Introduction
Liver cancer is the fifth most prevalent diagnosed
cancer and the second most common cause of cancer-associated
mortality worldwide (1). Of all
malignant types of liver cancer, hepatocellular carcinoma (HCC) is
the most common type in adults, which accounts for 70–85% of cases
(2), and the highest incidence
rates are reported to occur in Asia and Africa (3). HCC is an insidious disease without
symptoms of pain, which results in diagnosis at a late stage
(4). Although there has been
significant progress in HCC therapy, recurrence and metastasis
remain major obstacles in improving outcomes following surgical
resection and liver transplantation (5,6). The
5-year survival rate is limited to 30%. Therefore, the
investigation of novel effective biomarkers and targets to develop
novel effective strategies is urgently required.
The sirtuins are a family of orthologues, which
share extensive homologies with the silent information regulator 2
(Sir2) gene in yeast. In mammals, sirtuins comprise seven members,
termed sirtuin (SIRT)1-SIRT7, respectively, and are important in
the regulation of metabolism, cellular proliferation, aging,
survival, and oncogenesis or tumor suppression in the progression
of cancer (7–9).
Among the sirtuin family members, SIRT5 has been
investigated the least, and the role of SIRT5 in carcinogenesis
remains to be elucidated, which requires further detailed
investigation. In the present study, the expression status of SIRT5
in HCC specimens and cell lines were evaluated, and the functional
contribution of SIRT5 in the progression of HCC was
investigated.
Materials and methods
Cell culture
The human HCC SMMC-7721, HepG2, LO2 and Huh7 cell
lines, and HEK-293T cells were purchased from the American Type
Culture Collection (Manassas, VA, USA). Non-tumorigenic
immortalized liver cell line MiHA were purchased from the Cell bank
of the Institute of Biochemistry and Cell Biology (Academy of Life
Science; Shanghai, China). The cells were placed in Dulbecco's
modified Eagle's medium (DMEM) (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (FBS) (Invitrogen; Thermo Fisher Scientific, Inc.),
penicillin (100 U/ml) and streptomycin (100 µg/ml) (Invitrogen;
Thermo Fisher Scientific, Inc.), and were cultured in a 5%
CO2 incubator at 37°C.
Reagents and transfection
The relative small interfering (si)RNAs and the
negative control siRNA were purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). The siRNA sequences were as
follows: SIRT5 siRNA#1,
5′-CCGGGCGTGCCAGTGGCTGAATTTACTCGAGTAAATTCAGCCACTGGCACGCTTTTTG-3′;
SIRT5 siRNA#2,
5′-CCGGCTGAGTACTGAACAATCTAAACTCGAGTTTAGATTGTTCAGTACTCAGTTTTTG-3′;
E2F1 siRNA#1,
5′-CCGGCCCAACTACAAGCTGTGGATTCTCGAGAATCCACAGCTTGTAGTTGGGTTTTTG-3′;
E2F1 siRNA#2
(5′-CCGGGCCAAGAAGTCCAAGAATCATCTCGAGATGATTCTTGGACTTCTTGGCTTTTTG-3′);
and non-silencing siRNA
5′-CTAGCCCGGTTCTCCGAACGTGTCACGTATCTCGAGATACGTGACACGTTCGGAGAATTTTTTTAAT-3′.
The cells (2×105) were seeded in 6-well plates overnight
and transfected with 5 µM siRNAs using Lipofectamine®
RNAiMAX (Invitrogen Life Technologies; Thermo Fisher Scientific,
Inc.). After 1–2 days, the transfected cells were harvested for RNA
or protein extraction. The antibodies used in the present study
were as follows: Anti-SIRT5 antibody (cat. no. ab105040), anti-E2F
transcription factor 1 (E2F1) antibody (cat. no. ab179445), and
anti-β-actin antibody (cat. no. ab8226; all from Abcam, Cambridge,
UK).
Generation of lentivirus
HEK-293T cells were used to produce the lentivirus.
The HEK-293T cells (2×107) were grown to 50–60%
confluence in 15 cm dishes, and the purified shRNA plasmids and the
lentivirus GenePharm (Shanghai, China) were co-transfected into the
HEK-293T cells using Lipofectamine 2000 reagent (Invitrogen Life
Technologies; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. After 6 h, the medium was removed and
fresh medium was added. At 4 days-post transfection, the media were
harvested. The viruses were released by three freeze/thaw cycles
and stored at −80°C until use. For viral infection, 30 µl of viral
stock solution was added to the culture medium in each group. The
expression of SIRT5 was determined in the infected cells using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis.
Patients and tissue specimens
A total of 55 fresh HCC tissue samples and paired
non-cancerous lung tissue samples were obtained during surgery in
the first surgical procedure following the diagnosis of HCC between
January 2005 and December 2009 at the Department of Pathology at
Guangdong General Hospital (Guangzhou, China). None of the patients
had received radiotherapy or chemotherapy prior to surgery.
Tumor-node-metastasis (TNM) classification was defined according to
the American Joint Committee on International Union Against Cancer
(10). The stage IV samples were
obtained from patients with metastasis. The indication for surgery
was according to tumor size, age and patient aspirations. The
clinical information of patients are summarized in Table I. Pertinent follow-up information
was available for all patients. The cut-off value was decided by
the ROC curve. Written informed consent was obtained from each
patient and established study approval was obtained from the
Institutional Research Ethics Committee from Southern Medical
University.
 | Table I.Clinicopathologic variables in 55
patients with hepatocellular carcinoma. |
Table I.
Clinicopathologic variables in 55
patients with hepatocellular carcinoma.
|
|
| SIRT5 protein
expression |
|
|---|
|
|
|
|
|
|---|
| Variable | n | Low (n=20) | High (n=35) | P-value |
|---|
| Age (years) |
|
<50 | 32 | 12 | 20 | 0.836 |
| ≥50 | 23 | 8 | 15 |
|
| Sex |
| Male | 38 | 14 | 24 | 0.912 |
|
Female | 17 | 6 | 11 |
|
| Tumor size |
| Small (≤5
cm) | 20 | 11 | 9 | 0.030 |
| Large (≥5
cm) | 35 | 9 | 26 |
|
| Lymph node
metastasis |
| Yes | 27 | 5 | 22 | 0.007 |
| No | 28 | 15 | 13 |
|
| TNM stage |
| I+II | 27 | 14 | 13 | 0.019 |
|
III+IV | 28 | 6 | 22 |
|
| Differentiation |
|
Well/moderate | 29 | 11 | 18 | 0.799 |
| Poor | 26 | 9 | 17 |
|
RNA extraction and RT-qPCR
analysis
Total RNA was extracted from the frozen human
clinical tissues and cultured cell lines using TRIzol reagent
(Invitrogen Life Technologies; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The RNA (2 µg) was used
for first strand cDNA synthesis using the Revert Aid™ First Strand
cDNA Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.). In
order to quantify the transcripts of the genes, qPCR was performed
using a SYBR-Green mixture 2 µl cDNA, 1 µl primers, 10 µl mixture
buffer and 7 µl water (Qiagen, Inc., Valencia, CA, USA) on an ABI
7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The primers used were as follows: GAPDH forward,
5′-GAGAAGTATGACAACAGCCTC-3′ and reverse,
5′-ATGGACTGTGGTCATGAGTC-3′; SIRT5 forward,
5′-AAATAACTAAAGCCCGCCTC-3′ and reverse,
5′-TCCTGAGATGATGACTATGTG-3′; and E2F1 forward,
5′-GACTCTTCGGAGAACTTTCAG-3′ and reverse,
5′-GATCTGTGGTGAGGGATGAG-3′. The PCR amplification was 95°C for 5
min, followed by 35 cycles of 95°C for 20 sec, 60°C for 15 sec, and
72°C for 30 sec. The genes expression were normalized against that
of GAPDH and relative fold changes were calculated using the
formula 2−ΔΔCq (11).
Western blot analysis
The relative tissues or cells were lysed in 50 mM
Tris HCl (pH 7.5; Beyotime Institute of Biotechnology, Shanghai,
China) and 1% SDS with protease/phosphatase inhibitor cocktail
(Roche Diagnostics, Basel, Switzerland), and then heated at 95°C
for 10 min. The protein was quantified using a BCA kit (Thermo
Fisher Scientific, Inc.). Equal quantities of protein (30 µg) were
subjected to 10% SDS-PAGE, and transferred onto polyvinylidene
fluoride membranes (EMD Millipore, Billerica, MA, USA). Following
blocking in 5% nonfat milk with PBS containing 0.1% Tween-20, the
membranes were incubated with primary antibodies anti-SIRT5
antibody (1:1,000, cat. no. ab105040), anti-E2F1 antibody (1:1,000,
cat. no. ab179445) and anti-β-actin antibody (1:1,000, cat. no.
ab8226) at 4°C overnight, and then washed for least five times.
This was followed by incubation with horseradish peroxidase-labeled
secondary antibodies (Santa Cruz Biotechnology, Inc.) goat
anti-mouse IgG-horseradish peroxidase (sc-2005; 1:3,000) and
anti-rabbit IgG-horseradish peroxidase (sc-2004; 1:3,000) at room
temperature for 1 h. The membranes were detected using an enhanced
chemiluminescence-based method.
Quantitative chromatin
immunoprecipitation (qChIP) analysis
The qChIP assay was performed using the Chip-IT
Express kit (Active Motif, Carlsbad, CA, USA). The relative
cultured cells were cross-linked with formaldehyde, incubated with
ChIP lysis buffer for 30 min, and sonicated to generate 200-500-bp
DNA fragments at 4°C. Supernatant was incubated with 5 µg relative
antibody, or normal IgG as a negative control, overnight at 4°C.
Following being washed five times with lysis buffer, the DNA was
de-crosslinked, extracted using phenol chloroform and precipitated
with ethanol. Following resuspension in 50 µl of H2O,
qPCR was performed with 5 µl of the immunoprecipitated target DNA,
1 µl primers and 9 µl mixture (1 µl enzyme, 2 µl dNTP and 6 µl
solution buffer). The qChIP primers were as follows: E2F1 forward,
5′-CTGGGTGCAGGATTGGATA-3′ and reverse, 5′-TCAACCTGTAGCCCCCAAC-3′.
The PCR amplification was performed at 95°C for 5 min, followed by
35 cycles of 95°C for 20 sec, 55°C for 20 sec, and 72°C for 30 sec.
Bioinformatics was performed using PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)
to submit the promoter sequence of E2F1 to find the potential
binding of SIRT5.
Luciferase reporter gene assay
A luciferase reporter gene assay was performed to
validate the predicted target. Briefly, the promoter region of E2F1
containing the putative binding site was cloned into a PGL3
luciferase reporter vector (Promega, Madison, WI, USA). The HCC
cells (1×103) were seeded in 96-well plates, and the
E2F1 reporter plasmid was cotransfected into cells with 2 ng/well
Renilla luciferase construct in triplicate. After 36 h, the
activities of Firefly and Renilla luciferase were determined
using a dual luciferase detection kit (Promega).
Proliferation assay
A Cell Counting kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used to detect cell
proliferation. The relative transfected cells (5×103
cells) were plated on 48-well plates and cultured overnight to
allow attachment. Following serum-starvation for 8 h, FBS was added
into the medium. At 0, 12, 24, 36, 48 and 72 h, the cells were
incubated with 10 µl CCK-8 solution for 2 h, and analyzed at 450 nm
using a multi-well plate reader. The experiments were performed in
triplicate.
Invasion assay
An invasion assay was performed using Transwell
chambers with inserts of 8-mm pore size (Corning Costar; Corning
Inc., Corning, NY, USA). Briefly, the relative hepatoma cells
(1×105) were suspended in serum-free medium and seeded
into Matrigel invasion chambers at 37°C. Complete medium (DMEM with
10% FBS) was added to the lower chamber. Following cultured for 72
h, the cells on the lower surface were stained with crystal violet.
The cell numbers were counted under a microscope (magnification,
×40) (Leica DM4B; Leica Microsystems, Inc., Buffalo Grove, IL, USA)
in five randomly selected fields in each well.
Statistical analysis
Statistical analysis was performed using SPSS
version 17.0 (SPSS, Inc., Chicago, IL, USA). Data are expressed as
the mean ± standard deviation of three independent experiments.
Student's t-test or one-way analysis of variance were used, as
appropriate. A Kaplan-Meier plot was used to analyze the survival
curves and a log-rank test was used to compare the survival curves.
The association between expression levels and clinicopathological
characteristics were analyzed using a χ2 test and
Spearman's correlation. P<0.05 was considered to indicate a
statistically significant difference.
Results
SIRT5 is upregulated in HCC cell lines
and tumor tissues
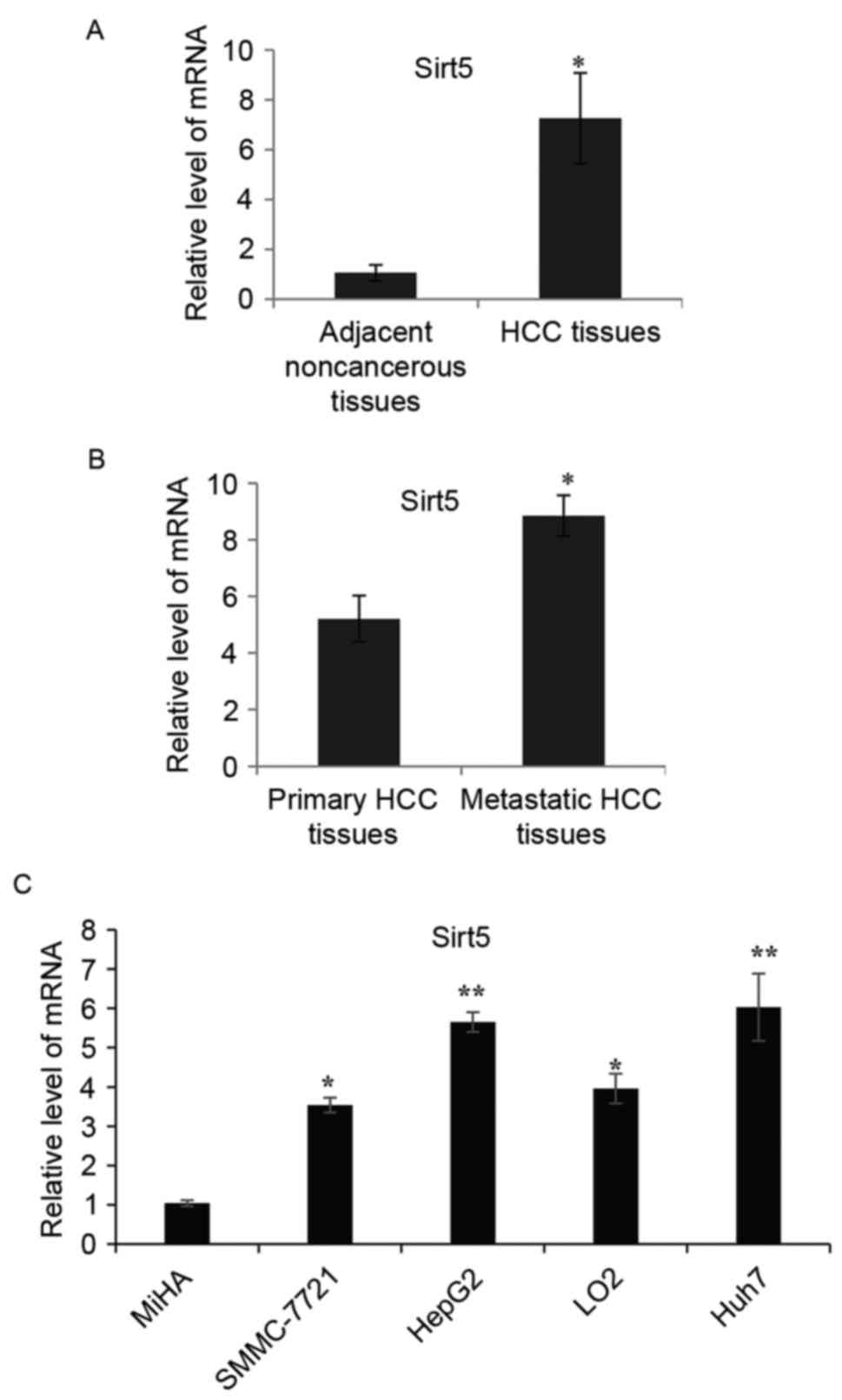
RT-qPCR analysis was used to analyze the expression
of SIRT5 in 55 surgical HCC specimens. As shown in Fig. 1A, the levels of SIRT5 were
significantly upregulated in the majority of the HCC tissues,
compared with those in the paired adjacent noncancerous tissues.
The upregulation of SIRT5 in HCC tissues suggested that SIRT5 may
be important in the progression of HCC. The expression of SIRT5 was
further examined in 20 metastatic and primary HCC tissues. As shown
in Fig. 1B, there was a
significantly higher expression of SIRT5 in the metastatic tissues,
compared with the primary HCC samples. In subsequent experiments,
RT-qPCR analysis was performed to detect the expression of SIRT5 in
different HCC cell lines (SMMC-7721, HepG2, LO2 and Huh7 cells). It
was found that the expression of SIRT5 was generally higher in
these HCC cell lines, compared with that in the non-tumorigenic
immortalized liver cell line (MiHA), as shown in Fig. 1C.
High expression of SIRT5 is associated
with a poor clinical prognosis in HCC
To further elucidate the clinical relevance of the
expression of SIRT5 in HCC, the present study investigated the
association between the expression of SIRT5 and clinicopathological
features. As summarized in Table
I, the higher expression level of SIRT5 in HCC was positively
correlated with tumor size, lymph node metastasis status and TNM
stage. However, no significant correlations were observed between
the expression levels of SIRT5 and the clinical features of sex,
age or degree of tumor differentiation. These data indicated that
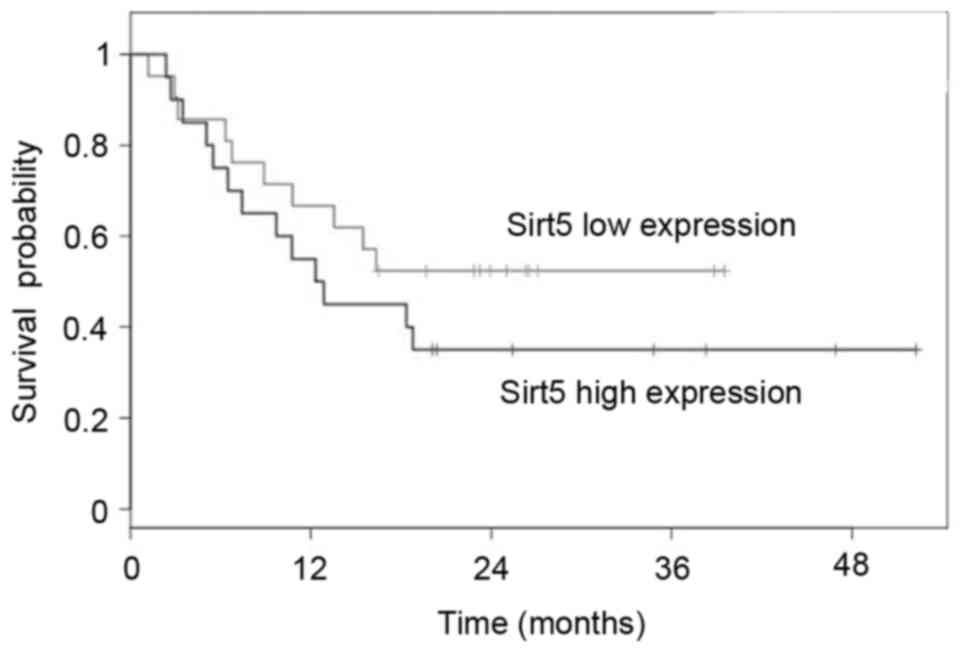
SIRT5 may be involved in the progression of HCC. Kaplan-Meier
analysis was also performed to analyze the association between
tumor expression and patient overall survival rates. As shown in
Fig. 2, patients with a high
expression of SIRT5 had a poorer OS, compared with patients with a
low expression of SIRT5 (log-rank test, P<0.001).
SIRT5 promotes HCC cell growth in
vitro
The high expression levels of SIRT5 in HCC suggested
that SIRT5 may be involved in the development and prognosis of HCC.
To determine whether SIRT5 is essential for HCC cell growth, two
types of lentiviral vector carrying various sh-RNA sequences to
knockdown SIRT5 (shSIRT5) were stably expressed in Huh7 and HepG2
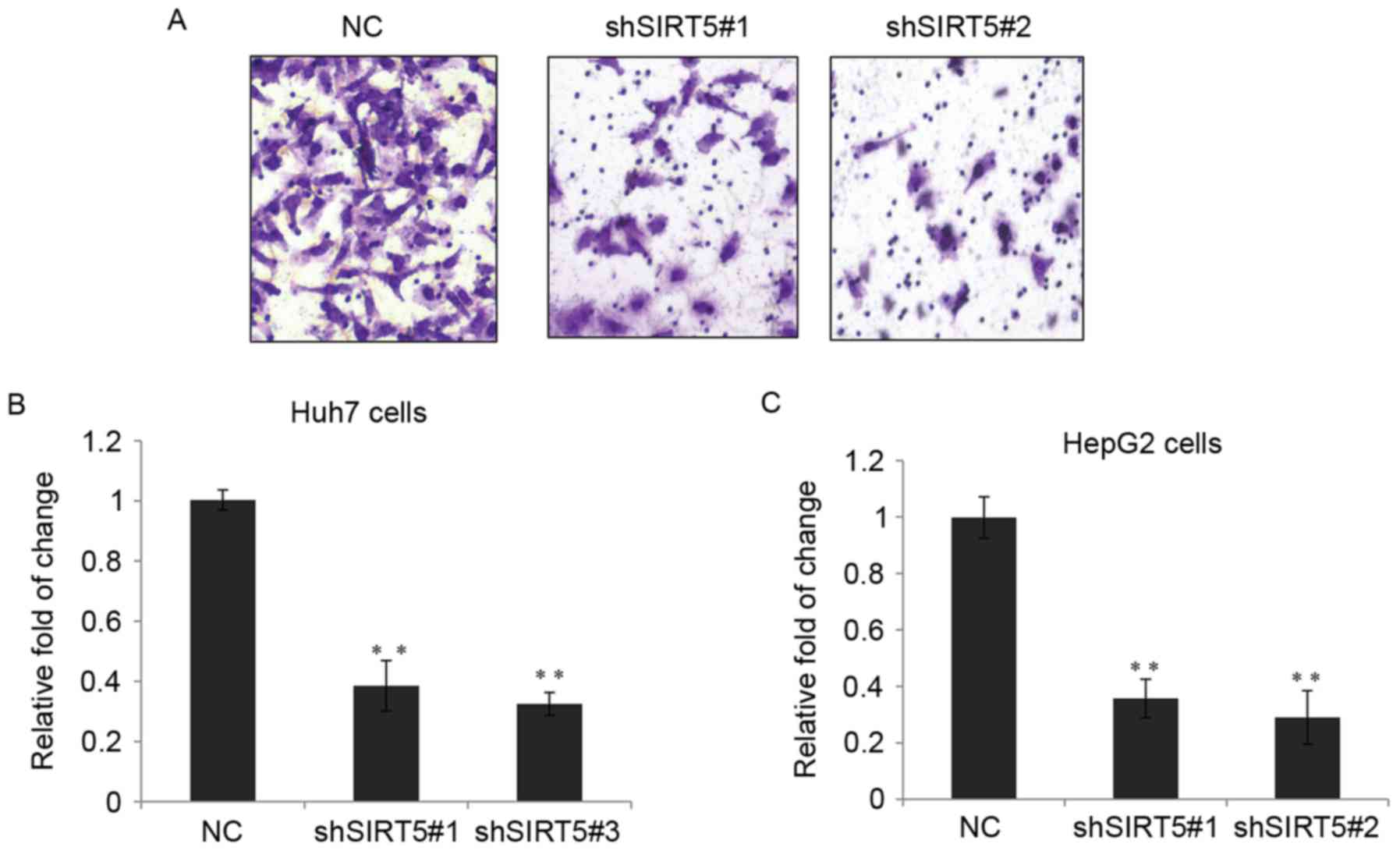
cells. As shown in Fig. 3A and B,
shSIRT5-1 and shSIRT5-2 were able to efficiently reduce the
expression of SIRT5 at the mRNA level. Cell proliferation was
detected using a CCK-8 assay in the two cell lines, and the cells
stably expressing shSIRT5-1 or shSIRT5-2 showed significantly
decreased cell proliferation, compared with the control (Fig. 3C and D).
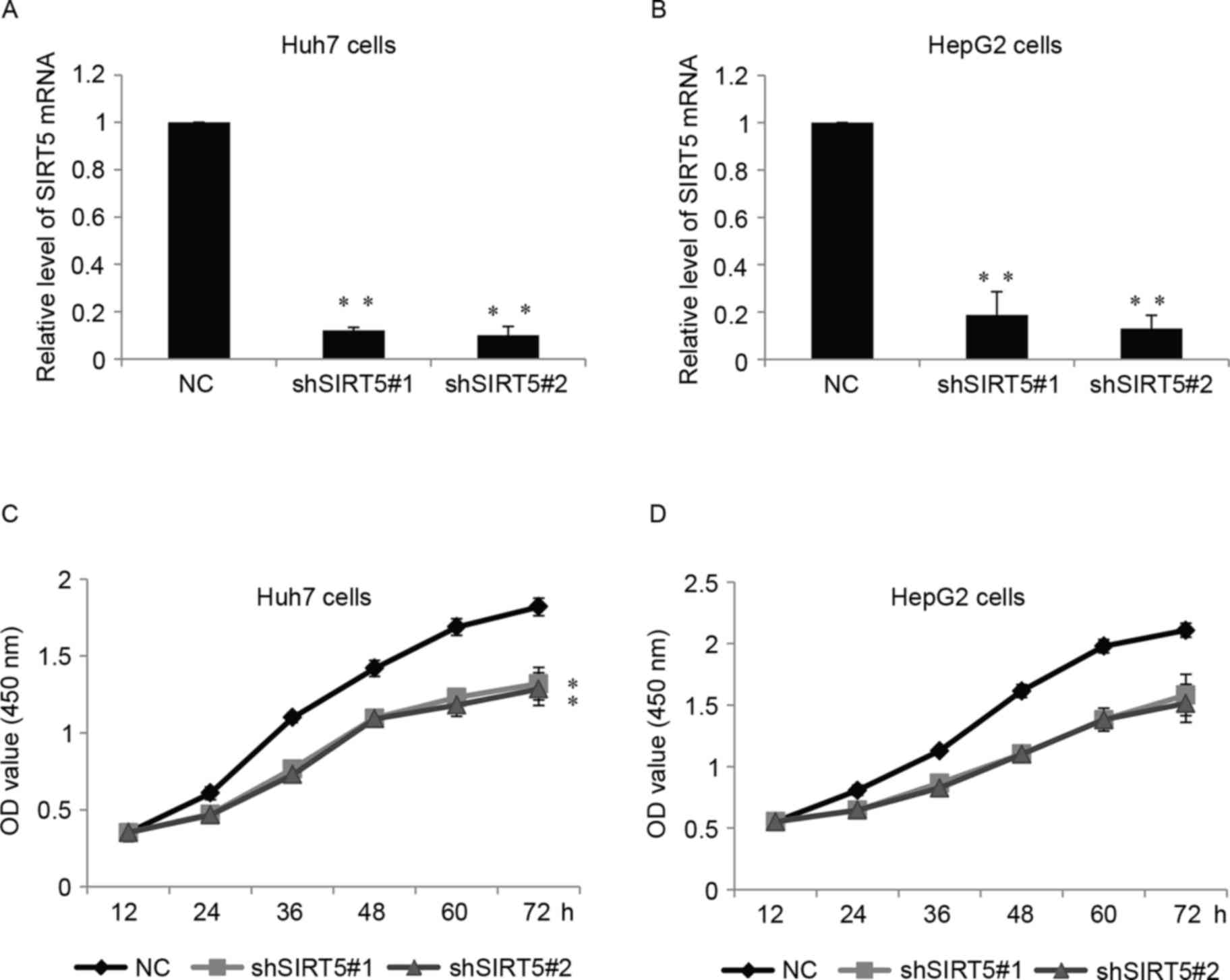 | Figure 3.SIRT5 promotes hepatocellular
carcinoma cell growth in vitro. (A) mRNA levels of SIRT5
were measured in Huh7 cells stably transfected with shSIRT5#1,
shSIRT5#2 or NC. Data are presented as the mean ± standard
deviation. **P<0.01. (B) Relative mRNA levels of SIRT5 in HepG2
cells. (C) CCK-8 assay of Huh7 cells, which were stably transfected
with either shSIRT5#1, shSIRT5#2 or NC, at 0, 12, 24, 36, 48 and 72
h. **P<0.01. (D) Relative CCK-8 assay in HepG2 cells. SIRT5,
sirtuin 5; sh, short hairpin RNA; NC, negative control; CCK-8, Cell
Counting kit-8; OD, optical density. |
SIRT5 regulates HCC cell invasion in
vitro
To better understand the functionality of SIRT5 in
the progression of HCC, the present study examined whether SIRT5
can exert an effect on HCC cell invasion. A Transwell assay was
performed and, as expected, the depletion of SIRT5 in Huh7 cells
resulted in a marked decrease in cell invasion, as shown in the
images in Fig. 4A and the
statistical analyses in Fig. 4B. A
Transwell assay was also performed for the HepG2 cells, and a
similar trend was observed (Fig.
4C).
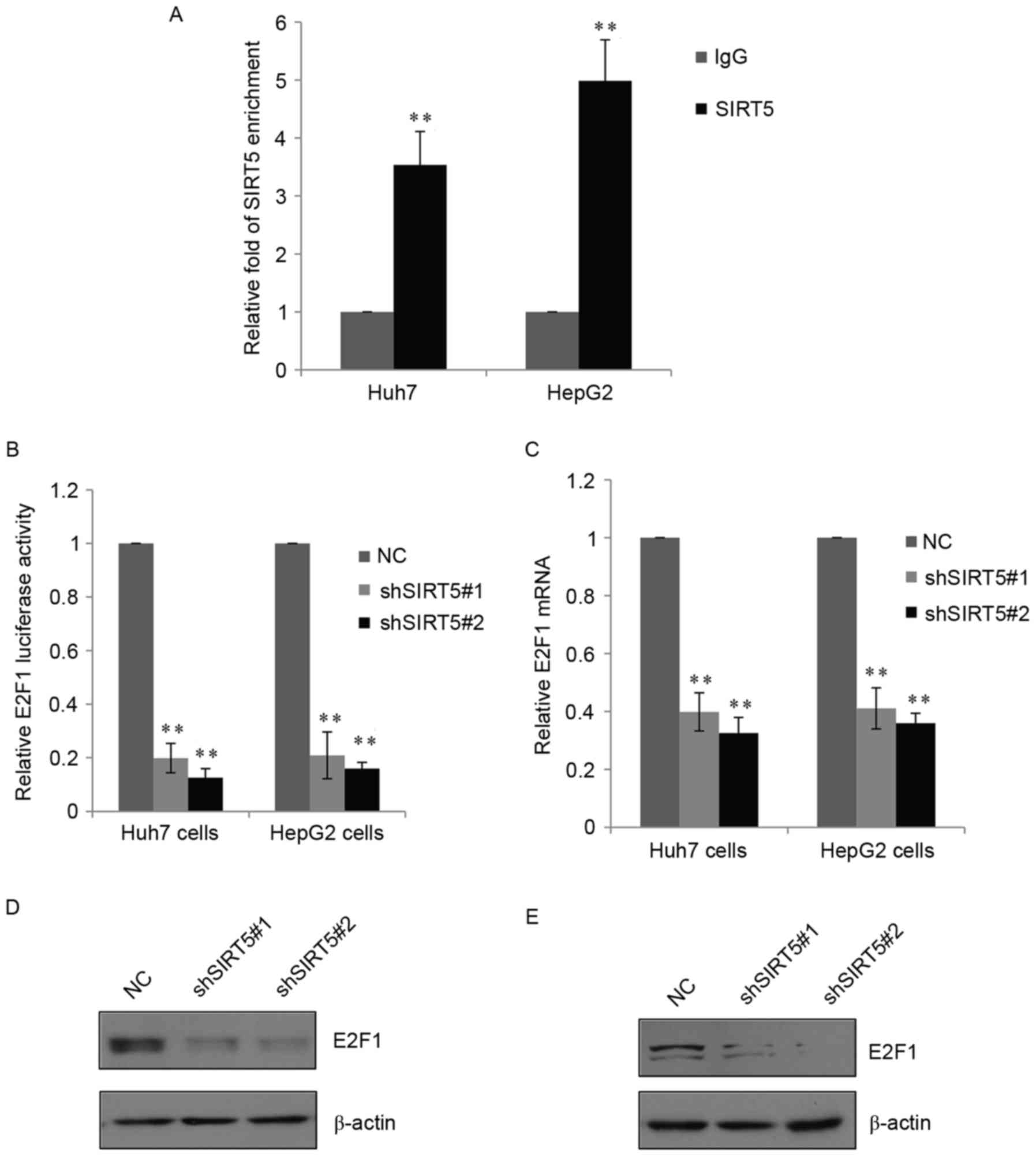
SIRT5 directly targets E2F1 and
activates its expression
Previous studies (12,13)
have shown that sirtuins can regulate the expression of several
genes at the transcriptional or post-transcriptional level. To
identify potential target genes of SIRT5, bioinformatics analysis
was used, which revealed that SIRT5 directly targeted E2F1, an
important regulator involved in tumor progression. To further
examine this, a qChIP assay was performed in the huh7 cells and
HepG2 cells (Fig. 5A). Compared
with the normal IgG group, there was binding enrichment of SIRT5 on
the promoter of E2F1. A luciferase reporter vector containing the
promoter of E2F1 was also constructed, and the reporter assay
showed that the knockdown of SIRT5 was able to significantly reduce
the expression of E2F1 luciferase in the two cell lines (Fig. 5B). Consistently, shSIRT5
significantly inhibited the mRNA levels and inhibited protein
levels of E2F1 in the Huh7 and HepG2 cells (Fig. 5C and D). These data demonstrated
that SIRT5 directly targeted E2F1 and activated the endogenous
expression of E2F1 in HCC cells.
SIRT5 promotes HCC cell proliferation
and invasion by targeting E2F1
The above results showed that SIRT5 acted primarily
as a promoter of HCC cell proliferation and invasion, and
identified E2F1 as a target gene. To evaluate the direct effect of
E2F1 in the growth and invasive potential of SIRT5, two shRNAs
(shE2F1-1 and shE2F1-2) targeting E2F1 were used to suppress its
expression. As shown in Fig. 6A and
B, the shRNAs efficiently reduced the expression of E2F1 at the
mRNA level, compared with the NC group and mock-treated group. As
shown in Fig. 6C and D, the
knockdown of SIRT5 inhibited Huh7 cell and HepG2 cell growth, and
the additional knockdown of E2F1 in the two cells partially
reversed the inhibitory effect of SIRT5-knockdown. Similarly, SIRT5
promoted Huh7 or HepG2 cell invasion by regulating the expression
of E2F1, as the knockdown of E2F1 partially reduced the effect of
SIRT5 (Fig. 6E and F).
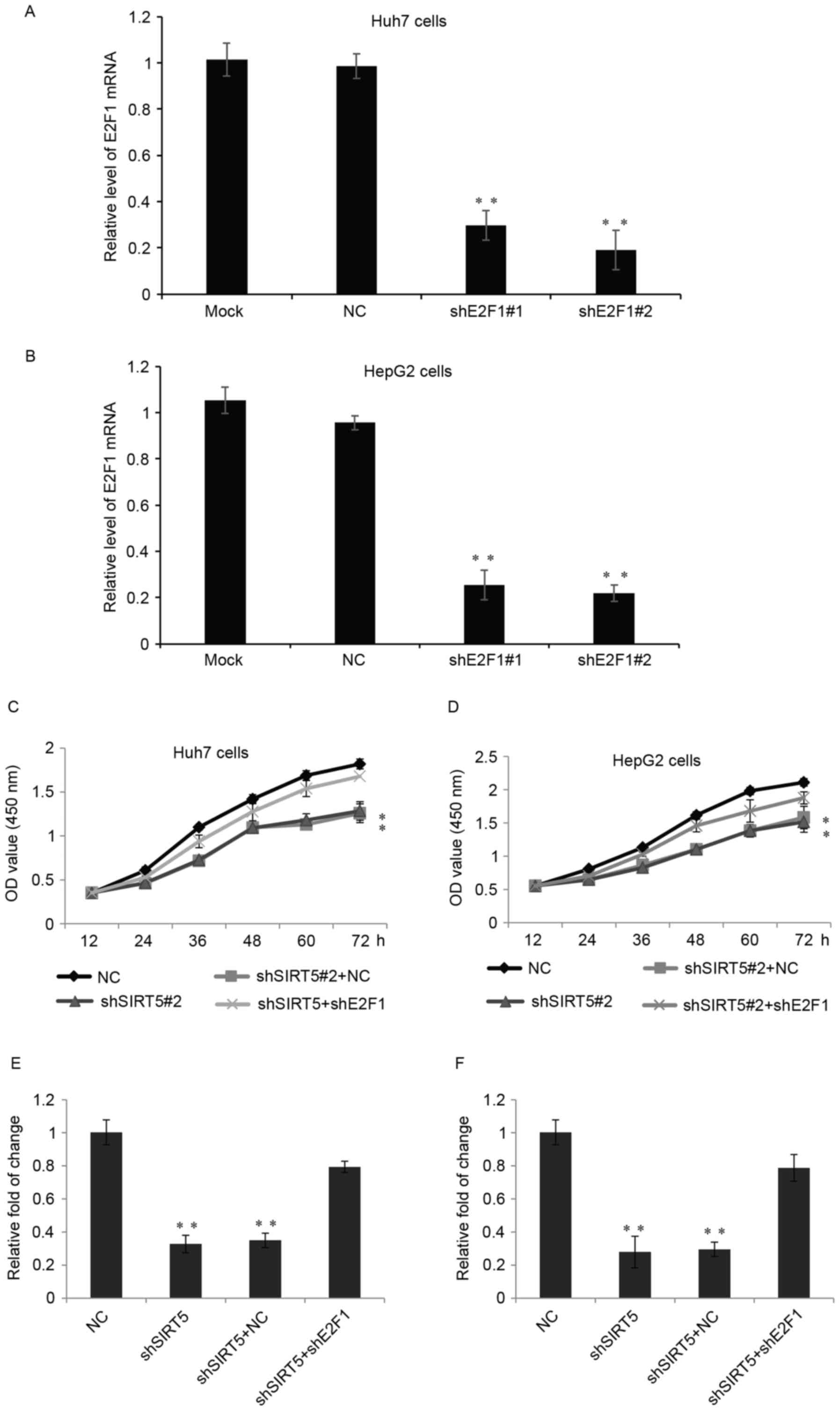 | Figure 6.SIRT5 promotes hepatocellular
carcinoma cell proliferation and invasion by targeting E2F1. (A)
mRNA levels of E2F1 were measured in Huh7 cells stably transfected
with shE2F1#1, shE2F1#2, NC and in untreated (Mock) cells. Data are
presented as the mean ± standard deviation. **P<0.01. (B)
Relative mRNA levels of E2F1 were measured in HepG2 cells. (C)
CCK-8 assay of Huh7 cells stably transfected with either NC,
shSIRT5+NC or shSIRT5+shE2F1 at 0, 12, 24, 36, 48 and 72 h.
**P<0.01. (D) Relative CCK-8 assay was performed in HepG2 cells.
(E) Cell invasion assay was performed in Huh7 cells transfected
with NC, shSIRT5, shSIRT5+NC or shSIRT5+shE2F1. Data are presented
as the mean ± standard deviation of at least three independent
experiments. (F) Relative Transwell assay in HepG2 cells. SIRT5,
sirtuin 5; E2F1, E2F transcription factor 1; sh, short hairpin RNA;
NC, negative control; OD, optical density; CCK-8, Cell Counting
kit-8. |
Discussion
In previous years, the high recurrence rate of HCC
has remained a major challenge in improving the outcome for
patients with HCC. Therefore, identifying early therapeutic targets
is important for improved clinical outcome. In the present study,
it was shown that SIRT5 was overexpressed in the majority of
primary HCC tumor tissues, compared with corresponding non-tumorous
liver tissues. The data revealed that the expression of SIRT5 was
significantly upregulated in HCC tissues, and that higher
expression levels of SIRT5 were observed in metastatic HCC samples.
The expression pattern of SIRT5 found in the present study was
consistent with previous investigations, in which SIRT5 was also
overexpressed in human non-small cell lung cancer, with a high
expression of SIRT5 predictive of poor survival rates (13,14).
Further statistical analysis found that a higher expression of
SIRT5 in the HCC tissues was significantly associated with
malignant tumor characteristics, including tumor size, lymph node
metastasis and TNM stage. Therefore, these results demonstrated
that SIRT5 is a potential promoter of metastasis in HCC.
Kaplan-Meier survival analysis also revealed that the higher
expression of SIRT5 was associated with poor prognosis following
surgical resection in patients with HCC.
The above findings suggested that SIRT5 may serve as
a novel prognostic predictor and therapeutic target in patients
with HCC. By obtaining in vitro evidence using HCC cell
lines, the present study demonstrated that SIRT5 facilitated cell
growth and invasion of the selected HCC cell lines.
Mechanistically, it was observed that SIRT5 regulated the
expression of E2F1.
As a founding member of the E2F transcription
factors, E2F1 is involved in regulating cell differentiation, DNA
repair and apoptosis (15,16). E2F1 also functions as a connector
and coordinator between cell proliferation and metabolic pathways
in mitochondria. Mitochondria are multifunctional organelles, which
are suggested to be actively involved in various aspects of
tumorigenesis (17). It has been
reported that E2F1 enhances glycolysis through suppressing the
transcription of Sirt6 in PC3 prostate cancer and UMUC3 bladder
cancer cells (18). In
vitro studies have shown that E2F1 may act either as an
oncogene or as a tumor suppressor (19). E2F1 is overexpressed and
pro-apoptotic in human HCC (20)
and acts as a growth-promoting factor in non-small cell lung
carcinoma, which is associated with adverse prognosis, and as a
promoter of invasion and metastasis (21,22).
The present study revealed that, through the activation of E2F1,
SIRT5 promotes HCC proliferation and invasion. However, the present
study was limited by the low quantity of patient tissue samples, as
there was insufficient tissue to measure protein expression levels
to determine their association with SIRT5 and E2F1.
As the most well-characterized member of the sirtuin
family, it has been reported that, in HCC, the overexpression of
SIRT1 can promote metastasis through epithelial-mesenchymal
transition (EMT) (23). This
suggests that SIRT5 may also regulate HCC cell invasion through the
EMT progress, which is reported to be associated with liver cancer
migration and metastasis (24,25)
EMT-associated transcription factors may be target genes of SIRT1
in HCC, therefore, additional target genes require investigation in
the future.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Forner A, Llovet JM and Bruix J:
Hepatocellular carcinoma. Lancet. 379:1245–1255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bosch FX, Ribes J, Díaz M and Cléries R:
Primary liver cancer: Worldwide incidence and trends.
Gastroenterology. 127 5 Suppl 1:S5–S16. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alazawi W, Cunningham M, Dearden J and
Foster GR: Systematic review: Outcome of compensated cirrhosis due
to chronic hepatitis C infection. Aliment Pharmacol Ther.
32:344–355. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bertino G, Demma S, Ardiri A, Proiti M,
Gruttadauria S, Toro A, Malaguarnera G, Bertino N, Malaguarnera M,
Malaguarnera M and Di Carlo I: Hepatocellular carcinoma: Novel
molecular targets in carcinogenesis for future therapies. Biomed
Res Int. 2014:2036932014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Faivre S, Bouattour M and Raymond E: Novel
molecular therapies in hepatocellular carcinoma. Liver Int. 31
Suppl 1:S151–S160. 2011. View Article : Google Scholar
|
|
7
|
Liang XJ, Finkel T, Shen DW, Yin JJ,
Aszalos A and Gottesman MM: SIRT1 contributes in part to cisplatin
resistance in cancer cells by altering mitochondrial metabolism.
Mol Cancer Res. 6:1499–1506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Michan S and Sinclair D: Sirtuins in
mammals: Insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dali-Youcef N, Lagouge M, Froelich S,
Koehl C, Schoonjans K and Auwerx J: Sirtuins: The ‘magnificent
seven’, function, metabolism and longevity. Ann Med. 39:335–345.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Varotti G, Ramacciato G, Ercolani G, Grazi
GL, Vetrone G, Cescon M, Del Gaudio M, Ravaioli M, Ziparo V, Lauro
A and Pinna A: Comparison between the fifth and sixth editions of
the AJCC/UICC TNM staging systems for hepatocellular carcinoma:
Multicentric study on 393 cirrhotic resected patients. Eur J Surg
Oncol. 31:760–767. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Carafa V, Rotili D, Forgione M, Cuomo F,
Serretiello E, Hailu GS, Jarho E, Lahtela-Kakkonen M, Mai A and
Altucci L: Sirtuin functions and modulation: From chemistry to the
clinic. Clin Epigenetics. 8:612016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kida Y and Goligorsky MS: Sirtuins, cell
senescence and vascular aging. Can J Cardiol. 32:634–641. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu W, Zuo Y, Feng Y and Zhang M: SIRT5
facilitates cancer cell growth and drug resistance in non-small
cell lung cancer. Tumour Biol. 35:10699–10705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iaquinta PJ and Lees JA: Life and death
decisions by the E2F transcription factors. Curr Opin Cell Biol.
19:649–657. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Polager S and Ginsberg D: E2F – at the
crossroads of life and death. Trends Cell Biol. 18:528–535. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mori K, Uchida T, Fukumura M, Tamiya S,
Higurashi M, Sakai H, Ishikawa F and Shibanuma M: Linkage of E2F1
transcriptional network and cell proliferation with respiratory
chain activity in breast cancer cells. Cancer Sci. 107:963–971.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu M, Seto E and Zhang J: E2F1 enhances
glycolysis through suppressing Sirt6 transcription in cancer cells.
Oncotarget. 6:11252–11263. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsantoulis PK and Gorgoulis VG:
Involvement of E2F transcription factor family in cancer. Eur J
Cancer. 41:2403–2414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Palaiologou M, Koskinas J, Karanikolas M,
Fatourou E and Tiniakos DG: E2F-1 is overexpressed and
pro-apoptotic in human hepatocellular carcinoma. Virchows Arch.
460:439–446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gorgoulis VG, Zacharatos P, Mariatos G,
Kotsinas A, Bouda M, Kletsas D, Asimacopoulos PJ, Agnantis N,
Kittas C and Papavassiliou AG: Transcription factor E2F-1 acts as a
growth-promoting factor and is associated with adverse prognosis in
non-small cell lung carcinomas. J Pathol. 198:142–156. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gu Y, Cheng Y, Song Y, Zhang Z, Deng M,
Wang C, Zheng G and He Z: MicroRNA-493 suppresses tumor growth,
invasion and metastasis of lung cancer by regulating E2F1. PLoS
One. 9:e1026022014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hao C, Zhu PX, Yang X, Han ZP, Jiang JH,
Zong C, Zhang XG, Liu WT, Zhao QD, Fan TT, et al: Overexpression of
SIRT1 promotes metastasis through epithelial-mesenchymal transition
in hepatocellular carcinoma. BMC Cancer. 14:9782014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mima K, Hayashi H, Kuroki H, Nakagawa S,
Okabe H, Chikamoto A, Watanabe M, Beppu T and Baba H:
Epithelial-mesenchymal transition expression profiles as a
prognostic factor for disease-free survival in hepatocellular
carcinoma: Clinical significance of transforming growth factor-β
signaling. Oncol Lett. 5:149–154. 2013.PubMed/NCBI
|
|
25
|
Li YM, Xu SC, Li J, Han KQ, Pi HF, Zheng
L, Zuo GH, Huang XB, Li HY, Zhao HZ, et al: Epithelial-mesenchymal
transition markers expressed in circulating tumor cells in
hepatocellular carcinoma patients with different stages of disease.
Cell Death Dis. 4:e8312013. View Article : Google Scholar : PubMed/NCBI
|