGCGTTCATTACGGAACATC-5′ |
| AXL | F:
3′-AGTGGATTGCTATCGAGAGTCTG-5′ |
|
| R:
3′-CCTTGACGTAGGTAGTCGTAAATCT-5′ |
Statistical analysis
Data are presented as the mean ± standard error of
the mean. A Student's t-test using SPSS software (version 10.0;
SPSS, Inc., Chicago, IL, USA) was used to compare the relative
expression levels of genes between the two groups. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were repeated three times.
Results
DEGs between AngII treatment group and
control group
In the present study, a total of 16,974 genes with
FPKM values >0 were detected. Based on the aforementioned
criteria, 266 DEGs were selected, including 126 upregulated and 140
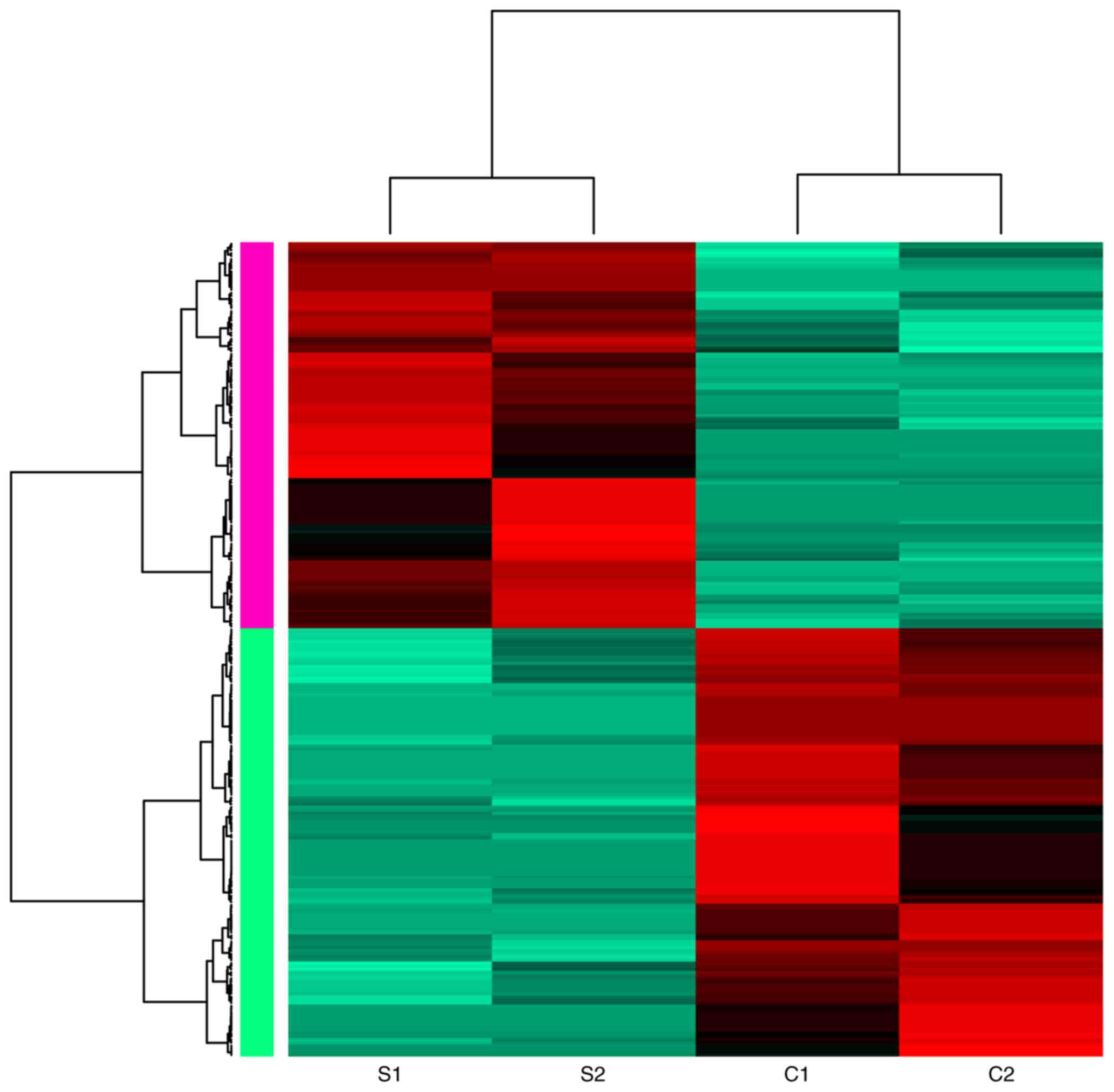
downregulated genes. A heat map of the gene expression is presented
in Fig. 1, demonstrating that
samples in the two groups were able to be distinguished by these
DEGs. The top 10 DEGs ranked by FC are presented in Table II, including five upregulated
genes [interleukin 1β (IL1B), nitric oxide synthase 2
(NOS2), dual specificity tyrosine phosphorylation regulated
kinase 3 (DYRK3), paired box 8 and matrix metallopeptidase 3
(MMP3)] and five downregulated genes [boule homolog RNA
binding protein (BOLL), troponin C2 fast skeletal type
(TNNC2), F-box protein 2, ankyrin repeat SAM and basic
leucine zipper domain containing 1, and MAP6 domain containing
1).
 | Table II.Top ten differentially-expressed
genes (ranked by fold-change). |
Table II.
Top ten differentially-expressed
genes (ranked by fold-change).
| Gene | Log2
fold change | P-value |
|---|
| IL1B | 5.210354306 | 0.052672383 |
| NOS2 | 4.874224035 | 0.000369813 |
| DYRK3 | 4.454414593 | 0.001057582 |
| PAX8 | 4.308293178 | 0.001274081 |
| MMP3 | 4.205044062 | 0.006835961 |
| BOLL | −4.227038973 | 0.002187013 |
| TNNC2 | −4.247495833 | 0.000384867 |
| FBXO2 | −4.285939067 | 0.003258329 |
| ASZ1 | −4.367008762 | 0.009252033 |
| MAP6D1 | −4.397909336 | 0.018432302 |
Enrichment analysis of the DEGs
According to the enrichment analysis, the
upregulated DEGs were significantly enriched in biological
processes (BPs) including response to bacterium [e.g. glutathione
peroxidase 2, histone cluster 1 H2B family member 1
(HIST1H2BL) and NOS2], defense response [e.g.,
cytochrome b-245 β chain (CYBB), HIST1H2BL and
NOS2], immune response (e.g. CYBB, linker for
activation of T-cells family member 2 and interleukin 20 receptor
subunit β), regulation of angiogenesis [e.g. CD36 molecule
(CD36), C-X3-X motif chemokine ligand 1 and interleukin 1α
(IL1A)] and superoxide metabolic process (CYBB, NADPH
oxidase organizer 1 and NOS2); and molecular functions (MFs)
including carboxylic acid binding [e.g. CD36, acetyl coA
carboxylase β (ACACB) and NOS2] (Table III). A total of two significant
pathways were enriched, including leukocyte transendothelial
migration [e.g. claudin 18 (CLDN18), claudin 11
(CLDN11) and myosin light chain 2] and cell adhesion
molecules (e.g. neuronal cell adhesion molecule, CLDN18 and
CLDN11) (Table IV).
 | Table III.Significant enriched functions and
pathways for upregulated genes. |
Table III.
Significant enriched functions and
pathways for upregulated genes.
| Category | Term | Count | Genes | P-value |
|---|
| GOTERM_BP_FAT | GO:0009617~response
to bacterium | 7 | GPX2, HIST1H2BL,
BCL3, NOS2, IRG1, IL1A, GCH1 |
1.61×10−3 |
| GOTERM_BP_FAT | GO:0006952~defense
response | 9 | CYBB, IL20RB,
HIST1H2BL, BCL3, NOS2, CFI, RT1-BA, IL1A, GCH1 |
2.47×10−3 |
| GOTERM_BP_FAT | GO:0006955~immune
response | 9 | CYBB, LAT2, IL20RB,
BCL3, CFI, CX3CL1, RT1-BA, IL1A, GCH1 |
3.11×10−3 |
| GOTERM_BP_FAT |
GO:0045765~regulation of angiogenesis | 4 | CD36, CX3CL1, IL1A,
RGD1565355 |
5.43×10−3 |
| GOTERM_BP_FAT |
GO:0006801~superoxide metabolic
process | 3 | CYBB, NOXO1,
NOS2 |
6.82×10−3 |
| GOTERM_MF_FAT |
GO:0031406~carboxylic acid binding | 5 | CD36, ACACB, NOS2,
RGD1565355, GCHFR |
1.82×10−2 |
| GOTERM_CC_FAT | GO:0005886~plasma
membrane | 27 | CLDN18, CD244,
LPPR4, SYT3, CLDN11 |
2.57×10−3 |
| GOTERM_CC_FAT | GO:0044459~plasma
membrane part | 17 | CLDN18, CD244,
LPPR4, NOXO1, SLC12A3 |
1.05×10−2 |
| GOTERM_CC_FAT | GO:0009986~cell
surface | 7 | CD244, CD36, LPPR4,
CD2, CX3CL1, RT1-BA, IL1A |
2.11×10−2 |
| GOTERM_CC_FAT | GO:0016021~integral
to membrane | 36 | CD244, CLDN18,
LPPR4, GCNT2, SYT3 |
4.96×10−2 |
| KEGG_PATHWAY | rno04670: Leukocyte
transendothelial migration | 5 | CLDN18, CYBB, MYL2,
TXK, CLDN11 |
1.04×10−2 |
| KEGG_PATHWAY | rno04514: Cell
adhesion molecules | 5 | NRCAM, CLDN18, CD2,
CLDN11, RT1-BA |
2.42×10−2 |
 | Table IV.Significant enriched functions and
pathways for downregulated genes. |
Table IV.
Significant enriched functions and
pathways for downregulated genes.
| Category | Term | Count | Genes | P-value |
|---|
| GOTERM_BP_FAT | GO:0007601~visual
perception | 5 | GABRR2, GNAT1,
OPN1SW, RGS9, SAG |
2.03×10−3 |
| GOTERM_BP_FAT | GO:0050953~sensory
perception of light stimulus | 5 | GABRR2, GNAT1,
OPN1SW, RGS9, SAG |
2.11×10−3 |
| GOTERM_BP_FAT | GO:0046460~neutral
lipid biosynthetic process | 3 | MOGAT1, AGPAT9,
PNPLA3 |
2.69×10−3 |
| GOTERM_BP_FAT |
GO:0046463~acylglycerol biosynthetic
process | 3 | MOGAT1, AGPAT9,
PNPLA3 |
2.69×10−3 |
| GOTERM_BP_FAT | GO:0046504~glycerol
ether biosynthetic process | 3 | MOGAT1, AGPAT9,
PNPLA3 |
3.59×10−3 |
| GOTERM_MF_FAT | GO:0005216~ion
channel activity | 9 | GABRR2, KCNS1,
KCNK15, FXYD4, TRPC5, SLC26A7, CLIC5, KCNIP4, KCNJ13 |
6.07×10−4 |
| GOTERM_MF_FAT |
GO:0022838~substrate specific channel
activity | 9 | GABRR2, KCNS1,
KCNK15, FXYD4, TRPC5, SLC26A7, CLIC5, KCNIP4, KCNJ13 |
7.35×10−4 |
| GOTERM_MF_FAT | GO:0015267~channel
activity | 9 | GABRR2, KCNS1,
KCNK15, FXYD4, TRPC5, SLC26A7, CLIC5, KCNIP4, KCNJ13 |
9.35×10−4 |
| GOTERM_MF_FAT | GO:0022803~passive
transmembrane transporter activity | 9 | GABRR2, KCNS1,
KCNK15, FXYD4, TRPC5, SLC26A7, CLIC5, KCNIP4, KCNJ13 |
9.35×10−4 |
| GOTERM_MF_FAT |
GO:0005267~potassium channel activity | 5 | KCNS1, KCNK15,
FXYD4, KCNIP4, KCNJ13 |
4.78×10−3 |
| GOTERM_CC_FAT | GO:0000800~lateral
element | 2 | REC8, SYCP3 |
4.64×10−2 |
The downregulated DEGs were significantly associated
with BPs such as visual perception [e.g. γ-aminobutyric acid type A
receptor rho2 subunit (GABRR2), G protein subunit α
transducin 1 (GNAT1) and opsin 1 short wave sensitive
(OPN1SW)], sensory perception of light stimulus (e.g.
GABRR2, GNAT1 and OPN1SW) and neutral lipid
biosynthetic process (e.g. monoacylglycerol O-acyltransferase 1,
glycerol-3-phosphate acyltransferase 3 and patatin like
phospholipase domain containing 3) and MFs including substrate
specific channel activity [e.g. GABRR2, potassium
voltage-gated channel modifier subfamily S member 1 (KCNS1)
and potassium two pore domain channel subfamily K member 15
(KCNK15)], channel activity (e.g. GABRR2,
KCNS1 and KCNK15) and passive transmembrane
transporter activity (e.g. GABRR2, KCNS1 and
KCNK15). However, no significant pathway was enriched.
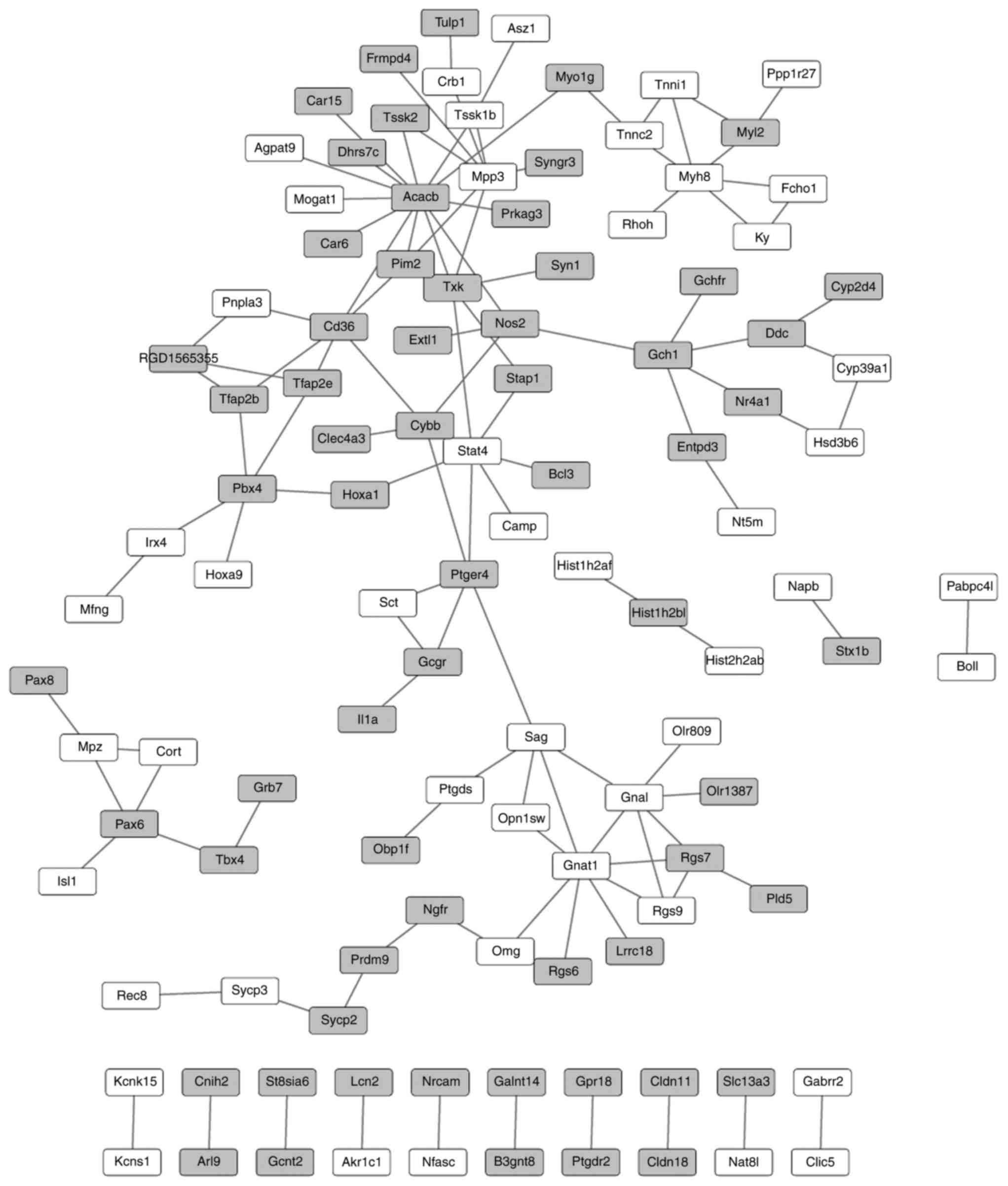
PPI network of the DEGs
Under the criterion of combined score >0.4, a PPI
network was established, comprising 112 nodes (representing the
protein product of a DEG) and 118 interactions. The top 5 nodes in
the PPI network were ACACB (degree=13), GNAT1 (degree=8), MPP3
(degree=7), signal transducer and activator of transcription 4
(degree=6) and myosin-8 (degree=6) (Fig. 2).
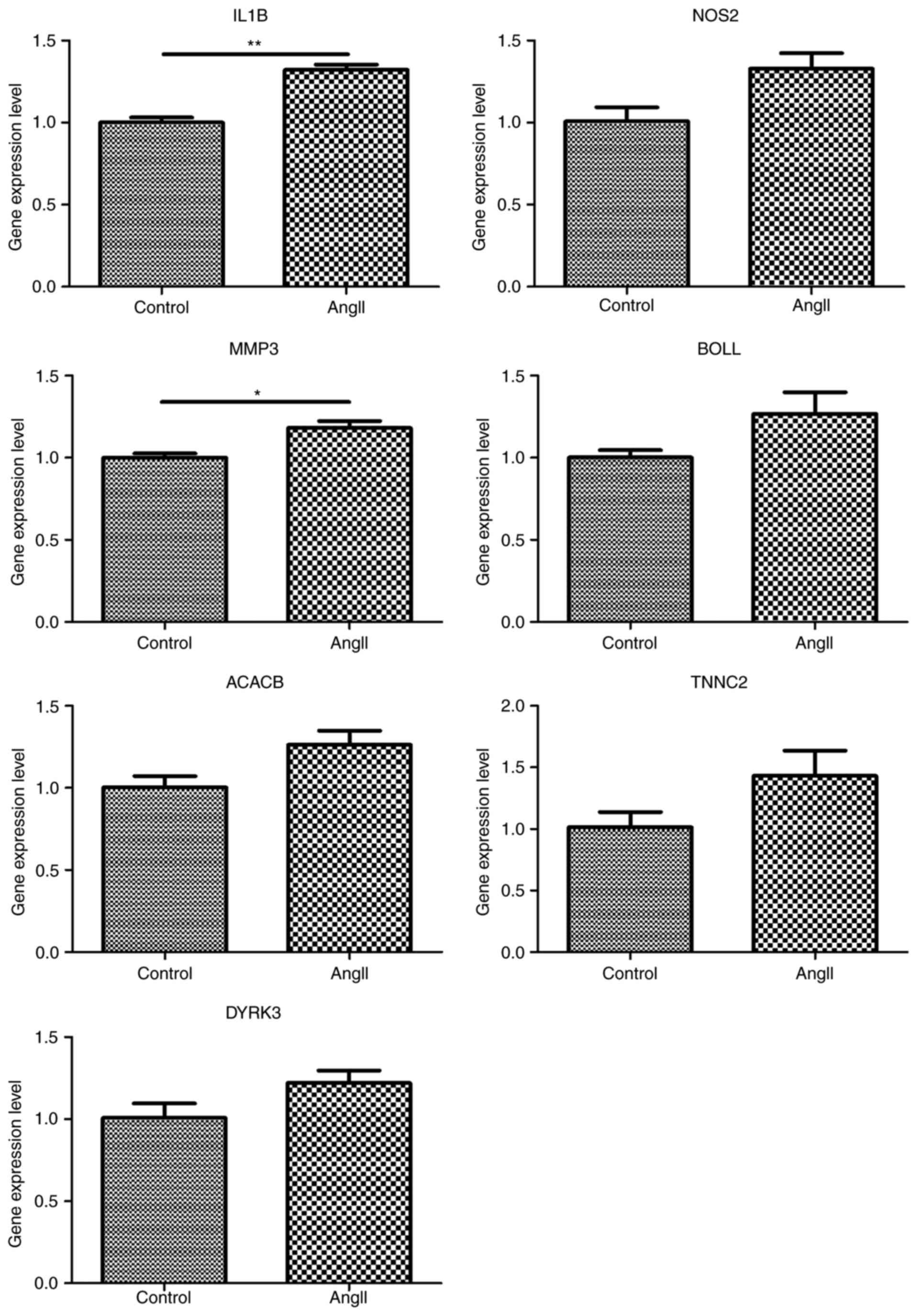
Gene expression validations
Reportedly, AXL, a novel receptor tyrosine
kinase, is closely associated with cardiac remodeling and its
expression level is altered by AngII stimulation in vascular smooth
muscle cells. Thus, the present study detected its expression
levels in CFs by RT-qPCR analysis. The results showed that the
expression of AXL was downregulated in AngII-treated CFs
compared with the control, but this difference was not significant
(Fig. 3). In addition, the
expression of genes of interest, listed in the top 10 DEGs ranked
by FC or top 5 nodes in the PPI network, were detected using
RT-qPCR analysis, including IL1B (reference no.
NM_031512.2), NOS2 (reference no. NM_012611.3), DYRK3
(reference no. NM_001024767.1), MMP3 (reference no.
NM_133523.3), BOLL (reference no. NM_001113370.1),
ACACB (reference no. NM_053922.1) and TNNC2
(reference no. NM_001037351.2). As hypothesized, IL1B and
MMP3 were significantly increased under treatment with
AngII, compared with the control (P<0.01 and P<0.05,
respectively). However, the expression of other genes did not
exhibit any significant alteration between the control and
treatment groups (P>0.05) (Fig.
3).
Discussion
Adrenergic receptors (ARs) belong to the family of G
protein-coupled receptors (GPCRs). Reportedly, ARs are involved in
aging and cardiovascular physiopathology (15). In response to stimulation of the
sympathetic nervous system, α1ARs (α1A,
α1B and α1D) are able to activate
Gαq, which contributes to cardiac remodeling (16). AngII is another GPCR that is
involved in cardiac remodeling (17). In the present study, AngII was used
to treat the rat CFs, and a number of important genes for
AngII-induced cardiac remodeling were identified, including
ACACB, IL1B, IL1A, NOS2 and MMP3. Among
them, IL1B and MMP3 were demonstrated to be
upregulated. The majority of the genes were associated with the
defense response, immune response, regulation of angiogenesis,
superoxide metabolic process and carboxylic acid binding BPs. In
addition, ACACB and MPP3 were two of the predominant nodes in the
PPI network.
The ACACB protein, additionally termed ACC2,
regulates fatty acid oxidation. It has been reported that
ACACB may be involved in upregulated pathways, including the
adipocytokine and type 2 diabetes signaling pathways (18). The knockout of ACC2 has been
demonstrated to exert a preventative effect on cardiac remodeling
(19). In addition, the
cardiac-specific deletion of ACC2 in rats resulted in
improved cardiac function (20).
These previous results suggested that the upregulation of
ACC2 may be the primary cause of cardiac dysfunction. In the
present study, associated with the carboxylic acid binding BP, ACC2
was identified to be an upregulated node in the PPI network of DEGs
in AngII-induced cardiac remodeling, further supporting the
causative role of ACC2 in cardiac remodeling. Additionally, ACC2
was significantly enriched in the carboxylic acid binding MF,
suggesting it may regulate this function to exert its role in
cardiac remodeling.
IL1B belongs to the interleukin 1 cytokine family.
It is important for the regulation of inflammatory responses.
Proinflammatory cytokines, including IL1B, are associated with
cardiac dysfunction (21). The
expression of transforming growth factor-β (TGF-β) is
induced by AngII in primary human CFs, and it has been reported to
downregulate a number of proinflammatory genes, including
IL1B (22). This suggested
that the expression of IL1B may be mediated by TGF-β.
Conversely, the present study did not detect the gene expression of
TGF-β, although IL1B was upregulated, which was
inconsistent with the previous study. It may be inferred that
without the regulation of TGF-β, IL1B may be
upregulated in cardiac remodeling. IL1A is another interleukin 1
cytokine family member that has important roles in the mediation of
immune responses and hematopoiesis (23). Adverse cardiac remodeling results
from an immune response driven by T helper 1 cells and type 1
macrophages (24). The results of
the present study demonstrated that IL1A was significantly enriched
in the immune response and regulation of angiogenesis BPs,
suggesting that this gene may be important in AngII-induced cardiac
remodeling via involvement in these above processes.
The NOS2 gene may be induced by a number of
cytokines in the liver. It has important roles in numerous
processes, including antitumoral activities and the regulation of
the defense response (25,26). Reportedly, suppression of NOS2
contributes to cardiac function recovery in rats by ameliorating
cardiac remodeling (27). In a
failing human heart, it has been demonstrated that NOS2
downregulates the muscular protein LIM/homeobox protein Lhx1, an
important molecule for cardiac hypertrophy, thereby promoting
adverse cardiac remodeling (28).
In support of these previous findings, the results of the present
study demonstrated that NOS2 was an upregulated gene that was
enriched in the response to bacterium and defense response BPs,
implying it may be an important regulator in AngII-induced cardiac
remodeling. Unfortunately, this upregulation was not validated by
the RT-qPCR results, which may be due to the small sample size.
MMP3 is a membrane-associated protein. Increased
expression of MMPs is frequently associated with cardiac remodeling
(29). As a well-known target of
osteopontin, MMP3 has been suggested to be a potential therapeutic
target to inhibit cardiac remodeling, as its expression contributes
to the progression of extensive fibrosis in acute myocarditis
(30). In accordance with these
previous results, the present study demonstrated that MMP3 was an
upregulated gene in AngII-induced cardiac remodeling. Notably, MMP3
was highlighted in the PPI network, suggesting that it serve roles
in the progression of AngII-induced cardiac remodeling by
interacting with other genes.
The AXL encoded protein is a family member of
the Tyro3-Axl-Mer (TAM) tyrosine receptors. Reportedly, the
expression of AXL is a potential genetic marker for
salt-dependent hypertension, and at early stage of this type of
hypertension, AXL is associated with kidney pathology
(31). In addition, in the
hematopoietic compartment of early hypertensive kidneys, AXL
mediates the expression of interferon γ (31) which controls the interactions
between different macrophages and T cells in AngII-stimulated
cardiac remodeling (32). The
ablation of AXL may cause degenerative alterations to the
hematopoietic system (33).
Transplantation of cardiac resident stem cells (CRSCs) is
considered to be a promising strategy for the treatment of heart
disease. CRSCs have a cardioprotective effect and W8B2+
CRSCs are reported to secrete cytokines that are potentially
involved in cell growth and survival, including AXL (34). These previous results collectively
indicated that increased expression of AXL may have a
protective effect in cardiac remodeling, while decreased expression
may cause or contribute to the progression of cardiac remodeling.
Based on the RNA-seq results of the present study, although the
expression of AXL was downregulated in AngII-treated CFs
compared with the control, the difference was not significant.
Further experiments are required to investigate the expression of
AXL in AngII-induced cardiac remodeling.
Despite comprehensive analyses and validation
experiments, a number of limitations remained in the present study.
The sample size was small as each group contained only two samples.
A gradient-concentration experiment to select the optimal AngII
concentration was not performed, with the optimum concentration
being inferred from a number of published articles. Although the
expression of a number of genes was validated, their interactions
as predicted in the PPI network were not validated. Therefore,
further samples and concentration-response experiments are required
in the future. However, the present study provided an insight into
the AngII-induced cardiac remodeling mechanism and may facilitate
the identification of potential therapeutic targets.
In conclusion, the dysregulation of 5 genes,
ACACB, IL1A, NOS2, IL1B and MMP3, may
be a primary cause of AngII-induced cardiac remodeling, and all of
the genes, particularly IL1B and MMP3, may be used as
candidate markers for prevention and treatment.
Acknowledgements
The present study was supported by the Changning
District of Shanghai Science and Technology Commission (grant no.
cnkw2014j03).
References
|
1
|
Takano H, Hasegawa H, Nagai T and Komuro
I: Implication of cardiac remodeling in heart failure: Mechanisms
and therapeutic strategies. Intern Med. 42:465–469. 2003.
View Article : Google Scholar
|
|
2
|
Bujak M and Frangogiannis NG: The role of
TGF-beta signaling in myocardial infarction and cardiac remodeling.
Cardiovasc Res. 74:184–195. 2007. View Article : Google Scholar
|
|
3
|
Iwanaga K, Takano H, Ohtsuka M, Hasegawa
H, Zou Y, Qin Y, Odaka K, Hiroshima K, Tadokoro H and Komuro I:
Effects of G-CSF on cardiac remodeling after acute myocardial
infarction in swine. Biochem Biophys Res Commun. 325:1353–1359.
2004. View Article : Google Scholar
|
|
4
|
Grobe JL, Mecca AP, Lingis M, Shenoy V,
Bolton TA, Machado JM, Speth RC, Raizada MK and Katovich MJ:
Prevention of angiotensin II-induced cardiac remodeling by
angiotensin-(1–7). Am J Physiol Heart Circ Physiol. 292:H736–H742.
2007. View Article : Google Scholar
|
|
5
|
Huang XR, Chung AC, Yang F, Yue W, Deng C,
Lau CP, Tse HF and Lan HY: Smad3 mediates cardiac inflammation and
fibrosis in angiotensin II-induced hypertensive cardiac remodeling.
Hypertension. 55:1165–1171. 2010. View Article : Google Scholar
|
|
6
|
Hu C, Dandapat A, Sun L, Marwali MR, Inoue
N, Sugawara F, Inoue K, Kawase Y, Jishage K, Suzuki H, et al:
Modulation of angiotensin II-mediated hypertension and cardiac
remodeling by lectin-like oxidized low-density lipoprotein
receptor-1 deletion. Hypertension. 52:556–562. 2008. View Article : Google Scholar
|
|
7
|
McCalmon SA, Desjardins DM, Ahmad S,
Davidoff KS, Snyder CM, Sato K, Ohashi K, Kielbasa OM, Mathew M,
Ewen EP, et al: Modulation of angiotensin II-mediated cardiac
remodeling by the MEF2A target gene Xirp2. Circ Res. 106:952–960.
2010. View Article : Google Scholar :
|
|
8
|
Yang D, Yuan J, Liu G, Ling Z, Zeng H,
Chen Y, Zhang Y, She Q and Zhou X: Angiotensin receptor blockers
and statins could alleviate atrial fibrosis via regulating
platelet-derived growth factor/Rac1/nuclear factor-kappa B Axis.
Int J Med Sci. 10:812–824. 2013. View Article : Google Scholar :
|
|
9
|
Chomczynski P and Sacchi N: The
single-step method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction: Twenty-something years
on. Nat Protoc. 1:581–585. 2006. View Article : Google Scholar
|
|
10
|
Ran M, Chen B and Li Z, Wu M, Liu X, He C,
Zhang S and Li Z: Systematic identification of long noncoding RNAs
in immature and mature porcine testes. Biol Reprod. 94:772016.
View Article : Google Scholar
|
|
11
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015. View
Article : Google Scholar :
|
|
12
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:(Web Server Issue). W169–W175. 2007.
View Article : Google Scholar :
|
|
13
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:(Database Issue). D561–D568. 2011. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
Relative Gene Expression Data Using Real-Time Quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Santulli G: Sympathetic nervous system
signaling in heart failure and cardiac aging. Pathophysiol Pharm
Cardio Disease. 83–105. 2015.
|
|
16
|
Perrino C and Rockman HA: Reversal of
cardiac remodeling by modulation of adrenergic receptors: A new
frontier in heart failure. Curr Opin Cardiol. 22:443–449. 2007.
View Article : Google Scholar
|
|
17
|
Karnik SS and Unal H: Angiotensin II
receptor-induced cardiac remodeling in mice without angiotensin II.
Hypertension. 59:542–544. 2012. View Article : Google Scholar :
|
|
18
|
Ables GP, Ouattara A, Hampton TG, Cooke D,
Perodin F, Augie I and Orentreich DS: Dietary methionine
restriction in mice elicits an adaptive cardiovascular response to
hyperhomocysteinemia. Sci Rep. 5:88862015. View Article : Google Scholar :
|
|
19
|
Hwang IW, Makishima Y, Kato T, Park S,
Terzic A and Park EY: Human acetyl-CoA carboxylase 2 expressed in
silkworm Bombyx mori exhibits posttranslational biotinylation and
phosphorylation. Appl Microbiol Biotechnol. 98:8201–8209. 2014.
View Article : Google Scholar :
|
|
20
|
Yan H, Li Y, Wang C, Zhang Y, Liu C, Zhou
K and Hua Y: Contrary microRNA expression pattern between fetal and
adult cardiac remodeling: Therapeutic value for heart failure.
Cardiovasc Toxicol. 17:267–276. 2017. View Article : Google Scholar
|
|
21
|
Salvador AM, Nevers T, Velázquez F,
Aronovitz M, Wang B, Molina Abadía A, Jaffe IZ, Karas RH, Blanton
RM and Alcaide P: Intercellular adhesion molecule 1 regulates left
ventricular leukocyte infiltration, cardiac remodeling, and
function in pressure overload-induced heart failure. J Am Heart
Assoc. 5:e0031262016. View Article : Google Scholar :
|
|
22
|
Kapoun AM, Liang F, O'young G, Damm DL,
Quon D, White RT, Munson K, Lam A, Schreiner GF and Protter AA:
B-Type natriuretic peptide exerts broad functional opposition to
transforming growth factor-beta in primary human cardiac
fibroblasts, myofibroblast conversion, proliferation, and
inflammation. Circ Res. 94:453–461. 2004. View Article : Google Scholar
|
|
23
|
Akula MK, Shi M, Jiang Z, Foster CE, Miao
D, Li AS, Zhang X, Gavin RM, Forde SD, Germain G, et al: Control of
the innate immune response by the mevalonate pathway. Nat Immunol.
17:922–929. 2016. View
Article : Google Scholar :
|
|
24
|
Bönner F, Borg N, Jacoby C, Temme S, Ding
Z, Flögel U and Schrader J: Ecto-5′-nucleotidase on immune cells
protects from adverse cardiac remodeling. Circ Res. 113:301–312.
2013. View Article : Google Scholar
|
|
25
|
Stych B: Regulation and role of RNA Pol II
CTD phosphorylation in the transcription cycle of the Nos2 gene
(unpublished PhD thesis). University of Vienna. 2014.
|
|
26
|
Sun J, Shi YH, Le GW and Ma XY: Distinct
immune response induced by peptidoglycan derived from lactobacillus
sp. World J Gastroenterol. 11:6330–6337. 2005. View Article : Google Scholar :
|
|
27
|
Zheng B, Cao LS, Zeng QT, Wang X, Li DZ
and Liao YH: Inhibition of NOS2 ameliorates cardiac remodeling,
improves heart function after myocardial infarction in rats. Basic
Res Cardiol. 99:264–271. 2004. View Article : Google Scholar
|
|
28
|
Kempf T and Wollert KC: Nitric oxide and
the enigma of cardiac hypertrophy. Bioessays. 26:608–615. 2004.
View Article : Google Scholar
|
|
29
|
Liao Y, Zhao H, Ogai A, Kato H, Asakura M,
Kim J, Asanuma H, Minamino T, Takashima S and Kitakaze M:
Atorvastatin slows the progression of cardiac remodeling in mice
with pressure overload and inhibits epidermal growth factor
receptor activation. Hypertens Res. 31:335–344. 2008. View Article : Google Scholar
|
|
30
|
Szalay G, Sauter M, Haberland M, Zuegel U,
Steinmeyer A, Kandolf R and Klingel K: Osteopontin: A
fibrosis-related marker molecule in cardiac remodeling of
enterovirus myocarditis in the susceptible host. Circ Res.
104:851–859. 2009. View Article : Google Scholar
|
|
31
|
Batchu SN, Hughson A, Gerloff J, Fowell DJ
and Korshunov VA: Role of Axl in early kidney inflammation and
progression of salt-dependent hypertension. Hypertension.
62:302–309. 2013. View Article : Google Scholar
|
|
32
|
Han YL, Li YL, Jia LX, Cheng JZ, Qi YF,
Zhang HJ and Du J: Reciprocal interaction between macrophages and T
cells stimulates IFN-γ and MCP-1 production in Ang II-induced
cardiac inflammation and fibrosis. PLoS One. 7:e355062012.
View Article : Google Scholar :
|
|
33
|
Arandjelovic S and Ravichandran KS: A
MERry response after myocardial infarction. Circ Res. 113:949–951.
2013. View Article : Google Scholar :
|
|
34
|
Zhang Y, Sivakumaran P, Newcomb AE,
Hernandez D, Harris N, Khanabdali R, Liu GS, Kelly DJ, Pébay A,
Hewitt AW, et al: Cardiac repair with a novel population of
mesenchymal stem cells resident in the human heart. Stem Cells.
33:3100–3113. 2015. View Article : Google Scholar
|