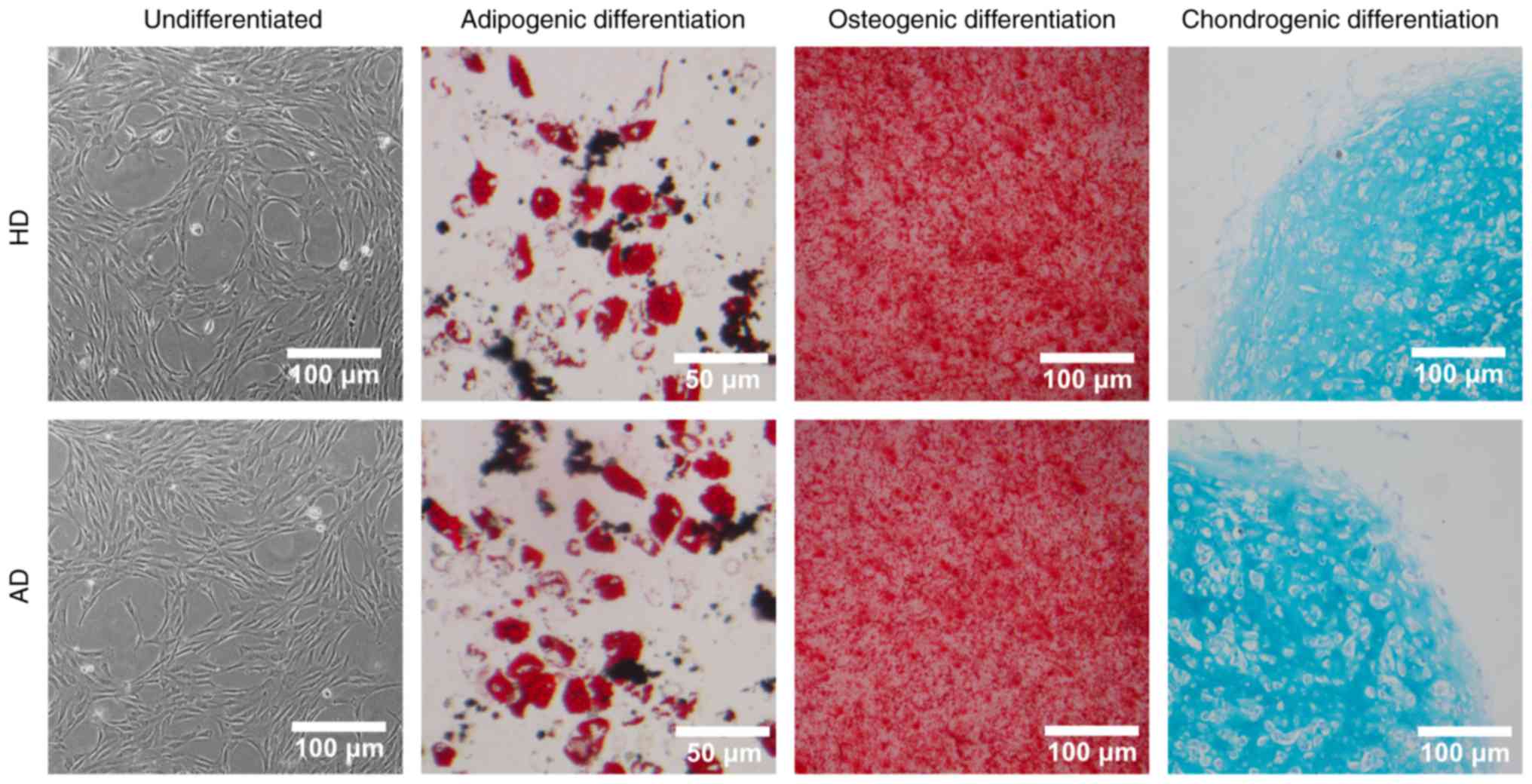
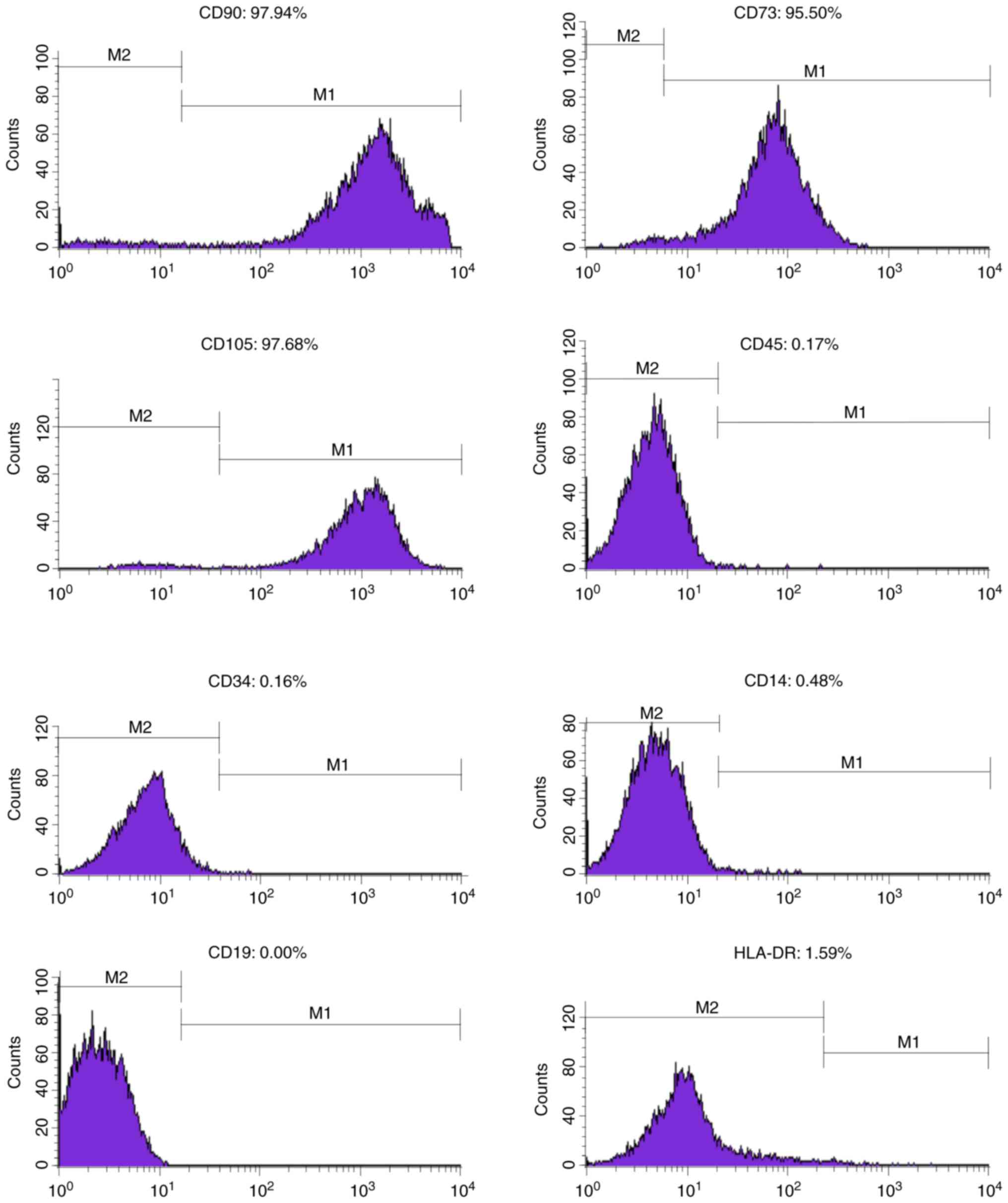
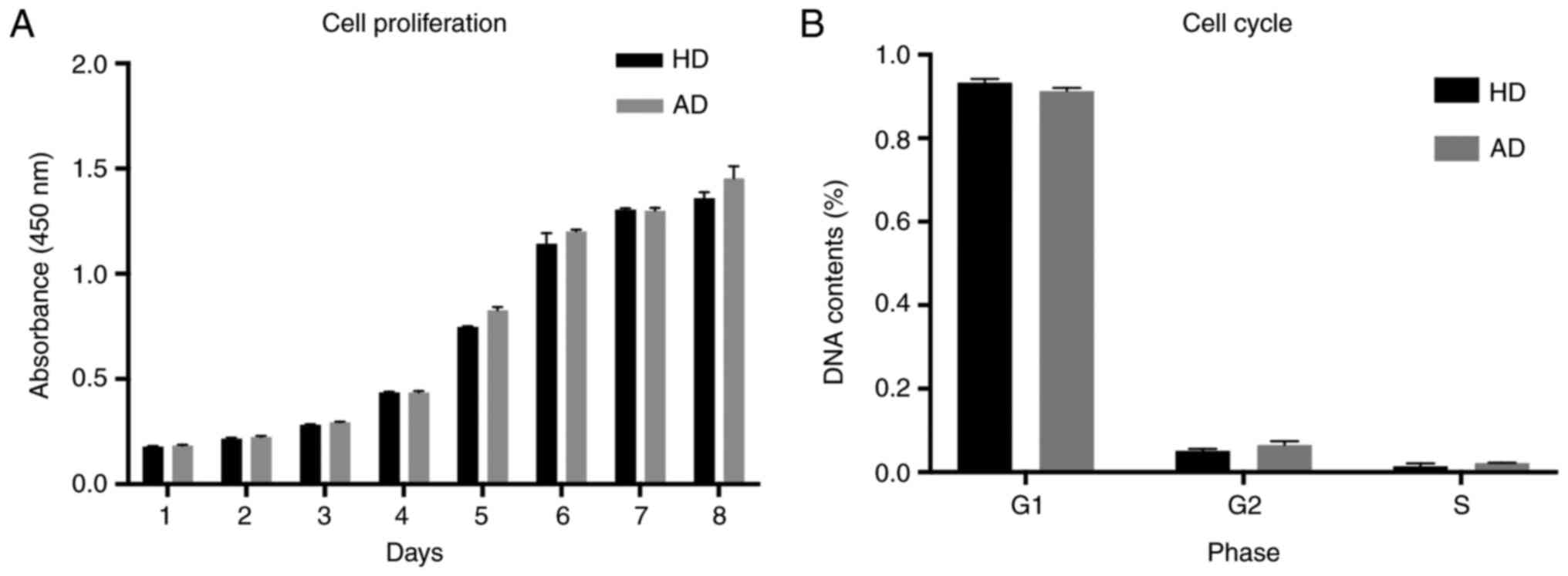
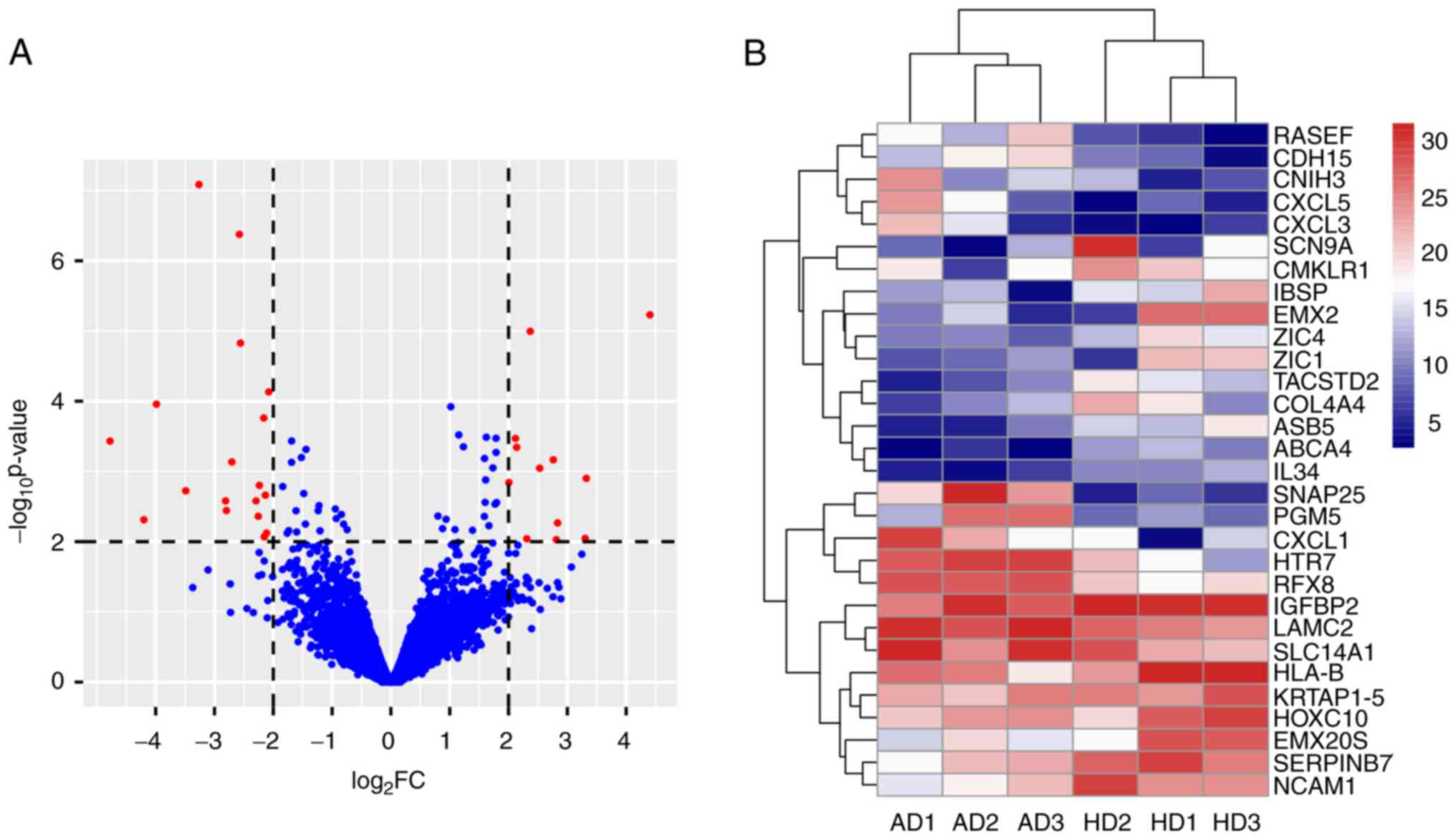
|
1
|
Hagan PG, Nienaber CA, Isselbacher EM,
Bruckman D, Karavite DJ, Russman PL, Evangelista A, Fattori R,
Suzuki T, Oh JK, et al: The International Registry of Acute Aortic
Dissection (IRAD): New insights into an old disease. JAMA.
283:897–903. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schlatmann TJ and Becker AE: Pathogenesis
of dissecting aneurysm of aorta. Comparative histopathologic study
of significance of medial changes. Am J Cardiol. 39:21–26. 1977.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
López-Candales A, Holmes DR, Liao S, Scott
MJ, Wickline SA and Thompson RW: Decreased vascular smooth muscle
cell density in medial degeneration of human abdominal aortic
aneurysms. Am J Pathol. 150:993–1007. 1997.PubMed/NCBI
|
|
4
|
De Bruin JL, Baas AF, Buth J, Prinssen M,
Verhoeven EL, Cuypers PW, van Sambeek MR, Balm R, Grobbee DE and
Blankensteijn JD: DREAM Study Group: Long-term outcome of open or
endovascular repair of abdominal aortic aneurysm. N Engl J Med.
362:1881–1889. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
SVN Task Force for Clinical Practice
Guideline: 2009 Clinical practice guideline for patients undergoing
endovascular repair of abdominal aortic aneurysms (AAA). J Vasc
Nurs. 27:48–63. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nabel EG, Yang Z, Liptay S, San H, Gordon
D, Haudenschild CC and Nabel GJ: Recombinant platelet-derived
growth factor B gene expression in porcine arteries induce intimal
hyperplasia in vivo. J Clin Invest. 91:1822–1829. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Phinney DG and Prockop DJ: Concise review:
Mesenchymal stem/multipotent stromal cells: The state of
transdifferentiation and modes of tissue repair-current views. Stem
Cells. 25:2896–2902. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hashizume R, Yamawaki-Ogata A, Ueda Y,
Wagner WR and Narita Y: Mesenchymal stem cells attenuate
angiotensin II-induced aortic aneurysm growth in apolipoprotein
E-deficient mice. J Vasc Surg. 54:1743–1752. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schneider F, Saucy F, de Blic R, Dai J,
Mohand F, Rouard H, Ricco JB, Becquemin JP, Gervais M and Allaire
E: Bone marrow mesenchymal stem cells stabilize already-formed
aortic aneurysms more efficiently than vascular smooth muscle cells
in a rat model. Eur J Vasc Endovasc Surg. 45:666–672. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Davis JP, Salmon M, Pope NH, Lu G, Su G,
Sharma AK, Ailawadi G and Upchurch GR Jr: Attenuation of aortic
aneurysms with stem cells from different genders. J Surg Res.
199:249–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zou S, Ren P, Zhang L, Azares AR, Zhang S,
Coselli JS, Shen YH and LeMaire SA: AKT2 promotes bone marrow
cell-mediated aortic protection in mice. Ann Thorac Surg.
101:2085–2096. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen YH, Hu X, Zou S, Wu D, Coselli JS and
LeMaire SA: Stem cells in thoracic aortic aneurysms and
dissections: Potential contributors to aortic repair. Ann Thorac
Surg. 93:1524–1533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie J, Jones TJ, Feng D, Cook TG, Jester
AA, Yi R, Jawed YT, Babbey C, March KL and Murphy MP: Human
adipose-derived stem cells suppress elastase-induced murine
abdominal aortic inflammation and aneurysm expansion through
paracrine factors. Cell Transplant. 26:173–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sharma AK, Salmon MD, Lu G, Su G, Pope NH,
Smith JR, Weiss ML and Upchurch GR Jr: Mesenchymal stem cells
attenuate NADPH oxidase-dependent high mobility group box 1
production and inhibit abdominal aortic aneurysms. Arterioscler
Thromb Vasc Biol. 36:908–918. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
del Moral Riera L, Largo C, Ramirez JR,
Vega Clemente L, Heredero Fernández A, de Cubas Riera L,
Garcia-Olmo D and Garcia-Arranz M: Potential of mesenchymal stem
cell in stabilization of abdominal aortic aneurysm sac. J Surg Res.
195:325–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ciavarella C, Alviano F, Gallitto E, Ricci
F, Buzzi M, Velati C, Stella A, Freyrie A and Pasquinelli G: Human
vascular wall mesenchymal stromal cells contribute to abdominal
aortic aneurysm pathogenesis through an impaired immunomodulatory
activity and increased levels of matrix metalloproteinase-9. Circ
J. 79:1460–1469. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop Dj and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The international society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nicoletti I, Migliorati G, Pagliacci MC,
Grignani F and Riccardi C: A rapid and simple method for measuring
thymocyte apoptosis by propidium iodide staining and flow
cytometry. J Immunol Methods. 139:271–279. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Longo GM, Xiong W, Greiner TC, Zhao Y,
Fiotti N and Baxter BT: Matrix metalloproteinases 2 and 9 work in
concert to produce aortic aneurysms. J Clin Invest. 110:625–632.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hackam DG, Thiruchelvam D and Redelmeier
DA: Angiotensin-converting enzyme inhibitors and aortic rupture: A
population-based case-control study. Lancet. 368:659–665. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schouten O, van Laanen JH, Boersma E,
Vidakovic R, Feringa HH, Dunkelgrün M, Bax JJ, Koning J, van Urk H
and Poldermans D: Statins are associated with a reduced infrarenal
abdominal aortic aneurysm growth. Eur J Vasc Endovasc Surg.
32:21–26. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamawaki-Ogata A, Hashizume R, Satake M,
Kaneko H, Mizutani S, Moritan T, Ueda Y and Narita Y: A doxycycline
loaded, controlled-release, biodegradable fiber for the treatment
of aortic aneurysms. Biomaterials. 31:9554–9564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baxter BT, Pearce WH, Waltke EA, Littooy
FN, Hallett JW Jr, Kent KC, Upchurch GR Jr, Chaikof EL, Mills JL,
Fleckten B, et al: Prolonged administration of doxycycline in
patients with small asymptomatic abdominal aortic aneurysms: Report
of a prospective (Phase II) multicenter study. J Vasc Surg.
36:1–12. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Walton LJ, Franklin IJ, Bayston T, Brown
LC, Greenhalgh RM, Taylor GW and Powell JT: Inhibition of
prostaglandin E2 synthesis in abdominal aortic aneurysms:
Implications for smooth muscle cell viability, inflammatory
processes, and the expansion of abdominal aortic aneurysms.
Circulation. 100:48–54. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshimura K, Aoki H, Ikeda Y, Fujii K,
Akiyama N, Furutani A, Hoshii Y, Tanaka N, Ricci R, Ishihara T, et
al: Regression of abdominal aortic aneurysm by inhibition of c-Jun
N-terminal kinase. Nat Med. 11:1330–1338. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bianco P, Riminucci M, Gronthos S and
Robey PG: Bone marrow stromal stem cells: Nature, biology, and
potential applications. Stem Cells. 19:180–192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsumura G, Miyagawa-Tomita S, Shin'oka
T, Ikada Y and Kurosawa H: First evidence that bone marrow cells
contribute to the construction of tissue-engineered vascular
autografts in vivo. Circulation. 108:1729–1734. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Allaire E, Muscatelli-Groux B, Guinault
AM, Pages C, Goussard A, Mandet C, Bruneval P, Méllière D and
Becquemin JP: Vascular smooth muscle cell endovascular therapy
stabilizes already developed aneurysms in a model of aortic injury
elicited by inflammation and proteolysis. Ann Surg. 239:417–427.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sata M, Saiura A, Kunisato A, Tojo A,
Okada S, Tokuhisa T, Hirai H, Makuuchi M, Hirata Y and Nagai R:
Hematopoietic stem cells differentiate into vascular cells that
participate in the pathogenesis of atherosclerosis. Nat Med.
8:403–409. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saiura A, Sata M, Hirata Y, Nagai R and
Makuuchi M: Circulating smooth muscle progenitor cells contribute
to atherosclerosis. Nat Med. 7:382–383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ryu JH, Park M, Kim BK, Ryu KH and Woo SY:
Tonsil-derived mesenchymal stromal cells produce CXCR2-binding
chemokines and acquire follicular dendritic cell-like phenotypes
under TLR3 stimulation. Cytokine. 73:225–235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Veréb Z, Póliska S, Albert R, Olstad OK,
Boratkó A, Csortos C, Moe MC, Facskó A and Petrovski G: Role of
human corneal stroma-derived mesenchymal-like stem cells in corneal
immunity and wound healing. Sci Rep. 6:262272016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Q, Guo Y, Chen F, Liu J and Jin P:
Stromal cell-derived factor-1 promotes human adipose tissue-derived
stem cell survival and chronic wound healing. Exp Ther Med.
12:45–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharma AK, Lu G, Jester A, Johnston WF,
Zhao Y, Hajzus VA, Saadatzadeh MR, Su G, Bhamidipati CM, Mehta GS,
et al: Experimental abdominal aortic aneurysm formation is mediated
by IL-17 and attenuated by mesenchymal stem cell treatment.
Circulation. 126 11 Suppl 1:S38–S45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iida Y, Xu B, Xuan H, Glover KJ, Tanaka H,
Hu X, Fujimura N, Wang W, Schultz JR, Turner CR and Dalman RL:
Peptide inhibitor of CXCL4-CCL5 heterodimer formation, MKEY,
inhibits experimental aortic aneurysm initiation and progression.
Arterioscler Thromb Vasc Biol. 33:718–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Anzai A, Shimoda M, Endo J, Kohno T,
Katsumata Y, Matsuhashi T, Yamamoto T, Ito K, Yan X, Shirakawa K,
et al: Adventitial CXCL1/G-CSF expression in response to acute
aortic dissection triggers local neutrophil recruitment and
activation leading to aortic rupture. Circ Res. 116:612–623. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tanios F, Pelisek J, Lutz B, Reutersberg
B, Matevossian E, Schwamborn K, Hösel V, Eckstein HH and Reeps C:
CXCR4: A potential marker for inflammatory activity in abdominal
aortic aneurysm wall. Eur J Vasc Endovasc Surg. 50:745–753. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Michineau S, Franck G, Wagner-Ballon O,
Dai J, Allaire E and Gervais M: Chemokine (C-X-C motif) receptor 4
blockade by AMD3100 inhibits experimental abdominal aortic aneurysm
expansion through anti-inflammatory effects. Arterioscler Thromb
Vasc Biol. 34:1747–1755. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ramos-Mozo P, Rodriguez C, Pastor-Vargas
C, Blanco-Colio LM, Martinez-Gonzalez J, Meilhac O, Michel JB, de
Ceniga Vega M, Egido J and Martin-Ventura JL: Plasma profiling by a
protein array approach identifies IGFBP-1 as a novel biomarker of
abdominal aortic aneurysm. Atherosclerosis. 221:544–550. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Razuvaev A, Folkersen L, Hedin E,
Roy J, Brismar K and Hedin U: The expression of IGFs and IGF
binding proteins in human carotid atherosclerosis, and the possible
role of IGF binding protein-1 in the regulation of smooth muscle
cell proliferation. Atherosclerosis. 220:102–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Giancotti FG and Ruoslahti E: Integrin
signaling. Science. 285:1028–1032. 1999. View Article : Google Scholar : PubMed/NCBI
|