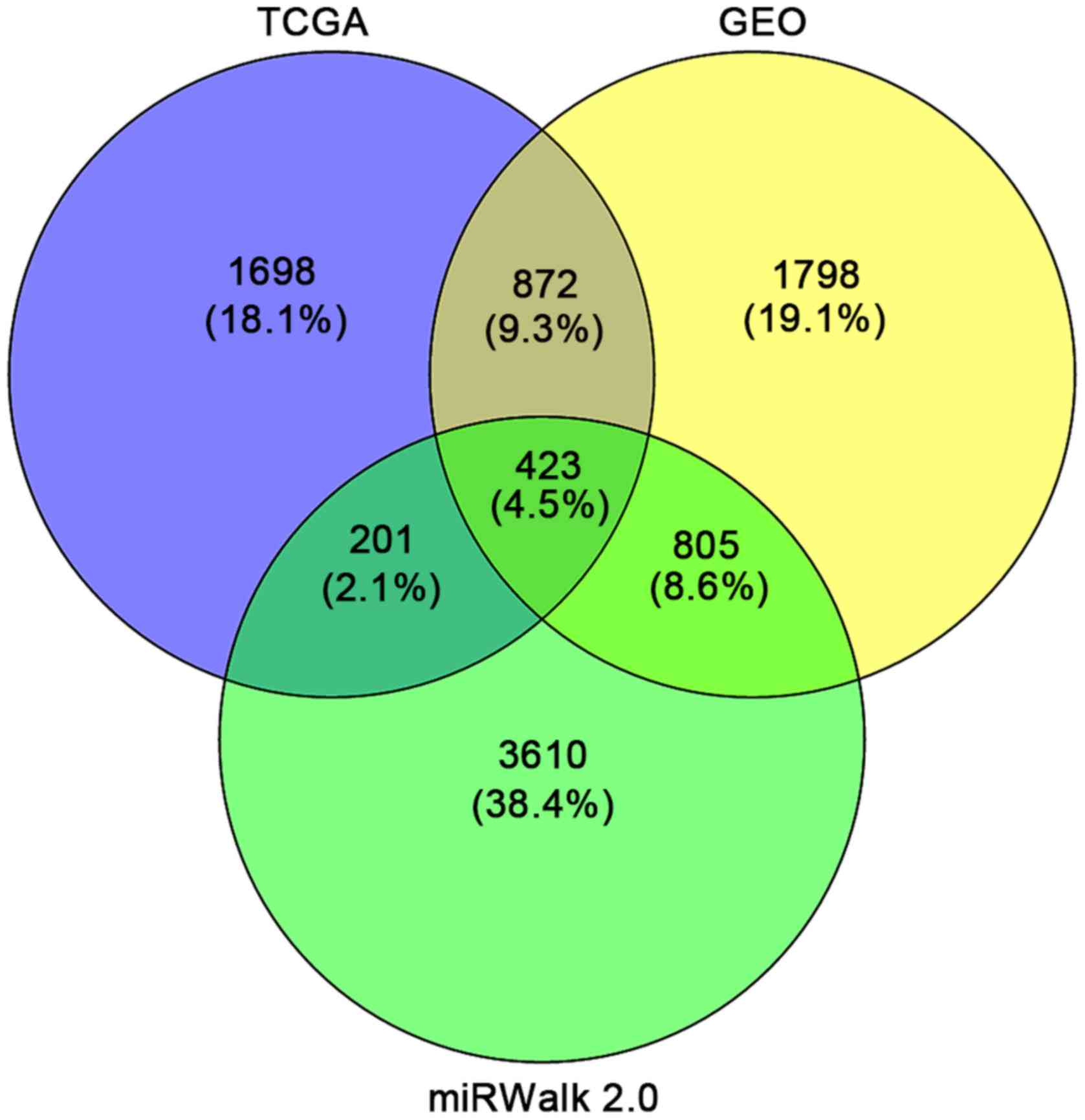
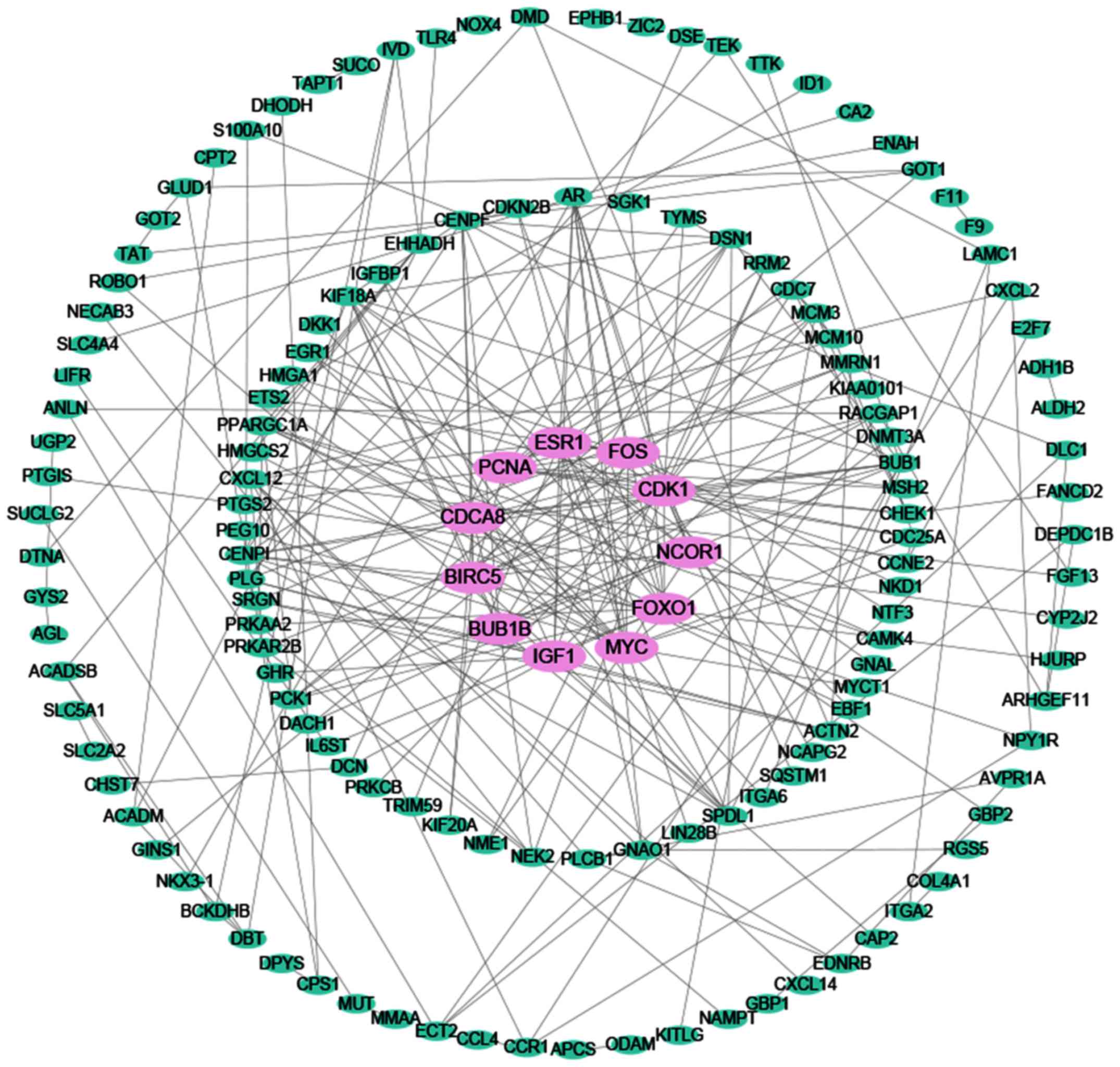
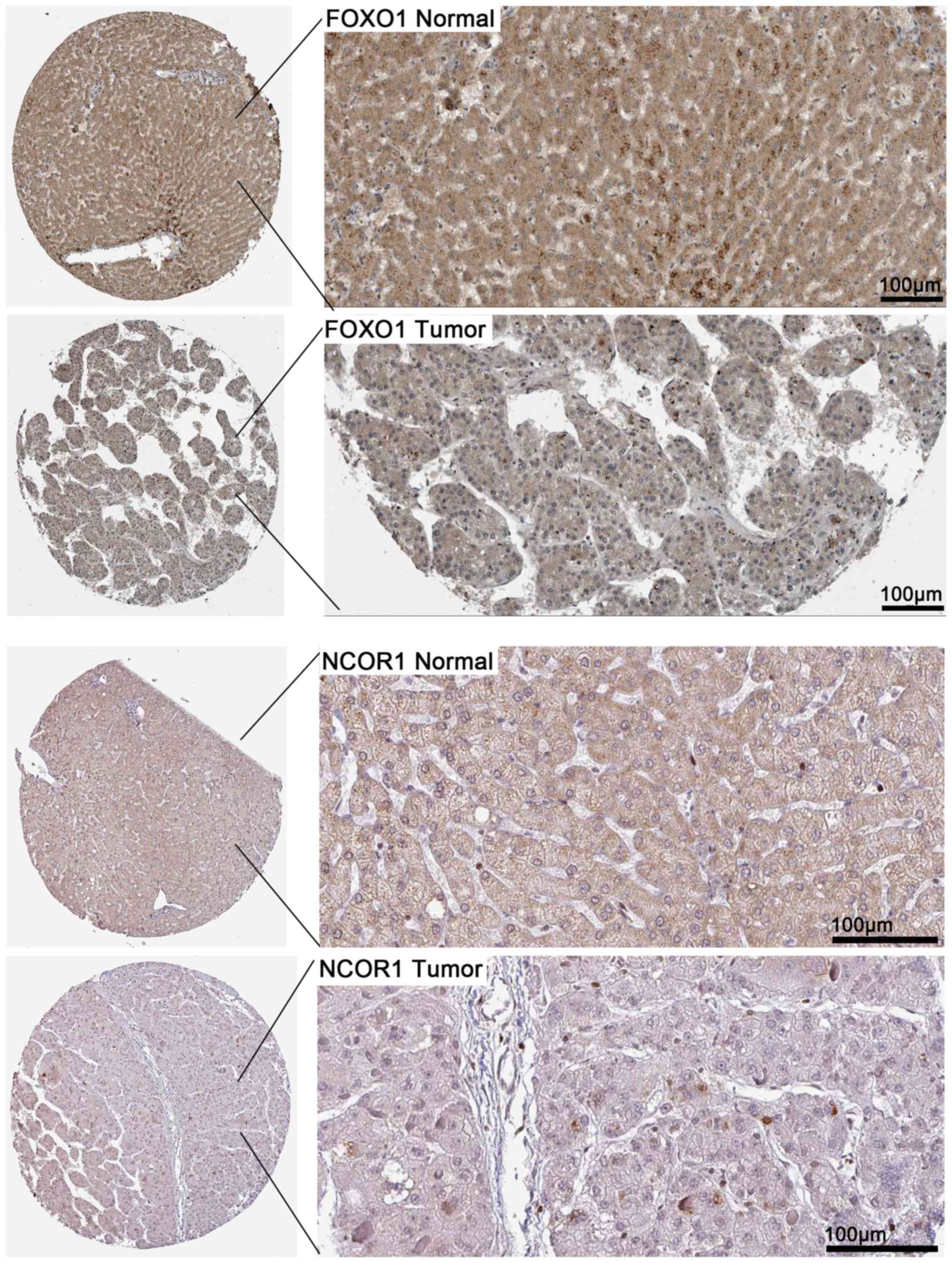
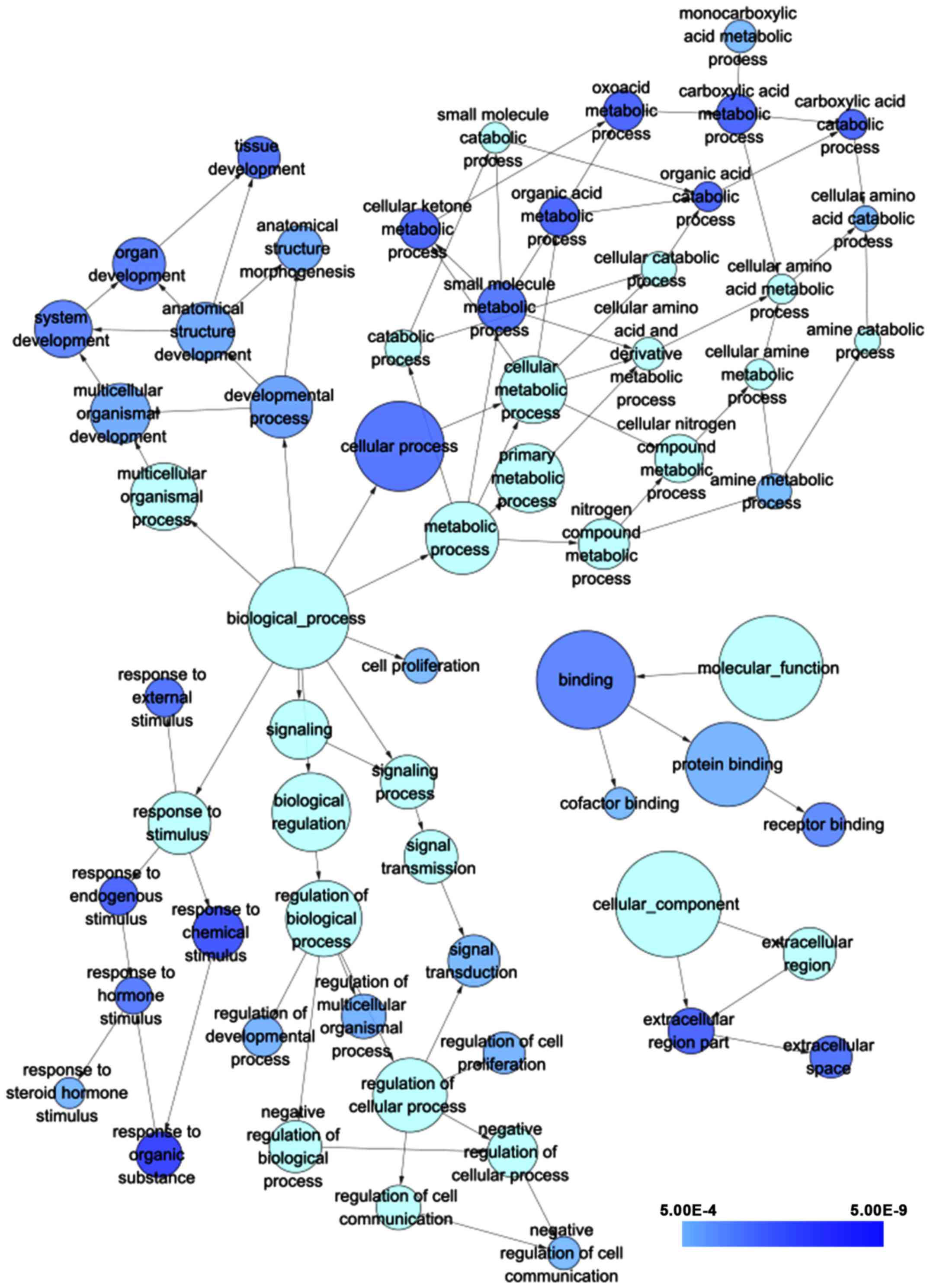
|
1
|
Nault JC, De Reyniès A, Villanueva A,
Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud
S, Rousseau F, et al: A hepatocellular carcinoma 5-gene score
associated with survival of patients after liver resection.
Gastroenterology. 145:176–187. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu AX: Molecularly targeted therapy for
advanced hepatocellular carcinoma in 2012: Current status and
future perspectives. Semin Oncol. 39:493–502. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Llovet JM, Villanueva A, Lachenmayer A and
Finn RS: Advances in targeted therapies for hepatocellular
carcinoma in the genomic era. Nat Rev Clin Oncol. 12:408–424. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lages E, Ipas H, Guttin A, Nesr H, Berger
F and Issartel JP: MicroRNAs: Molecular features and role in
cancer. Front Biosci (Landmark Ed). 17:2508–2540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Neilson JR and Sharp PA: Small RNA
regulators of gene expression. Cell. 134:899–902. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner
MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ and Schmittgen
TD: Expression profiling identifies microRNA signature in
pancreatic cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA, Liu CG, Sevignani C, Ferracin M,
Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, et al:
MicroRNA profiling reveals distinct signatures in B cell chronic
lymphocytic leukemias. Proc Natl Acad Sci USA. 101:pp. 11755–11760.
2004; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong D, Zhao L, He K, Sun H, Cai D, Ni L,
Sun R, Chang S, Song T, Huang C, et al: MECP2 promotes the growth
of gastric cancer cells by suppressing miR-338-mediated
antiproliferative effect. Oncotarget. 7:34845–34859. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen JS, Liang LL, Xu HX, Chen F, Shen SL,
Chen W, Chen LZ, Su Q, Zhang LJ, Bi J, et al: miR-338-3p inhibits
epithelial-mesenchymal transition and metastasis in hepatocellular
carcinoma cells. Oncotarget. 8:71418–71429. 2016.PubMed/NCBI
|
|
15
|
Chen X, Wei L and Zhao S: miR-338 inhibits
the metastasis of lung cancer by targeting integrin β3. Oncol Rep.
36:1467–1474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weng HL and Wang MJ: Effects of
microRNA-338-3p on morphine-induced apoptosis and its underlying
mechanisms. Mol Med Rep. 14:2085–2092. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhuang Y, Dai J and Wang Y, Zhang H, Li X,
Wang C, Cao M, Liu Y, Cai H, Zhang D and Wang Y: miR-338*
suppresses fibrotic pathogenesis in pulmonary fibrosis through
targeting LPA1. Am J Transl Res. 8:3197–3205. 2016.PubMed/NCBI
|
|
18
|
Xing Z, Yu L, Li X and Su X: Anticancer
bioactive peptide-3 inhibits human gastric cancer growth by
targeting miR-338-5p. Cell Biosci. 6:532016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yong FL, Law CW and Wang CW: Potentiality
of a triple microRNA classifier: miR-193a-3p, miR-23a and
miR-338-5p for early detection of colorectal cancer. BMC Cancer.
13:2802013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Besse A, Sana J, Lakomy R, Kren L, Fadrus
P, Smrcka M, Hermanova M, Jancalek R, Reguli S, Lipina R, et al:
miR-338-5p sensitizes glioblastoma cells to radiation through
regulation of genes involved in DNA damage response. Tumour Biol.
37:7719–7727. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Y, Chen J, Liu Y, Li S and Huang P:
Plasma miR-15b-5p, miR-338-5p, and miR-764 as biomarkers for
hepatocellular carcinoma. Med Sci Monit. 21:1864–1871. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
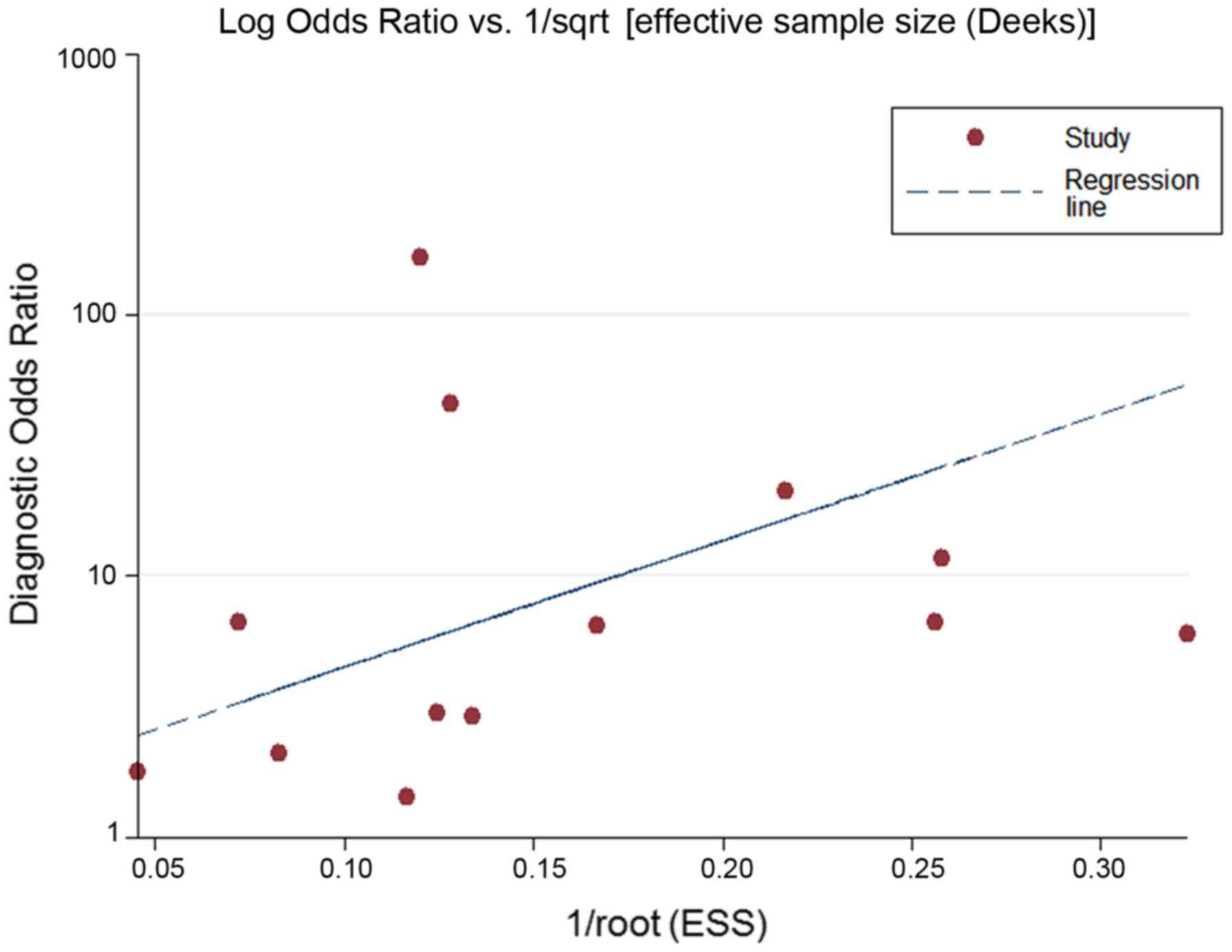
Deeks JJ, Macaskill P and Irwig L: The
performance of tests of publication bias and other sample size
effects in systematic reviews of diagnostic test accuracy was
assessed. J Clin Epidemiol. 58:882–893. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Glas AS, Lijmer JG, Prins MH, Bonsel GJ
and Bossuyt PM: The diagnostic odds ratio: A single indicator of
test performance. J Clin Epidemiol. 56:1129–1135. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Harbord RM, Deeks JJ, Egger M, Whiting P
and Sterne JA: A unification of models for meta-analysis of
diagnostic accuracy studies. Biostatistics. 8:239–251. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jackson D, White IR and Thompson SG:
Extending DerSimonian and Laird's methodology to perform
multivariate random effects meta-analyses. Stat Med. 29:1282–1297.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Uhlen M, Oksvold P, Fagerberg L, Lundberg
E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S,
et al: Towards a knowledge-based Human protein atlas. Nat
Biotechnol. 28:1248–1250. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Y, Huang P, Chen J, Liu Y, Li SL,
Wang Z and Song D: Plasma circulating miR-338-5p, miR-21-5p and
miR-15b-5p are potential biomarkers for screening hepatocellular
carcinoma. J Third Mil Med Univ. 37:1720–1726. 2015.(In
Chinese).
|
|
29
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Burchard J, Zhang C, Liu AM, Poon RT, Lee
NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al:
microRNA-122 as a regulator of mitochondrial metabolic gene network
in hepatocellular carcinoma. Mol Syst Biol. 6:4022010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Diaz G, Melis M, Tice A, Kleiner DE,
Mishra L, Zamboni F and Farci P: Identification of microRNAs
specifically expressed in hepatitis C virus-associated
hepatocellular carcinoma. Int J Cancer. 133:816–824. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen J, Wang A, Wang Q, Gurvich I, Siegel
AB, Remotti H and Santella RM: Exploration of genome-wide
circulating microRNA in hepatocellular carcinoma: miR-483-5p as a
potential biomarker. Cancer Epidemiol Biomarkers Prev.
22:2364–2373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
Intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martinez-Quetglas I, Pinyol R, Dauch D,
Torrecilla S, Tovar V, Moeini A, Alsinet C, Portela A,
Rodriguez-Carunchio L, Solé M, et al: IGF2 is up-regulated by
epigenetic mechanisms in hepatocellular carcinomas and is an
actionable oncogene product in experimental models.
Gastroenterology. 151:1192–1205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC,
Huang CH, Lee YS, Yen TC and Hsieh SY: MicroRNA-214 downregulation
contributes to tumor angiogenesis by inducing secretion of the
hepatoma-derived growth factor in human hepatoma. J Hepatol.
57:584–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: miR-145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sato F, Hatano E, Kitamura K, Myomoto A,
Fujiwara T, Takizawa S, Tsuchiya S, Tsujimoto G, Uemoto S and
Shimizu K: MicroRNA profile predicts recurrence after resection in
patients with hepatocellular carcinoma within the Milan Criteria.
PLoS One. 6:e164352011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma
X, Wei Q and Han L: Serum microRNA panel for early diagnosis of the
onset of hepatocellular carcinoma. Medicine (Baltimore).
96:e56422017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Okajima W, Komatsu S, Ichikawa D, Miyamae
M, Kawaguchi T, Hirajima S, Ohashi T, Imamura T, Kiuchi J, Arita T,
et al: Circulating microRNA profiles in plasma: Identification of
miR-224 as a novel diagnostic biomarker in hepatocellular carcinoma
independent of hepatic function. Oncotarget. 7:53820–53836. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yan SR, Liu ZJ, Yu S and Bao YX:
Investigation of the value of miR-21 in the diagnosis of early
stage HCC and its prognosis: A meta-analysis. Genet Mol Res.
14:11573–11586. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lei D, Zhang F, Yao D, Xiong N, Jiang X
and Zhao H: miR-338-5p suppresses proliferation, migration,
invasion, and promote apoptosis of glioblastoma cells by directly
targeting EFEMP1. Biomed Pharmacother. 89:957–965. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang W, Cho H, Shin HY, Chung JY, Kang ES,
Lee EJ and Kim JH: Accumulation of cytoplasmic Cdk1 is associated
with cancer growth and survival rate in epithelial ovarian cancer.
Oncotarget. 7:49481–49497. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tsaur I, Makarević J, Hudak L, Juengel E,
Kurosch M, Wiesner C, Bartsch G, Harder S, Haferkamp A and Blaheta
RA: The cdk1-cyclin B complex is involved in everolimus triggered
resistance in the PC3 prostate cancer cell line. Cancer Lett.
313:84–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sung WW, Lin YM, Wu PR, Yen HH, Lai HW, Su
TC, Huang RH, Wen CK, Chen CY, Chen CJ and Yeh KT: High
nuclear/cytoplasmic ratio of Cdk1 expression predicts poor
prognosis in colorectal cancer patients. BMC Cancer. 14:9512014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Willder JM, Heng SJ, McCall P, Adams CE,
Tannahill C, Fyffe G, Seywright M, Horgan PG, Leung HY, Underwood
MA and Edwards J: Androgen receptor phosphorylation at serine 515
by Cdk1 predicts biochemical relapse in prostate cancer patients.
Br J Cancer. 108:139–148. 2013.PubMed/NCBI
|
|
47
|
Banerjee SK, Weston AP, Zoubine MN,
Campbell DR and Cherian R: Expression of cdc2 and cyclin B1 in
Helicobacter pylori-associated gastric MALT and MALT lymphoma:
Relationship to cell death, proliferation, and transformation. Am J
Pathol. 156:217–225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hongo F, Takaha N, Oishi M, Ueda T,
Nakamura T, Naitoh Y, Naya Y, Kamoi K, Okihara K, Matsushima T, et
al: CDK1 and CDK2 activity is a strong predictor of renal cell
carcinoma recurrence. Urol Oncol. 32:1240–1246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tian J, Hu X, Gao W, Zhang J, Chen M,
Zhang X, Ma J and Yuan H: Identification a novel tumor-suppressive
hsa-miR-599 regulates cells proliferation, migration and invasion
by targeting oncogenic MYC in hepatocellular carcinoma. Am J Transl
Res. 8:2575–2584. 2016.PubMed/NCBI
|
|
50
|
Cao L, Li C, Shen S, Yan Y, Ji W, Wang J,
Qian H, Jiang X, Li Z, Wu M, et al: OCT4 increases BIRC5 and CCND1
expression and promotes cancer progression in hepatocellular
carcinoma. BMC Cancer. 13:822013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Le Coz V, Zhu C, Devocelle A, Vazquez A,
Boucheix C, Azzi S, Gallerne C, Eid P, Lecourt S and Giron-Michel
J: IGF-1 contributes to the expansion of melanoma-initiating cells
through an epithelial-mesenchymal transition process. Oncotarget.
7:82511–82527. 2016.PubMed/NCBI
|
|
52
|
Yang XW, Shen GZ, Cao LQ, Jiang XF, Peng
HP, Shen G, Chen D and Xue P: MicroRNA-1269 promotes proliferation
in human hepatocellular carcinoma via downregulation of FOXO1. BMC
Cancer. 14:9092014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ko YS, Cho SJ, Park J, Kim Y, Choi YJ, Pyo
JS, Jang BG, Park JW, Kim WH and Lee BL: Loss of FOXO1 promotes
gastric tumour growth and metastasis through upregulation of human
epidermal growth factor receptor 2/neu expression. Br J Cancer.
113:1186–1196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ren JW, Li ZJ and Tu C: miR-135
post-transcriptionally regulates FOXO1 expression and promotes cell
proliferation in human malignant melanoma cells. Int J Clin Exp
Pathol. 8:6356–6366. 2015.PubMed/NCBI
|
|
55
|
Martinez-Iglesias OA, Alonso-Merino E,
Gómez-Rey S, Velasco-Martín JP, Martín Orozco R, Luengo E, García
Martín R, Ibáñez de Cáceres I, Fernández AF, Fraga MF, et al:
Autoregulatory loop of nuclear corepressor 1 expression controls
invasion, tumor growth, and metastasis. Proc Natl Acad Sci USA.
113:pp. E328–E337. 2016; View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yu JJ, Wu YX, Zhao FJ and Xia SJ: miR-96
promotes cell proliferation and clonogenicity by down-regulating of
FOXO1 in prostate cancer cells. Med Oncol. 31:9102014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang B, Gui LS, Zhao XL, Zhu LL and Li
QW: FOXO1 is a tumor suppressor in cervical cancer. Genet Mol Res.
14:6605–6616. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li K, Chen MK, Situ J, Huang WT, Su ZL, He
D and Gao X: Role of co-expression of c-Myc, EZH2 and p27 in
prognosis of prostate cancer patients after surgery. Chin Med J
(Engl). 126:82–87. 2013.PubMed/NCBI
|
|
59
|
Liu Z, Jiang Y, Hou Y, Hu Y, Cao X, Tao Y,
Xu C, Liu S, Wang S, Wang L, et al: The IκB family member Bcl-3
stabilizes c-Myc in colorectal cancer. J Mol Cell Biol. 5:280–282.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Duffy MJ, O'Donovan N, Brennan DJ,
Gallagher WM and Ryan BM: Survivin: A promising tumor biomarker.
Cancer Lett. 249:49–60. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Grimberg A: Mechanisms by which IGF-I may
promote cancer. Cancer Biol Ther. 2:630–635. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Xie L, Ushmorov A, Leithäuser F, Guan H,
Steidl C, Färbinger J, Pelzer C, Vogel MJ, Maier HJ, Gascoyne RD,
et al: FOXO1 is a tumor suppressor in classical Hodgkin lymphoma.
Blood. 119:3503–3511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chun YS, Huang M, Rink L and Von Mehren M:
Expression levels of insulin-like growth factors and receptors in
hepatocellular carcinoma: A retrospective study. World J Surg
Oncol. 12:2312014. View Article : Google Scholar : PubMed/NCBI
|