Introduction
Glioblastoma (GBM) has become the most aggressive
and common type of central nervous system malignancy worldwide
(1). However, the causes of the
carcinogenesis and progression of GBM remain to be elucidated.
Several studies have indicated that the aberrant expression of the
Forkhead Box (FOX) family protein has a key function in tumor
growth, metastasis and response to cancer therapy (2–6).
The FOXO family includes FOXO1, FOXO3, FOXO4 and
FOXO6. They all have a crucial function in gene transcription,
which mediates cell processes, including DNA damage repair,
apoptotic cell death, glucose metabolism, cell cycle control and
carcinogenesis (7). FOXO members
consist of a conserved domain, which binds the DNA sequence
TTGTTTAC at target gene (8,9).
Several reports have demonstrated that FOXOs function in autophagy
(10–12). In oxidative stress, FOXO1 can
promote autophagy, which may due to its tumor suppressor activity
(13).
Sirtuin 1 (SIRT1) is the mammalian orthologue of
Sir2. It has been reported that SIRT1 has an important function in
cell senescence (14,15). Nicotinamide adenine dinucleotide
and nicotinamide can regulate the activity of SIRT1. The activation
of SIRT1 protects cardiomyocytes from death and promotes the
survival of neurons. In different tissues, SIRT1 has specific
functions. For example, SIRT1 promotes gluconeogenesis and fatty
acid oxidation under nutrient deprivation in the liver (16). SIRT1 is also involved in pancreatic
β-cell survival and insulin secretion through interacting with the
FOXO family (17) and inhibiting
of uncoupling protein 2 (18),
respectively.
Senescence is a state in which cells undergo
specific alterations in gene expression and cellular morphology,
accompanied by loss of the ability to proliferate. During cell
senescence, β-galactosidase, which is associated with senescence,
is activated, followed by cell cycle arrest at the
G0/G1 phase and a marked increase in the
expression of cyclin-dependent kinase inhibitor (19,20).
In the present study, it was revealed that FOXO1 was
significantly downregulated in the GBM tissues and GBM cell lines.
FOXO1 inhibited cell proliferation via arresting the cell cycle at
the G0/G1 phase. In addition, FOXO1
facilitated cell senescence through regulation of sirtuin 1
expression. In addition, FOXO1 suppressed epithelial mesenchymal
transition and metastasis. These findings suggested a novel
mechanism of FOXO1 in the suppression of tumorigenesis and
metastasis of GBM cells and suggested that FOXO1 may be a potential
therapeutic target for treating GBM.
Materials and methods
Cell culture
Human U87, T98G and LN18 GBM cell lines were
purchased from the America Type Culture Collection (Manassas, VA,
USA) and cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% FBS (Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany) in 5% CO2 at 37°C.
Plasmid constructs and
transfection
The human U87, T98G and LN18 GBM cells were grown in
a 6-well plate to almost 70% confluence, follow by transfection
with 3 µg of plasmids. The empty vector pcDNA3.1 plasmid and the
FOXO1-containing plasmid 1 (pcDNA3.1-FOXO) were used (VigeneBio,
Shandong, China). Transfection was performed using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol, for
U87, T98G and LN18 transfection.
Cell cycle analysis
The cells were detatched using 0.25% trypsin and
collected when the cells had reached almost 70–80% confluence,
following which they were fixed with 75% ethanol overnight.
~3×104 cells were treated with 1 mg/ml RNase A
(Sigma-Aldrich; Merck Millipore) at 37°C for 40 min, resuspended in
PBS and stained with propidium iodide for 30 min in the dark.
Finally, the DNA contents were measured using a FACScan flow
cytometry system. All experiments were performed three times.
Tissue samples
The 143 pairs' human GBM tissues and adjacent normal
tissues were obtained from patients at The First Affiliated
Hospital of China Medical University (Shenyang, China) between 2011
and 2015. All patients were diagnosed by pathological examination
as having GBM. A total of 85 were male and 58 were female patients.
The present study was approved by the Institutional Research Ethics
Committee of The First Affiliated Hospital of China Medical
University. All patients were informed and provided written
informed consent. All tissue samples were collected and frozen in
liquid nitrogen and stored at −80°C until processing. The
expression of FOXO1 in tissues was determined by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR). The
mean value of FOXO1 content in tumor cells acted as the standard.
Therefore, higher values than the standard value was denoted as
high expression and lower values than the standard value was
denoted as low expression.
MTT assay
An MTT assay was used to detect cell proliferation,
as described previously (21). In
brief, the cells were seeded at 3×103 per well in a
96-well plate and incubated with 20 µl MTT (10 mg/ml in PBS;
Sigma-Aldrich; Merck Millipore) at 37°C for 3 h, follow by
dissolving in DMSO. Finally, the density of cells was detected at
570 nm. Each assay was performed three times and all data presented
as the mean ± standard deviation of the three experiments.
Western blot analysis
The cells were lysed in RIPA buffer containing 1%
NP-40, 50 mM Tris-HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, 1 mM NaF,
0.25% Na-deoxycholate and 0.2 mM sodium orthovanadate, supplemented
with protease inhibitor cocktail. The protein concentration was
determined using BCA Protein Assay Reagent (Pierce; Thermo Fisher
Scientific, Inc.). Subsequently, 50 µg of total protein was
subjected to 10% SDS-PAGE and transferred onto PVDF membranes (EMD
Millipore, Billerica, MD, USA). The membranes were then blocked in
5% skimmed milk at room temperature for 1 h. Subsequently, the
membranes were incubated with primary antibodies at 4°C overnight,
washed with TBST at least three times and then incubated with
HRP-conjugated secondary antibodies (1:6,000; cat. nos. ab6721 and
ab6789; Abcam, Cambridge, UK) for 2 h at room temperature. This was
followed by washing with TBST three times. The primary antibodies
used were as follows: FOXO1 (1:3,000; cat. no. ab39670; Abcam);
β-actin (1:5,000; cat. no. ab8227; Abcam); FLAG (1:5,000; cat. no.
F7425; Sigma-Aldrich); EMT antibody sampler kit (1:2,000; cat. no.
9782; Cell Signaling Technology, Inc., Danvers, MA, USA); p16
(1:2,000; cat. no. ab118459; Abcam); p33 (1:3,000; cat. no.
ab124893; Abcam); Lsh (1:2,000; cat. no. 7798; Cell Signaling
Technology, Inc.); SIRT1 (1:5,000; cat. no. HPA006295;
Sigma-Aldrich). β-actin served as a loading control. Finally, the
proteins were visualized using ECL detection reagent (Beyotime
Institute of Biotechnology, Dalian, China) according to the
manufacturer's protocol. The blots were quantified using Image J
software version 1.0 (National Institutes of Health, Bethesda, MD,
USA).
RT-qPCR and data analyses
Total cellular RNAs were isolated using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), following
which a reverse transcription system (Promega A3500; Thermo Fisher
Scientific, Inc.) was used for the synthesis of first strand cDNA.
Quantitation of the indicated gene transcripts was performed using
qPCR. The mRNA level of GAPDH was used as the internal control. A
total of 1 µl cDNA sample, 1.5 µl primer (10 nM), 7.5 µl 2X MIX
buffer and 5 µl RNase-free water were added. The qPCR conditions
were as follows: 5 min at 95°C, denaturation at 95°C for 30 sec,
annealing at 57°C for 30 sec and extension at 72°C for 40 sec,
performed for 30 cycles. The primer pairs used were as follows:
E-cadherin, forward 5′-AATCTCAAGCTCATGGATAACC-3′ and reverse
5′-GCAGAATCAGAATTAGCAAAGC-3′; α-catenin, forward
5′-GCTGAAAGTTGTGGAAGATGG-3′ and reverse 5′-TTATAGGCTGCGACATCAGG-3′;
N-cadherin, forward 5′-TCAAAGCCTGGAACATATGTG-39 and reverse
5′-TGTTTGAAAGGCCATATGTGG-3′; fibronectin, forward
5′-GAGTAAACCTGAAGCTGAAGAG-3′ and reverse
5′-TCACCAATCTTGTAGGACTG-3′; GAPDH, forward
5′-ATGAGAAGTATGACAACAGCCT-3′ and reverse
5′-ATGGACTGTGGTCATGAGTC-3′; and SIRT1, forward
5′-GCACTAATTCCAAGTTCCATACC-3′ and reverse
5′-GCAAAGTTTGGCATATTCACC-3′. Relative mRNA expression was
calculated by the 2−ΔΔCq method (22).
Luciferase activity assay
The cells were seeded at 5×104 in 6-well
plates, followed by transfection with plasmids when the cells
reached 70–80% confluence. The activities of luciferase following
~48 h transfection were quantified using an illuminometer (Centro
LB 960; Berthold Technologies GmbH, Bad Wildbad, Germany). Each
experiment was performed at least three times.
ChIP assay
A ChIP assay was performed using an EZ-ChIP kit (EMD
Millipore) according to the manufacturer's protocol with minor
modifications. In brief, the LN18 cells (~3×104) were
used at a density of 80–90% confluence, and ChIP assays were
performed using 5 µl anti-FOXO1 antibody (cat. no. ab39670; Abcam)
at 4 at 44ntibod. The DNA was isolated from the immunoprecipitates
and quantified using RT-qPCR analysis with the specific primer
pairs: p16, forward 5′-CTCTTATACCAGGCAATGTA-3′; and reverse
5′-GTACGACTAGAAAGTGTCCC-3′; SIRT1, forward
5′-CAGATGGATTTCAGAGGGAT-3′ and reverse 5′-GAAGGCTGAGGCAGGAGAAT-3′.
A total of 2 µl sample, 1 µl primer (10 nM), 10 µl 2X MIX buffer
and 7 µl RNase-free water were added. The qPCR conditions were as
follows: 5 min at 95°C, denaturation at 95°C for 30 sec, annealing
at 57°C for 30 sec and extension at 72°C for 40 sec, performed for
30 cycles. Input DNA and DNA immunoprecipitated by anti-IgG served
as a positive and negative control, respectively.
Statistical analysis
All results were analyzed using SPSS v. 18.0 (SPSS,
Inc., Chicago, IL, USA) and reported as the mean ± standard
deviation. A paired samples t-test was used to compare between
adjacent normal tissue and cancer tissues. P<0.05 was considered
to indicate a statistically significant difference.
Results
FOXO1 is downregulated in human GBM
cell lines and tissues
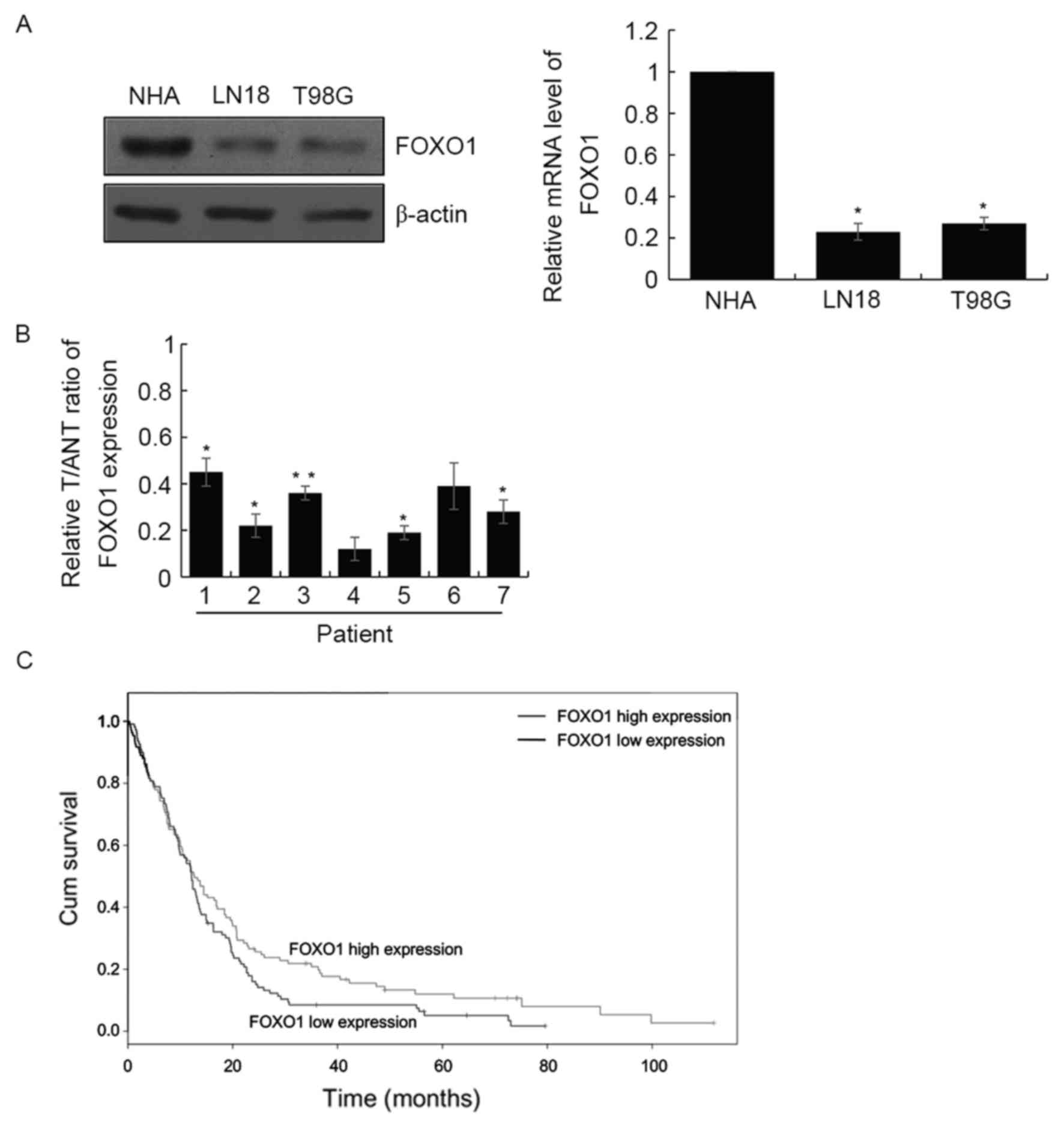
In order to examine the function of FOXO1 in GBM in
the present study, the expression levels of FOXO1 were determined
in the LN18 and T98G GBM cell lines, and normal human astrocytes.
The results demonstrated that the expression of FOXO1 was decreased
at the protein and mRNA levels in these GBM cell lines, compared
with the normal human astrocytes (Fig.
1A). In addition, RT-qPCR analysis was performed to examine the
mRNA levels of FOXO1 in seven paired human GBM tissues and adjacent
normal tissues. Similar results were obtained, which showed the
expression of FOXO1 was significantly downregulated in the GBM
tissues, compared with that in the adjacent normal tissues
(Fig. 1B). Together, these results
showed that FOXO1 was downregulated in GBM cell lines and primary
tumors. Subsequently, the present study investigated the expression
levels of FOXO1 in 143 patients with GBM. The RT-qPCR analysis
revealed that the expression of FOXO1 was significantly
downregulated in GBM. The expression of FOXO1 was negatively
correlated with pathological grade (P<0.001), tumor size
(P=0.003) and neck lymph node metastasis (P=0.01). However,
differentiation, age and gender were not correlated with FOXO1
(Table I). In addition, the
present study investigated prognosis in the downregulation of FOXO1
in GBM. As expected, the survival curve indicated that the survival
rates of patients with a high expression of FOXO1 were
significantly higher, compared with those with low expression
levels of FOXO1 (hazard ratio=1.49; P=0.024; Fig. 1C).
 | Table I.Clinicopathologic variables in 143
patients with glioblastoma. |
Table I.
Clinicopathologic variables in 143
patients with glioblastoma.
|
|
| FOXO1 expression |
|
|---|
|
|
|
|
|
|---|
| Variable | No. (n=143) | Low (n=90) | High (n=53) | P-value |
|---|
| Gender |
| Male | 85 | 56 | 29 | 0.377 |
|
Female | 58 | 34 | 24 |
|
| Age (years) |
|
<50 | 72 | 45 | 27 | 0.913 |
| ≥50 | 71 | 45 | 26 |
|
| Tumor diameter |
| Large (≥2
cm) | 61 | 47 | 14 | 0.003 |
| Small
(<2 cm) | 82 | 43 | 39 |
|
| Pathological
grade |
| I–II | 70 | 33 | 37 | <0.001 |
|
III–IV | 73 | 57 | 16 |
|
| Neck lymph node
metastasis |
| No | 69 | 36 | 33 | 0.01 |
| Yes | 74 | 54 | 20 |
|
| Differentiation |
|
Well/moderate | 70 | 49 | 21 | 0.087 |
|
Poor | 73 | 41 | 32 |
|
FOXO1 inhibits GBM cell
proliferation
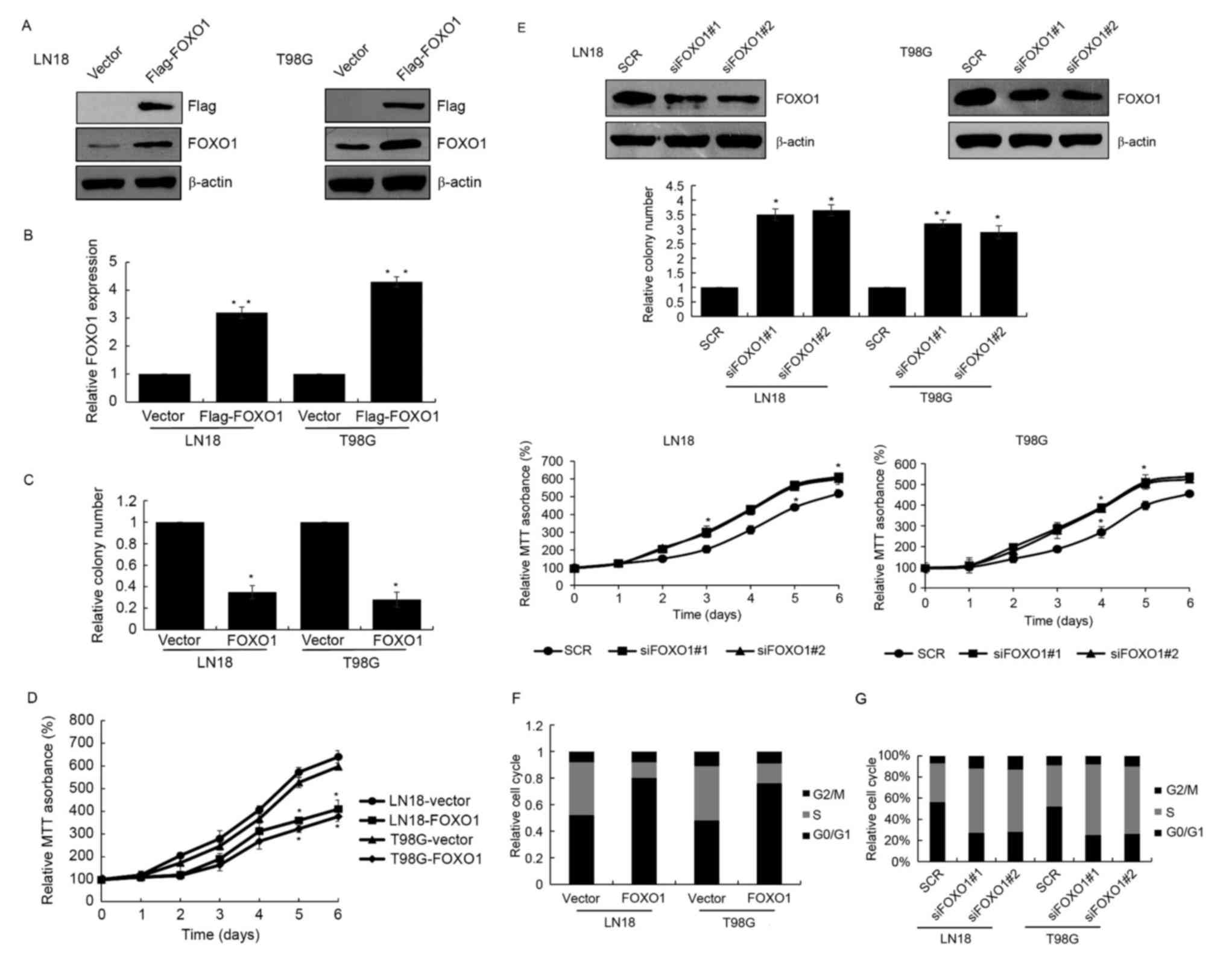
To further examine the role of FOXO1 in GBM,
FLAG-FOXO1 was overexpressed in GBM cell lines, LN18 and T98G
cells. RT-qPCR and western blot analyses were used to detect the
transfection efficiency (Fig. 2A and
B). Colony formation and MTT assays were also performed to
investigate the function of FOXO1 in cell growth in GBM. The colony
formation and MTT assays revealed that FOXO1 significantly
decreased colony forming ability and cell growth in the LN18 and
T98G cells (Fig. 2C and D). FOXO1
siRNA was also used to knock down FOXO1 in the LN18 and T98G cells,
followed by analysis using colony formation and MTT assays. The
depletion of FOXO1 increased the colony forming ability in cell
proliferation of the LN18 and T98G cells (Fig. 2E). To examine whether FOXO1
affected cell cycle progression, flow cytometry was used to
detected cell cycle. The results revealed that the overexpression
of FOXO1 arrested the cell cycle in the G0/G1
phase, suppressing cells from entering the S phase (Fig. 2F). By contrast, FOXO1 knockdown
resulted in a higher number of cells entering the S phase, follow
by promoted cell growth (Fig. 2G).
In brief, these findings demonstrated that FOXO1 arrested cells at
the G0/G1 phase and suppressed cell growth in
GBM.
FOXO1 promotes senescence via the
transcriptional inhibition of SIRT1
Following the induced overexpression of FOXO1,
p16INK4a was also expressed at a high level (Fig. 3A). p16INK4a has been
reported to be expressed at high levels in cell senescence, in
which it has a crucial function. Therefore, it was hypothesized
that FOXO1 may be involved in cell senescence of GBM. There are
several proteins associated with cell senescence, including histone
deacetylase (HDAC)4, SIRT1, lymphoid specific helicase (Lsh) and
p33ING1b. To confirm this hypothesis, the present study
investigated whether FOXO1 regulated these proteins. As shown in
Fig. 3A, in LN18 cells
overexpressing FOXO1, the expression of SIRT1 was suppressed,
whereas HDAC4, Lsh and p33ING1b were not affected. The
opposite results were obtained in the FOXO1-depleted cells, which
showed SIRT1 was increased and p16INK4a was decreased
(Fig. 3B). The mRNA levels of
SIRT1 and p16INK4a were also regulated by FOXO1, which
suggested that FOXO1 regulated SIRT1 and p16INK4a at the
transcriptional level (Fig. 3C).
The present study then examined whether FOXO1 affected the
transcription p16INK4a or SIRT1. A ChIP assay was
performed with FOXO1 antibody, which revealed that the SIRT1
promoter region interacted with FOXO1, whereas p16IN4a
did not interact with FOXO1. Although the mRNA and protein levels
of p16IN4a were affected by FOXO1, FOXO1 did not
interact with its promoter region, therefore, FOXO1 may be
indirectly regulated p16IN4a by other proteins (Fig. 3D). The luciferase reporter assay
also confirmed that the recruitment of wide-type FOXO1 (wt-FOXO1)
interacted with the SIRT1 promoter region and negatively regulated
its transcription. However, FOXO1 (K245A) failed to regulate the
transcription of SIRT1 (Fig.
3E).
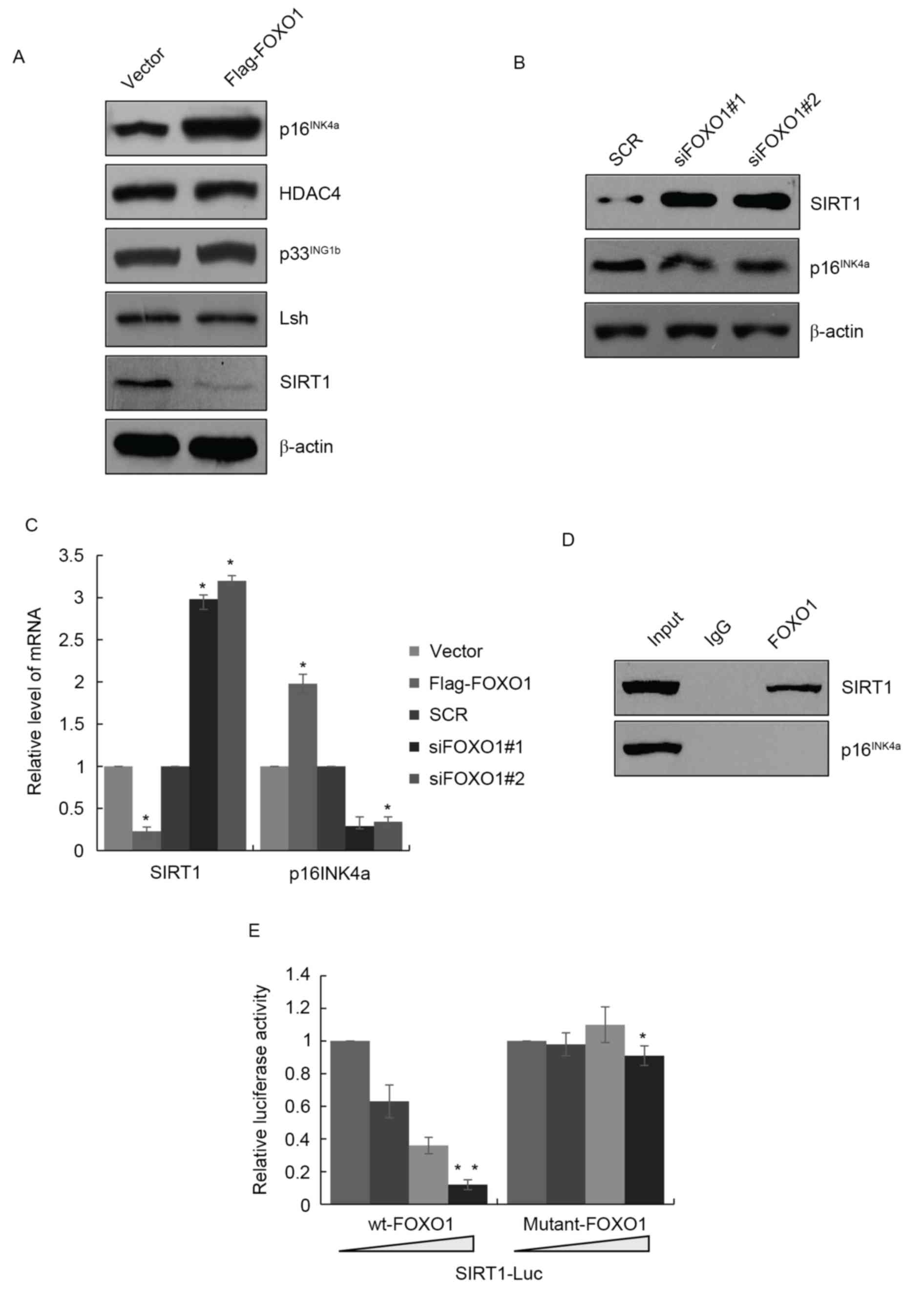 | Figure 3.FOXO1 promotes senescence via the
transcriptional inhibition of SIRT1. (A) Validation of marker of
senescence of p16INK4a, HDAC4, p33INK1b, Lsh
and SIRT1 in LN18 cells overexpressong FOXO1. (B) Knockdown of
FOXO1 by FOXO1 siRNA in LN18 cells, followed by the detection of
expression levels of SIRT1 and p16INK4a. (C) Reverse
transcription-quantitative polymerase chain reaction analysis
revealed mRNA levels of SIRT1 and p16INK4a were altered
in LN18 cells overexpressing FOXO1 or in those with FOXO1
knockdown. *P<0.05. (D) ChIP assay in LN18 cells using
anti-FOXO1 antibody. (E) Luciferase reporter assays in LN18 cells
co-transfected with SIRT1-Luc and wt FOXO1 or FOXO1 (K245A) as
indicated. Data are presented as the mean ± standard deviation of
the three experiments. *P<0.05; **P<0.01. FOXO1, Forkhead Box
O1; SIRT1, sirtuin 1; siRNA, small interfering RNA; HDAC4, histone
deacetylase 4; Lsh, lymphoid specific helicase; wt, wild-type. |
FOXO1 inhibits epithelial-mesenchymal
transition (EMT) and metastasis in GBM
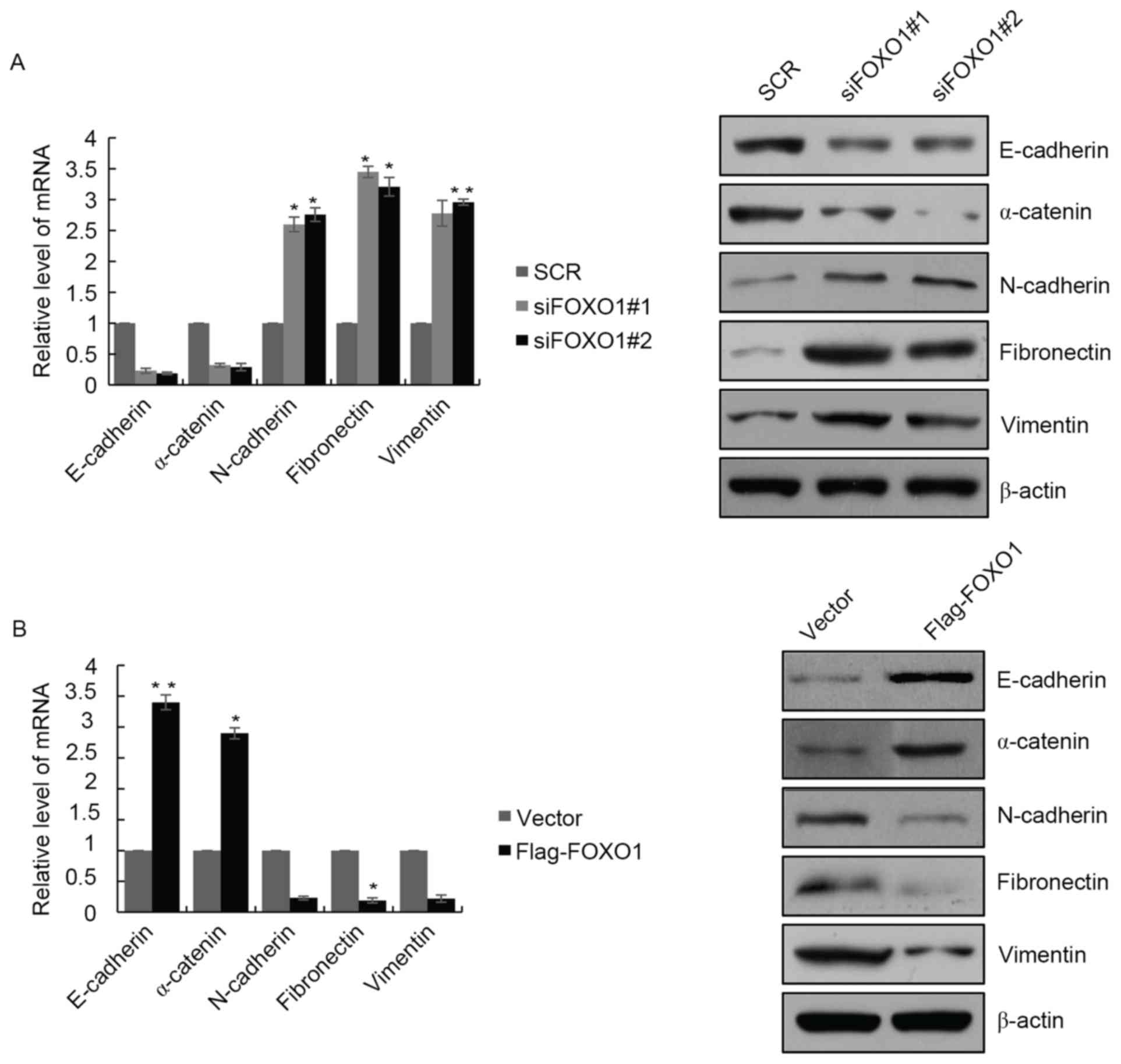
To determine the role of FOXO1 in the EMT of GBM
cells, the present study examined the expression of EMT markers in
LN18 cells with FOXO1 depletion. The results showed that, at the
mRNA and protein levels, the expression of epithelial markers,
E-cadherin and α-catenin were decreased, and the expression of
mesenchymal markers, N-cadherin, fibronectin and vimentin were
increased (Fig. 4A). The opposite
results were observed in cells overexpressing FOXO1, where the
expression of epithelial markers E-cadherin and α-catenin were
significantly increased, and expression of mesenchymal markers
N-cadherin, fibronectin and vimentin were decreased (Fig. 4B). These results indicated that
FOXO1 regulated EMT programming in the GBM cells. There are several
reports indicating that EMT promotes cell invasion. In the present
study, it was found that FOXO1 was negatively correlated with lymph
node metastasis, therefore, FOXO1 may suppress GBM cell invasion.
Transwell assays were performed to confirm this hypothesis; in the
highly invasive human U87 GBM cancer cell line, the results
indicated that, following knockdown of FOXO1, the number of invaded
cells through Matrigel was almost three times as high as in the
control, which revealed that the invasion ability of the U87 cells
was enhanced when FOXO1 was knocked down (Fig. 4C). By contrast, when FOXO1 was
overexpressed, the number of invaded cells was decreased, compared
with the number in the control group (Fig. 4C). Therefore, FOXO1 affected the
invasion ability of U87 cells. The present study also investigated
the role of FOXO1 in anchorage-independent cell growth. As shown in
Fig. 4D, the knockdown of FOXO1 in
U87 cells significantly promoted colony formation. Therefore, FOXO1
has a key function in several cancer development processes,
including EMT and cell invasion, and it also suppresses the
anchorage-independent grow ability of cancer cell.
Discussion
In the present study, it was found that FOXO1 was
significantly downregulated in GBM primary tumors and cell lines.
The expression of FOXO1 was negatively correlated with clinical
pathology, including neck lymph node metastasis (P=0.01) and tumor
size (P=0.003). The K-M plot analysis survival curve also indicated
that a high expression of FOXO1 in GBM was positively correlated
with increased survival rates. Colony formation and MTT assays
revealed that, following the ectopic expression of FOXO1, FOXO1
suppressed GBM cell growth. When FOXO1 siRNA was used to knock down
FOXO1 in the GBM cells, the opposite results were obtained. The
present study also found that the overexpression of FOXO1 arrested
GBM cells at the G0/G1 phase and inhibited
cell proliferation, whereas FOXO1 knockdown released cells at the
G0/G1 phase for entry into the S phase.
The present study also found that the ectopic
expression of FOXO1 resulted in p16INK4a being expressed
at a high level. The tumor suppressor p16INK1a has been
reported to be crucial in cell senescence (23). Various types of stress result in
decreased cell proliferation, following regulation by tumor
suppressor genes, including p53, p16INK4a and p21
(23,24). Several proteins have been found to
be involved in cell senescence, including HDAC4, SIRT1,
p33ING1b and LSH (25–27).
The present study investigated whether FOXO1 affected the
expression of these genes. The results showed that FOXO1 regulated
the expression of SIRT1, but had no effect on the expression of
HDAC, LSH or p33ING1b. The ChIP assay demonstrated that
FOXO1 bound to the promoter region of SIRT1 in GBM cells, and
decreased the mRNA and protein levels of SIRT1 in GBM cells. These
results suggested that FOXO1 decreased the expression of SIRT1 at
the transcriptional level and that FOXO1 promoted senescence via
the transcriptional inhibition of SIRT1.
The data obtained in the present study demonstrated
that FOXO1 inhibited EMT. EMT is a complex process in cancer cell
development, which involves cancer cell growth, different,
metastasis and other cell processes. The ectopic expression of
FOXO1 markedly increased epithelial markers, including E-cadherin
and α-catenin, whereas mesenchymal markers, N-cadherin, fibronectin
and vimentin were all decreased. The opposite results were obtained
when FOXO1 was silenced. Metastasis is a characteristic of
malignant cancer and results in poor prognosis. EMT enhances the
metastasis of cancer cells. As the present study found that FOXO1
was negatively correlated with neck lymph node metastasis, whether
FOXO1 regulated cancer metastasis was determined using a Transwell
assay. The results revealed FOXO1 suppressed GBM cell
metastasis.
Taken together, the results of the present study
revealed that FOXO1 was downregulated in GBM and, as a tumor
suppressor, FOXO1 inhibited tumor growth and senescence via
arresting cell cycle at the G0/G1 phase and inhibiting the
transcription of SIRT1, respectively. FOXO1 also suppressed EMT and
metastasis. These results provide evidence that FOXO1 may be a
potential biomarker and therapeutic target for patients with
GBM.
References
|
1
|
Sullivan PR: Brain tumors. N Engl J Med.
344:14782001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fu Z and Tindall DJ: FOXOs, cancer and
regulation of apoptosis. Oncogene. 27:2312–2319. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Paik JH, Kollipara R, Chu G, Ji H, Xiao Y,
Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al: FoxOs are
lineage-restricted redundant tumor suppressors and regulate
endothelial cell homeostasis. Cell. 128:309–323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sunayama J, Sato A, Matsuda K, Tachibana
K, Watanabe E, Seino S, Suzuki K, Narita Y, Shibui S, Sakurada K,
et al: FoxO3a functions as a key integrator of cellular signals
that control glioblastoma stem-like cell differentiation and
tumorigenicity. Stem Cells. 29:1327–1337. 2011.PubMed/NCBI
|
|
5
|
Morin RD, Mendez-Lago M, Mungall AJ, Goya
R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field
M, et al: Frequent mutation of histone-modifying genes in
non-Hodgkin lymphoma. Nature. 476:298–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Trinh DL, Scott DW, Morin RD, Mendez-Lago
M, An J, Jones SJ, Mungall AJ, Zhao Y, Schein J, Steidl C, et al:
Analysis of FOXO1 mutations in diffuse large B-cell lymphoma.
Blood. 121:3666–3674. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang H and Tindall DJ: Dynamic FoxO
transcription factors. J Cell Sci. 120:2479–2487. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Furuyama T, Nakazawa T, Nakano I and Mori
N: Identification of the differential distribution patterns of
mRNAs and consensus binding sequences for mouse DAF-16 homologues.
Biochem J. 349:629–634. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gilley J, Coffer PJ and Ham J: FOXO
transcription factors directly activate bim gene expression and
promote apoptosis in sympathetic neurons. J Cell Biol. 162:613–622.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ishikura S, Iwaihara Y, Tanaka Y, Luo H,
Nishi K, Doi K, Koyanagi M, Okamura T, Tsunoda T and Shirasawa S:
The nuclear zinc finger protein Zfat maintains FoxO1 protein levels
in peripheral T cells by regulating the activities of autophagy and
the Akt signaling pathway. J Biol Chem. 291:15282–15291. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang S, Xia P, Huang G, Zhu P, Liu J, Ye
B, Du Y and Fan Z: FoxO1-mediated autophagy is required for NK cell
development and innate immunity. Nat Commun. 7:110232016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Milan G, Romanello V, Pescatore F, Armani
A, Paik JH, Frasson L, Seydel A, Zhao J, Abraham R, Goldberg AL, et
al: Regulation of autophagy and the ubiquitin-proteasome system by
the FoxO transcriptional network during muscle atrophy. Nat Commun.
6:66702015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Y, Yang J, Liao W, Liu X, Zhang H,
Wang S, Wang D, Feng J, Yu L and Zhu WG: Cytosolic FoxO1 is
essential for the induction of autophagy and tumour suppressor
activity. Nat Cell Biol. 12:665–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guarente L and Picard F: Calorie
restriction-the SIR2 connection. Cell. 120:473–482. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Longo VD and Kennedy BK: Sirtuins in aging
and age-related disease. Cell. 126:257–268. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X, Zhang S, Blander G, Tse JG, Krieger
M and Guarente L: SIRT1 deacetylates and positively regulates the
nuclear receptor LXR. Mol Cell. 28:91–106. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lemieux ME, Yang X, Jardine K, He X,
Jacobsen KX, Staines WA, Harper ME and McBurney MW: The Sirt1
deacetylase modulates the insulin-like growth factor signaling
pathway in mammals. Mech Ageing Dev. 126:1097–1105. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ramsey KM, Mills KF, Satoh A and Imai S:
Age-associated loss of Sirt1-mediated enhancement of
glucose-stimulated insulin secretion in beta cell-specific
Sirt1-overexpressing (BESTO) mice. Aging Cell. 7:78–88. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paroni G, Mizzau M, Henderson C, Del Sal
G, Schneider C and Brancolini C: Caspase-dependent regulation of
histone deacetylase 4 nuclear-cytoplasmic shuttling promotes
apoptosis. Mol Biol Cell. 15:2804–2818. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Paroni G, Fontanini A, Cernotta N, Foti C,
Gupta MP, Yang XJ, Fasino D and Brancolini C: Dephosphorylation and
caspase processing generate distinct nuclear pools of histone
deacetylase 4. Mol Cell Biol. 27:6718–6732. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dole MG, Jasty R, Cooper MJ, Thompson CB,
Nunez G and Castle VP: Bcl-xL is expressed in neuroblastoma cells
and modulates chemotherapy-induced apoptosis. Cancer Res.
55:2576–2582. 1995.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lundberg AS, Hahn WC, Gupta P and Weinberg
RA: Genes involved in senescence and immortalization. Curr Opin
Cell Biol. 12:705–709. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Campisi J: Cellular senescence as a
tumor-suppressor mechanism. Trends Cell Biol. 11:S27–S31. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou R, Han L, Li G and Tong T: Senescence
delay and repression of p16INK4a by Lsh via recruitment of histone
deacetylases in human diploid fibroblasts. Nucleic Acids Res.
37:5183–5196. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han X, Niu J, Zhao Y, Kong Q, Tong T and
Han L: HDAC4 stabilizes SIRT1 via sumoylation SIRT1 to delay
cellular senescence. Clin Exp Pharmacol Physiol. 43:41–46. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li N, Li Q, Cao X, Zhao G, Xue L and Tong
T: The tumor suppressor p33ING1b upregulates p16INK4a expression
and induces cellular senescence. FEBS Lett. 585:3106–3112. 2011.
View Article : Google Scholar : PubMed/NCBI
|