Introduction
Miscarriage or missed abortion is the spontaneous
loss of a pregnancy between conception and 20 weeks into the
pregnancy, which affects between ~15–20% women of childbearing age
(1,2). The miscarriage until the 12th week of
the pregnancy is termed early pregnancy loss (EPL) which occurs in
~15% of the cases (3,4), and the ones which occur between 12th
and 20th weeks of the pregnancy are termed late pregnancy loss
(LPL) which has an incidence rate of 1–5% (5,6). It
has been previously estimated ~80% of miscarriages take place in
the first 12 weeks (the first trimester) of pregnancy as EPL. The
etiological factors during a miscarriage are generally defined as
multiple factors (7,8). The factors include abnormalities
inherited from the father (paternal abnormalities), fetal
anatomical abnormalities, the age of the mother during the
pregnancy, abortion history and environmental risk factors, such as
caffeine, alcohol, cigarettes. In addition, factors such as
genetics, infection, hematology and autoimmunity have been
previously identified as important for the occurrence of
miscarriages (5,7,8). The
pathogenesis of EPL remains to be elucidated (2). Previous studies based on advanced
molecular genetics methods have investigated EPL pathogenesis. The
development of the human placenta is critical for embryonic
development and successful pregnancy. One of the important factors
for the development and function of the placenta is epigenetic
mechanism in gene expression (3,9).
Epigenetic factors are defined as DNA methylation (CpG), chromatin
histone modification and noncoding regulatory RNAs (8). The growth and progress of a fetus
depend on the placental function and vein development; therefore,
placental samples may be used for genomic and epigenetic research
in miscarriage cases (4).
MicroRNAs (miRNAs) have been identified as, the important molecules
in genetic and genomic-based studies. It has been estimated that
there are >2,000 miRNAs in humans, which are single stranded RNA
molecules with ~20-nucleotide length, which are not protein coding
(9). One miRNA may regulate ~200
mRNA functions. miRNA expression levels may change from one tissue
type to another and also in different diseases. It has been
previously established that miRNAs contribute to various biological
events, such as development, differentiation, fertility and
canceration (10–12). A previous study summarized that
abnormal miRNA expression may be associated with ~400 diseases to
human diseases (13). During
pregnancy miRNA expression profiles in placenta and maternal blood
have an important epigenetic role during the growth and development
of fetus (14,15). Additionally, miRNA expression in
the placenta is regulated by environmental factors, signaling
pathways and epigenetic modifications (16). There are three aims for
genetics/genomics researches when investigating miscarriage
patients. Firstly, to determine a DNA/RNA-based biomarker which may
determine the direct predictive value of a couple's risk for
miscarriage. Secondly, to establish gene/protein expression
profiles, pathways and connections, which occur at the
(un)successful establishment of pregnancy. Thirdly, apply
hypothesis-based and hypothesis-free studies to pinpoint loci
coding for novel non-invasive biomarkers applicable in clinical
conditions for early pregnancy complications (8). Fetal nucleic acids that occur in the
maternal plasma and serum are used by specific isolation methods
for early diagnosis of genetic-based diseases such as preeclampsia,
detection of fetal gender in women with a high risk of
gender-associated disorders, fetal Rhesus (Rh) status and trisomies
in pregnancies with a high risk for hemolytic disease of newborn
(HDN) (9,17,18).
Fetal nucleic acids and miRNAs in maternal blood are analyzed by
advanced molecular methods, such as quantitative polymerase chain
reaction (qPCR), DNA/RNA microarrays and sequencing methods
(18).
The miRNA microarray findings in the present study
revealed that 4 miRNAs, hsa-miR-125a-3p, hsa-miR-3663-3p,
hsa-miR-423-5p and hsa-miR-575, were upregulated in tissue samples.
In maternal plasma, 2 miRNAs (hsa-let-7c, hsa-miR-122) were
upregulated and one miRNA (hsa-miR-135a) was downregulated. A total
of 6 out of 7 dysregulated miRNAs were validated by reverse
transcription (RT)-qPCR. The target genes of these dysregulated
miRNAs were identified using the GeneSpring database. The present
study aimed to quantify miRNA expression levels in villous tissue
and maternal plasma in EPL cases, to determine the miRNAs which
exhibit changes in expression levels, and to detect the target
genes of dysregulated miRNAs and the associated pathways which may
establishing the association between miRNAs and the factors leading
to EPL.
Materials and methods
Sample processing
Villous tissue samples which produce placenta and
blood were collected from a total of 24 patients, 16 patients with
EPL in 6th-8th gestational weeks (GWs) and 8 abortion cases
(control group) in 6th-8th GW. Samples were collected from
Department of Obstetrics and Gynecology, Cerrahpasa Medical
Faculty, Istanbul University (Istanbul, Turkey) and Medicus Health
Center (Istanbul, Turkey) between January 2013 and May 2015. All
patients and volunteers have been asked to sign written informed
consent, the present study was also approved by the Ethical
Committee. All informed consent forms are documented with the
signature of the patients. Women in the control group had requested
a surgical termination of pregnancy for non-medical reasons, and
women in the EPL group had undergone surgical uterine evacuation.
Villous tissue samples were collected immediately after the
surgical procedure in sterile collection containers with
physiological saline solution and were stored at −80°C until used.
The blood samples were collected immediately prior the surgical
procedure in ethylenediaminetetraacetic acid (EDTA) -containing
tubes (BD Biosciences, Franklin Lakes, NJ, USA) and immediately
centrifuged at 1,400 × g for 10 min at 4°C to separate the plasma.
Plasma samples were carefully transferred into new tubes and stored
at −80°C until used. Samples were stored in Istanbul University,
Department of Molecular Biology and Genetics for experimental
research.
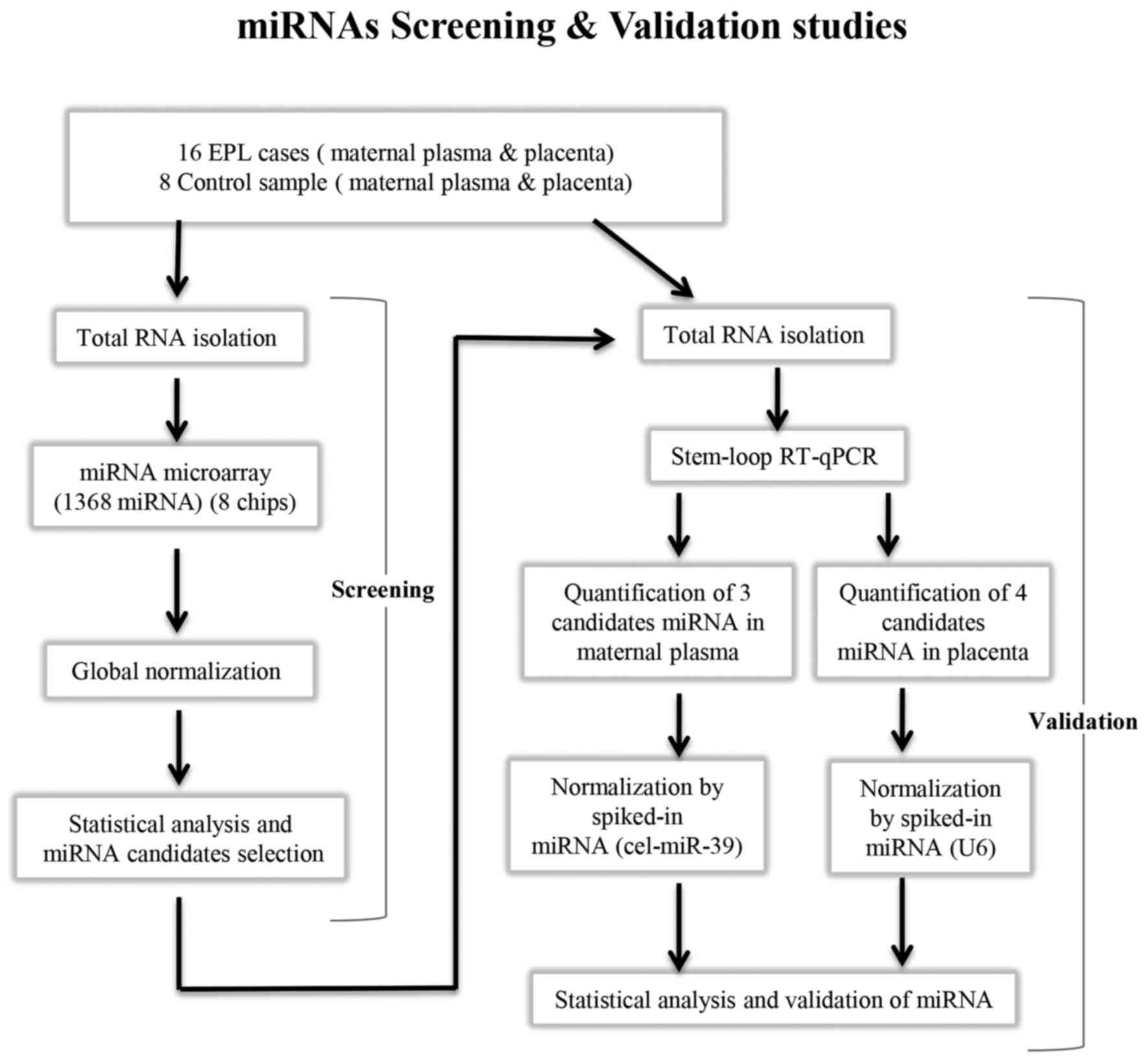
The present study was divided into two stages: i)
MiRNA profiling of villous samples and maternal plasma using
microarrays; and ii) validation of dysregulated miRNAs using
RT-qPCR (Fig. 1).
Total RNA isolation from villous
tissue samples
Total RNA, including miRNAs was extracted from the
villous tissue samples with the mirVana miRNA Isolation kit
(Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the manufacturer's protocol. Total RNA was eluted with
50 µl mirVana elution solution. The quantity and quality of
obtained total RNA was determined using NanoDrop ND-2000
spectrophotometer (Thermo Fisher Scientific, Inc.) and with the
Agilent RNA Nano Chip kit on Agilent Bioanalyzer 2100 system
(Agilent Technologies, Inc., Santa Clara, CA, USA). High quality
(RNA integrity number ≥7) samples from isolated total RNA were
selected for microarray analysis. The isolated total RNAs were
stored at −80°C during laboratory preparation.
Total RNA isolation from maternal
plasma samples
Total RNA, including miRNAs was isolated from 500 µl
maternal plasma sample using the mirVana miRNA Isolation kit
(Ambion; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Synthetic Caenorhabditis elegans (C.
elegans) miRNA, cel-miR-39 (working solution 1.6×108
copies/µl; Qiagen miRNeasy Serum/Plasma Spike-in control Qiagen,
Inc., Valencia, CA, USA), was spiked into each sample (following
the addition of a lysis buffer) to determine the purification
quality and to normalize variation. Synthetic cel-miR-39 was
selected as a spike-in due to the absence of homologous sequences
in humans. Total RNA was eluted with 30 µl mirVana elution
solution. The quantity total RNA obtained was determined using
NanoDrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.)
and subsequently by the Agilent Small RNA Chip kit on Agilent
Bioanalyzer 2100 system (Agilent Technologies, Inc.). The isolated
total RNA was stored at −80°C during laboratory preparation.
miRNA microarray analysis
In total, 32 EPL samples which were 16 maternal
plasma and 16 villous tissue, furthermore 16 normal samples which
were 8 maternal plasma and 8 villous tissue were analyzed by
Agilent miRNA microarray chips (G4870A SurePrint G3 Human v16 miRNA
8×60K microarray; Agilent Technologies, Inc.). The miRNA Labeling
and Hybridization kit (Agilent Technologies, Inc.) was used
according to the manufacturer's protocol. Total RNA used for each
villous sample was 100 ng/µl and maternal plasma sample of ~50
ng/µl RNA (which included small RNAs). Each sample was labeled with
cyanine 3-cytidine bisphosphate (pCp-Cy3) and the labeled RNA was
hybridized at 55°C for 24 h to Human miRNA Microarray version 16
(Agilent Technologies, Inc.) slides, which included 1,368 miRNAs
encoded by genes located across all chromosomes. After washing the
microarray slides with Gene Expression Wash buffers (Agilent
Technologies, Inc.) in order to increase the stringency, the slides
were scanned by an Agilent SureScan Microarray scanner (model
G2600D).
Agilent Feature Extraction version 11.0 was used for
image analysis. The raw data was statistically analyzed using
GeneSpring GX version 12.6 (Agilent Technologies, Inc.). In this
database, the raw data produced was normalized by quantile
normalization and probes with <50% coefficient of variation were
filtered. From these probes, a Student's t-test with a Bonferroni
correction for the familywise error rate was performed and the
following thresholds were used P<0.05 and fold-change (FC) >2
were identified to be significantly dysregulated. The dysregulated
miRNA results were obtained from the GeneSpring database (19) and gene targets and pathways
associated with these genes have been listed in the present study
according to this database. The target genes of significantly
differentially expressed miRNAs were predicted and pathway analysis
in villous and maternal plasma samples was performed using
GeneSpring version 12.6, a nominal P<0.05 was used to select
significant pathways. RefSeq Protein ID, RefSeq Transcript ID,
Entrez Gene ID, and UniGene ID annotations were selected for the
present study experiment. Pathway sources used in the analysis,
included WikiPathways-Analysis, WikiPathways-Reactome, and
WikiPathways-Other, which are valid for Homo sapiens. The
results of the pathway analysis were obtained from the ‘Single
Experiment Analysis (SEA)’ heading in the ‘GeneSpring’ database.
Calculation of the P-value is indicated in the subsequent
formula:
P(X≥k)=1(un)∑i=kiMax(mi)(u–mn–i)
Where u stands for the total number of entities in
the technology which have same ID for annotations selected in SEA;
m, number of entities in the entity list which have same ID for the
annotation selected; n, number of pathway entities that matched
with the technology; and k, number of pathway entities that matched
with the entity list.
miRNA validation by stem-loop
RT-qPCR
The validation of miRNA microarray results on the
EPL and control in maternal plasma and tissue samples was performed
using RT-qPCR. Dysregulated miRNAs (P<0.05 and FC>2) obtained
from the microarray bioinformatics analysis were subsequently
validated in 24 (16 EPL & 8 control) tissue samples and 24 (16
EPL & 8 control) maternal plasma samples using stem-loop
RT-qPCR.
The present study synthetic cel-miR-39 for
normalization and detection of extraction efficiency in maternal
plasma, and U6 for normalization in tissue samples of TaqMan
RT-qPCR in dysregulated miRNAs. miRNAs from each sample were
reverse-transcribed into cDNA using TaqMan microRNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.) and miRNA
specific 5X stem-loop primers (TaqMan™ MicroRNA Assay; Applied
Biosystems; Thermo Fisher Scientific, Inc.) hsa-miR-125a-3p (cat.
no. 4427975, RT: 002199), hsa-miR-3663-3p (cat. no. 4427975; RT:
465775_mat), hsa-miR-575 (cat. no. 4427975; RT: 001617),
hsa-miR-425-5p (cat. no. 4427975; RT: 002302), has-let-7c (cat. no.
4427975; RT: 000379), hsa-miR-122 (cat. no. 4427975; RT: 002245),
hsa-miR-135a (cat. no. 4427975; RT: 000460) in total reaction
volume of 15 µl according to the manufacturer's protocol.
Transcription reaction was preformed using an 8800 PCR thermal
cycler system (Agilent Technologies, Inc.) by heating to 16°C for
30 min, followed by 42°C for 60 min and 85°C for 5 min. The qPCR
was performed using TaqMan miRNA qPCR Universal PCR Master mix
assay (Thermo Fisher Scientific, Inc.), and 20X primer (Thermo
Fisher Scientific, Inc.), which included fluorescein dye-specific
probes for each previously stated miRNA in a 20 µl total reaction
mixture. Every batch of amplifications included two water blanks
and primers as no template negative controls for each cDNA product
and qPCR steps. The following thermocycling conditions were used
for the PCR: 50°C for 2 min, 95°C for 10 min; 40 cycles of 95°C for
15 sec and 60°C for 1 mins. The qPCR was replicated 2 times for
each sample. RT-qPCR results were analyzed as follows: ΔCt=Ct
sample-Ct cel-miR-39; ΔΔCt=ΔCt case-ΔCt control. Finally, the ΔΔCt
was normalized against cel-miR-39. All reactions were analyzed by
using the 2−ΔΔCt method (20).
Results
Characteristics of samples
There were two experimental groups in the present
study, including the patient group (the mean pregnancy age is ~30)
and the control group (the mean pregnancy age is ~29). In both
groups, none of the participants had anatomic abnormalities,
infections, endocrine or metabolic disorders, autoimmune diseases,
paternal or maternal chromosomal abnormalities, or smoked. Table I presents the characteristics of
the two experimental groups.
 | Table I.Clinical summary of EPL and normal
pregnancy control groups. |
Table I.
Clinical summary of EPL and normal
pregnancy control groups.
| Groups | Age (years) | Mean gravidity
(orders) | Mean of
miscarriage | Gestational age
(weeks) |
|---|
| EPL | 30.23±5.81 | 1.81 | 2.75 | 7.22±2.8 |
| Control | 28.62±5.24 | 2.12 | 0 | 7.11±5.2 |
miRNA microarray results
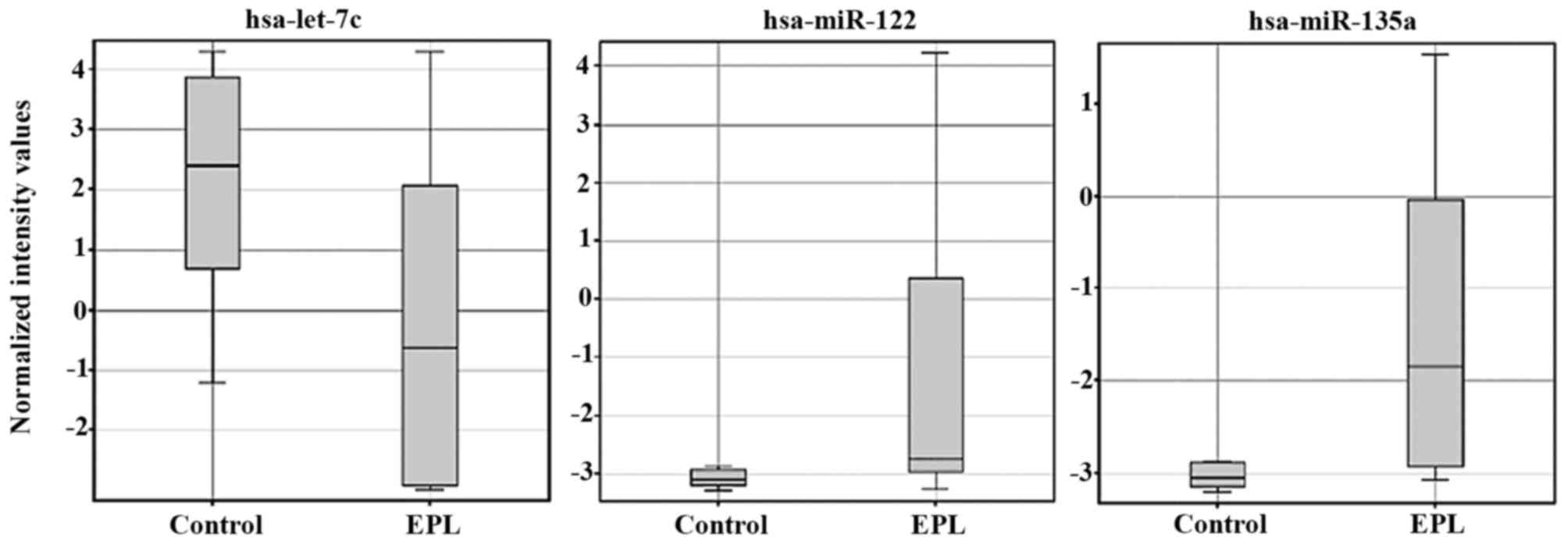
Statistical analysis of microarray results in
maternal plasma identified 3 dysregulated miRNAs. From the three
miRNAs in maternal plasma samples two were upregulated and one was
downregulated (Table II).
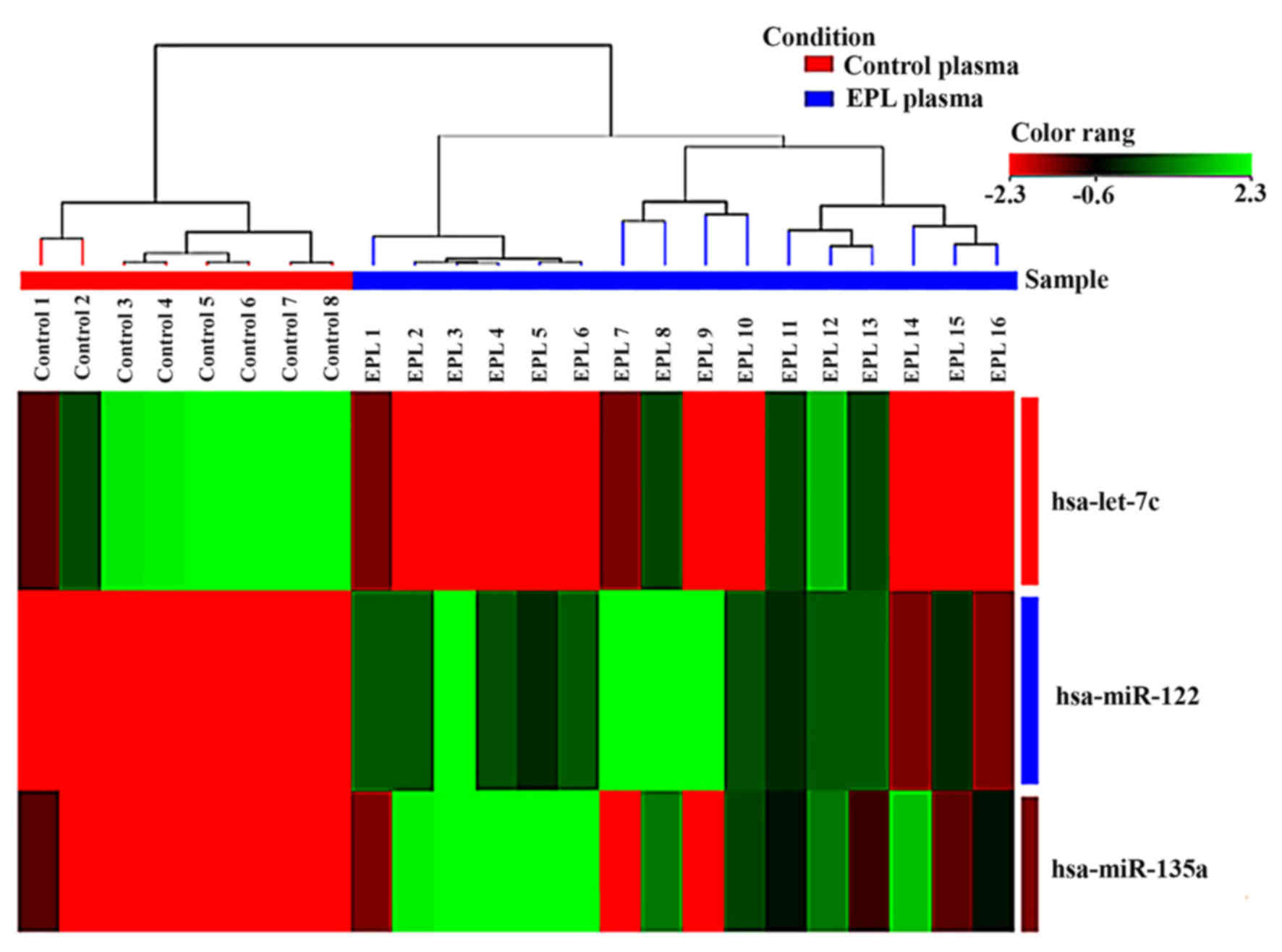
Expression levels for each miRNA in maternal plasma have been
presented as box-whisker plots (Fig.
2) and hierarchical clustering analysis revealed that the
subgroups were well-differentiated from the unified set of
differentially expressed miRNAs (Fig.
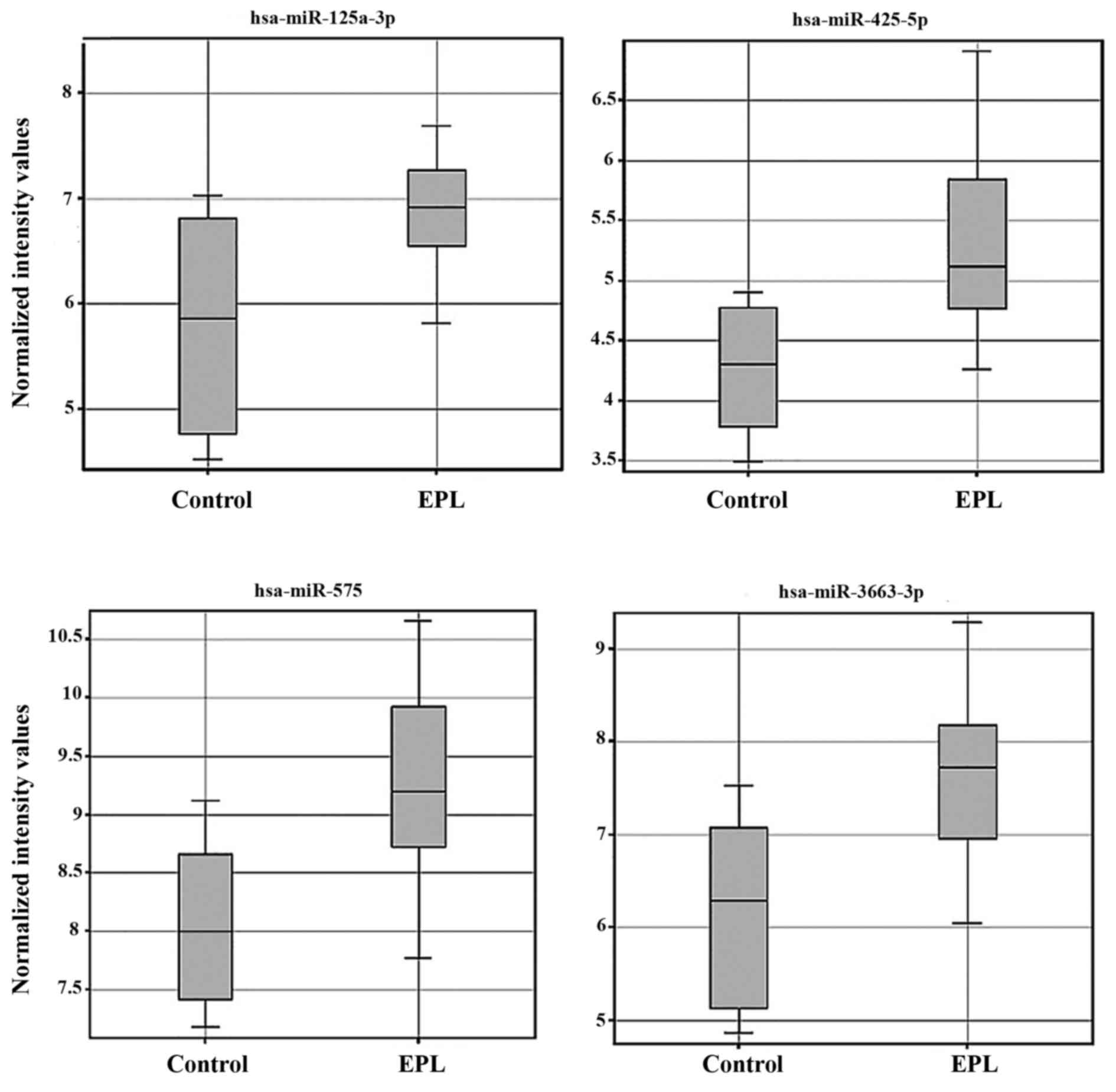
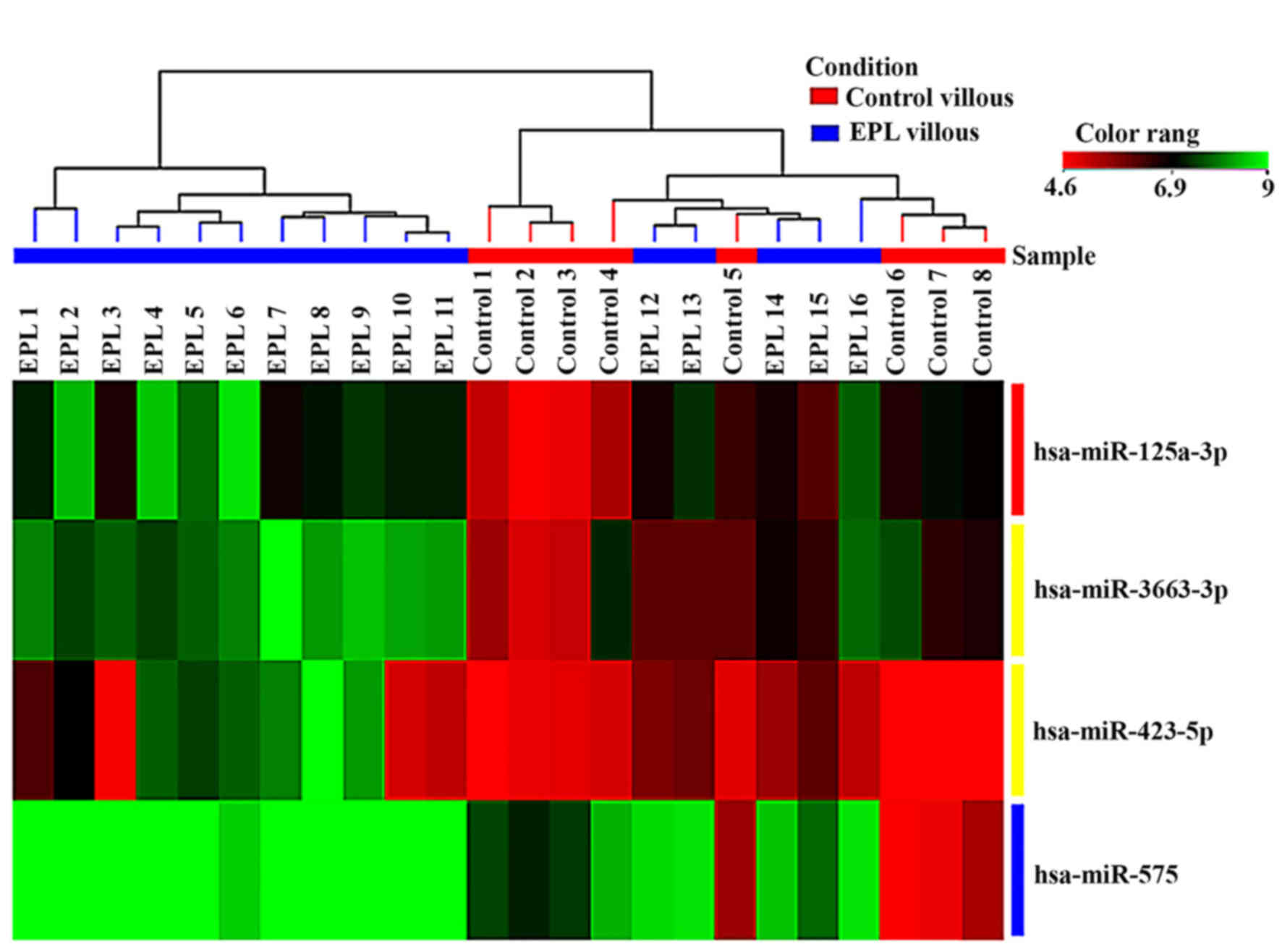
3). In the villous tissue samples, all 4 miRNAs were
upregulated (Table III) and have
been found to be significantly dysregulated (P<0.05 and FC>2)
in the EPL group compared with the control group. In villous
samples dysregulated miRNAs have been represented as box-whisker
plots (Fig. 4) and hierarchical
clustering (Fig. 5).
 | Table II.Dysregulated miRNAs for early
pregnancy loss and control maternal plasma samples. Corrected
P<0.05 and FC>2. |
Table II.
Dysregulated miRNAs for early
pregnancy loss and control maternal plasma samples. Corrected
P<0.05 and FC>2.
| Systematic
name | Regulation | P-value | FC | Active sequence
(5′→3′) | Chromosome |
|---|
| hsa-let-7c | Down | 0.0041 | −5.16 |
AACCATACAACCTACTACC | 21 |
| hsa-miR-122 | Up |
0.00039 |
4.72 |
CAAACACCATTGTCACACT | 18 |
| hsa-miR-135a | Up |
0.00294 |
2.62 | CGCCACGGCTCCA | 3 |
 | Table III.Dysregulated miRNAs for early
pregnancy loss and control villous sample Corrected P<0.05 and
FC>2. |
Table III.
Dysregulated miRNAs for early
pregnancy loss and control villous sample Corrected P<0.05 and
FC>2.
| Systematic
name | Regulation | P-value | FC | Active sequence
(5′→3′) | Chromosome |
|---|
|
hsa-miR-125a-3p | Up | 0.000407 | 2.15 |
GGCTCCCAAGAACCTCA | 19 |
|
hsa-miR-3663-3p |
| 0.000337 | 2.73 | GCGCCCGGCCT | 10 |
| hsa-miR-423-5p |
| 0.0332 | 2.01 |
AAAGTCTCGCTCTCTG | 17 |
| hsa-miR-575 |
| 0.00275 | 2.33 |
GCTCCTGTCCAACTGGCT | 4 |
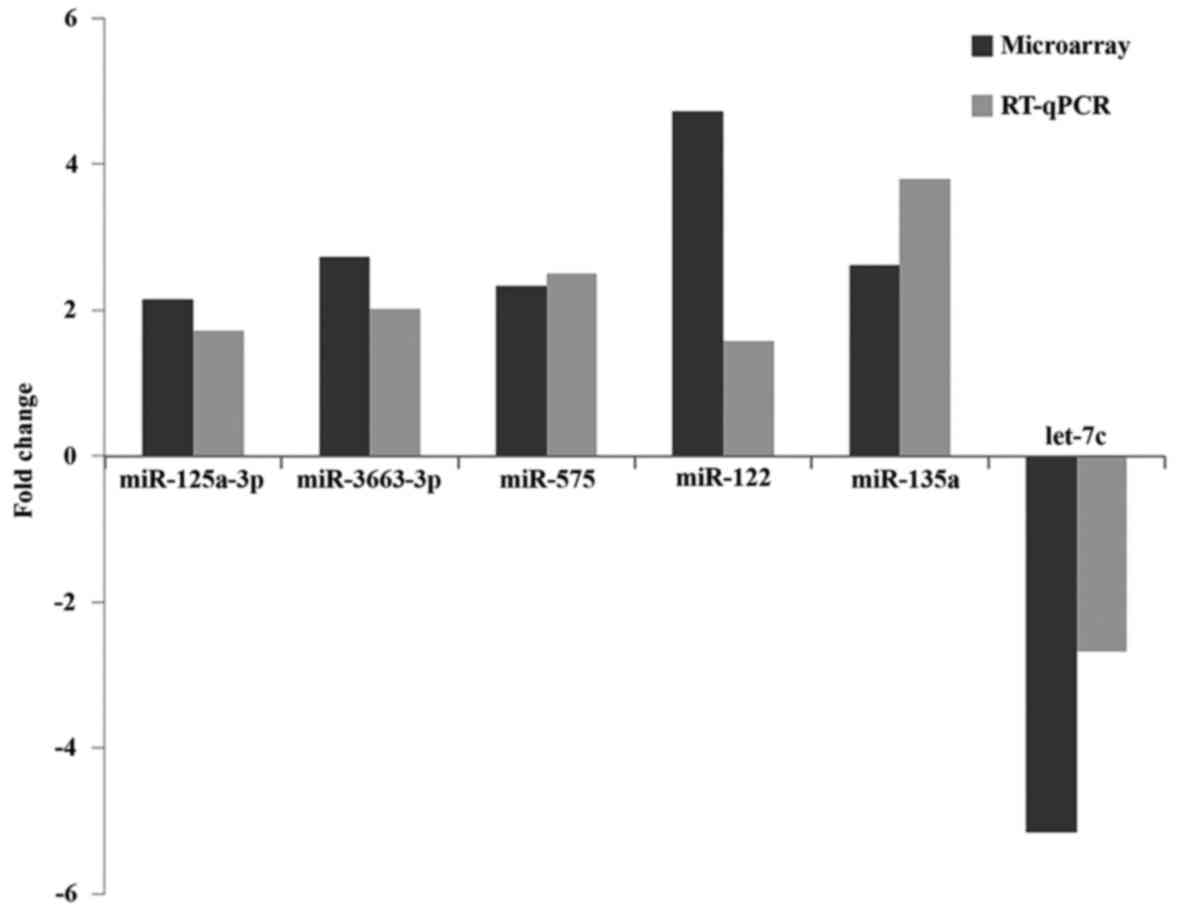
Stem-loop RT-qPCR findings
Stem-loop RT-qPCR was performed to validate the 7
differentially expressed miRNAs identified in the miRNA microarray
analysis. A total of 24 plasma samples, consisting of 16 EPL
maternal plasma and 8 normal plasma samples, were used for maternal
plasma validation and the same quantity of EPL and normal villous
tissues were used for validation by stem-loop RT-qPCR. The present
study identified 3 miRNAs (hsa-let-7c, hsa-miR-122 and
hsa-miR-135a) in maternal plasma samples and 3 miRNAs, including
hsa-miR-125a-3p, hsa-miR-3663-3p and hsa-miR-575 in villous samples
were validated to be upregulated in EPL samples. However,
miR-425-5p did not demonstrate any marked expression alterations in
villous tissues by RT-qPCR. Consistent with miRNA microarray
analysis, the changes of all 6 upregulated miRNAs in EPL and
control samples in villous tissues and maternal plasma are
presented in Table IV and
Fig. 6.
 | Table IV.Differential expression of validated
signature miRNAs for early pregnancy loss and control samples using
microarray and TaqMan RT-qPCR platforms. |
Table IV.
Differential expression of validated
signature miRNAs for early pregnancy loss and control samples using
microarray and TaqMan RT-qPCR platforms.
|
|
|
| miRNA
microarray | RT-qPCR |
|---|
|
|
|
|
|
|
|---|
| Sample | Regulation | miRNA | FC | P-value | FC | P-value |
|---|
| Maternal
plasma | Up | hsa-miR-122 |
4.7 | 0.00039 | 1.57 | 0.0241 |
|
|
| hsa-miR-135a |
2.62 | 0.00294 | 3.8 | 0.0249 |
|
| Down | hsa-let-7c | −5.16 | 0.00412 | −2.68 | 0.0323 |
| Villous tissue | Up |
hsa-miR-125a-3p |
2.15 | 0.000407 | 1.72 | 0.0258 |
|
|
|
hsa-miR-3663-3p |
2.73 | 0.000337 | 2.02 | 0.0075 |
|
|
| hsa-miR-423-5p |
2.01 | 0.03323 | N | N |
|
|
| hsa-miR-575 |
2.33 | 0.000275 | 2.5 | 0.00029 |
Target genes and pathway analysis
Target analysis for validated dysregulated miRNAs in
GeneSpring version 12.6 identified 432 target genes for the villous
sample dysregulated miRNAs and 274 target genes for maternal plasma
dysregulated miRNAs, where P<0.05. Pathway analysis of targeted
genes of villous sample and maternal plasma samples with P<0.05
by GeneSpring identified 233 and 130 pathways, respectively. The
top significant pathways (sorted by lowest P-value) of the target
genes was predicted by differentially expressed miRNAs in villous
samples and maternal plasma are presented in Tables V and VI, respectively.
 | Table V.Top pathways involved by number of
genes targeted by dysregulated miRNAs, including miR-125a-3p,
miR-3663-3p and miR-575, obtained from villous samples. |
Table V.
Top pathways involved by number of
genes targeted by dysregulated miRNAs, including miR-125a-3p,
miR-3663-3p and miR-575, obtained from villous samples.
| Pathway | Nominal
P-value | Matched
entities | Pathway
entities |
|---|
| Hs EGF EGFR
signaling pathway WP437-72106 | 0.000000001015 | 7 | 162 |
| Hs TSLP signaling
pathway WP2203-72104 | 0.0000003804 | 3 | 47 |
| Hs T cell receptor
and co-stimulatory signaling WP2583-75360 | 0.0000003804 | 3 | 30 |
| Hs ErbB signaling
pathway WP673-69914 | 0.0000003804 | 3 | 54 |
| Hs apoptosis
WP254-72110a | 0.0000003804 | 3 | 84 |
| Hs calcium
regulation in the cardiac cell WP536-75219a | 0.000001417 | 6 | 150 |
| Hs IL-7 signaling
pathway WP205-70018 | 0.00005262 | 2 | 27 |
| Hs MAPK cascade
WP422-72129a | 0.00005262 | 2 | 29 |
| Hs IL-2 signaling
pathway WP49-75383 | 0.00005262 | 2 | 42 |
| Hs
integrin-mediated cell adhesion WP185-71391 | 0.00005262 | 2 | 99 |
| Hs Wnt signaling
pathway and pluripotency WP399-74897a | 0.00005262 | 2 | 100 |
| Hs IL-3 signaling
pathway WP286-72139 | 0.00005262 | 2 | 49 |
| Hs focal adhesion
WP306-71714a | 0.0001974 | 5 | 188 |
| Hs myometrial
relaxation and contraction pathways WP289-72107a | 0.0001974 | 5 | 156 |
| Hs MAPK signaling
pathway WP382-72103a | 0.0027438 | 4 | 168 |
| Hs GPCRs class A
rhodopsin-like WP455-74420a | 0.0027438 | 4 | 261 |
| Hs oxidative stress
WP408-69029 | 0.007262 | 1 | 30 |
| Hs IL-5 signaling
pathway WP127-70017 | 0.007262 | 1 | 40 |
| Hs JAK-STAT
WP2593-74127a | 0.007262 | 1 | 44 |
 | Table VI.Top pathways involved by number of
genes targeted by dysregulated miRNAs, including miR-122, miR-135a
and let-7c, obtained from maternal plasma samples. |
Table VI.
Top pathways involved by number of
genes targeted by dysregulated miRNAs, including miR-122, miR-135a
and let-7c, obtained from maternal plasma samples.
| Pathway | Nominal
P-value | Matched
entities | Pathway
entities |
|---|
| Hs calcium
regulation in the cardiac cell WP536-75219a | 0.00002112 | 2 | 150 |
| Hs MAPK cascade
WP422 −72129a | 0.00002112 | 2 | 29 |
| Hs inflammatory
response pathway WP453-63217 | 0.00002112 | 2 | 33 |
| Hs focal adhesion
WP306-71714a | 0.00002112 | 2 | 188 |
| Hs myometrial
relaxation and contraction pathways WP289-72107a | 0.004421 | 4 | 156 |
| Hs GPCRs, Class C
Metabotropic glutamate, pheromone WP501-63205 | 0.004632 | 1 | 15 |
| Hs Regulation of
toll like receptor signaling pathway WP1449-72114 | 0.004632 | 1 | 150 |
| Hs Toll-like
receptor signaling pathway WP75-72133 | 0.004632 | 1 | 102 |
| Hs IL-1 signaling
pathway WP195-74010 | 0.004632 | 1 | 55 |
| Hs Wnt signaling
pathway and pluripotency WP399-74897a | 0.004632 | 1 | 100 |
| Hs JAK STAT
WP2593-74127a | 0.004632 | 1 | 44 |
| Hs MAPK signaling
pathway WP382-72103a | 0.009645 | 3 | 168 |
| Hs Apoptosis
Modulation and Signaling WP1772-63162 | 0.009645 | 3 | 93 |
| Hs apoptosis
WP254-72110a | 0.009645 | 3 | 84 |
| Hs GPCRs, class A
rhodopsin-like WP455-74420a | 0.009645 | 3 | 261 |
Discussion
The formation of placenta is of high importance in a
healthy pregnancy process and in embryo development. Mechanisms
such as differentiation, migration, invasion, angiogenesis,
proliferation and apoptosis have a significant role in placenta
formation (8,9). It has been estimated that >500
miRNA expression levels specific to placenta have been identified
during pregnancy. The expression levels of these miRNAs are
regulated by environmental factors such as hypoxia, signaling
pathways and epigenetic modifications (9,12).
Abnormal expression levels of miRNAs were also identified in
gynecological diseases, including ovarian carcinoma, endometriosis,
benign gynecological conditions and infertility (21). In the present study, maternal blood
and villous samples, which have an important role in the formation
of placenta were collected from EPL cases and patients voluntarily
having abortion. MiRNA expressions were quantified in the samples.
In EPL patients, significant miRNA expression changes were
identified compared to the control patients. According to the
microarray findings, a total of 4 miRNAs were identified in the
tissue samples (Table III) and 4
miRNAs in maternal plasma samples (Table II) with evident expression
differentiations. These findings were confirmed for 7 miRNAs, using
RT-qPCR as presented in Table IV.
MiR-125a-3p, miR-3663-3p, miR-575, let-7c, miR-122 and hsa-miR-135a
obtained from villous and maternal plasma samples have been
validated following RT-qPCR analysis. In the present study EPL
cases between 6th-8th GWs, for which the reason for the occurrence
of the EPL were unknown, have been investigated. Tissue and blood
samples have been collected from each subject and miRNA analyses
were performed for each case. To the best of our knowledge, based
on EPL grouping and the performed analyses, the present study may
be defined as first in this area.
Dysregulated miRNAs obtained from
villous tissue samples
The comparison between EPL and control group samples
by microarray and RT-qPCR revealed that miR-125a-3p, miR-3663-3p
and miR-575 were significantly upregulated in tissue samples.
The miR-125 family is consists of miR-125a,
miR-125b-1 and miR-125-2 homologues which have a wide variety of
functions, such as control of proliferation of the hematopoietic
stem cells, control of genes associated with carcinomas, whose
expression patterns are protected in the genome (22,23).
The present study identified that expression of miR-125a-3p, which
is the mature form of miR-125a, was increased in villous tissue
samples of EPL group compared with the control. Li and Li
demonstrated that miR-125a expression was increased in Unexplained
Recurrent Spontaneous Abortion (URSA) samples compared with control
samples (22). Therefore, miR-125a
expression may be associated with recurrent miscarriages (24). A previous study revealed that
expression levels of ErbB2 receptor tyrosine kinase 2
(ERBB2), which is the receptor of heparin-binding epidermal
growth factor and leukemia inhibitory factor receptor
(LIFR), and leukemia inhibitory factor have an effect on the
implantation of embryo on the uterus wall which is increased in
implantation stage of menstrual cycle (25). ERBB2 and LIFR are
among the genes targeted by miR-125a-3p, therefore, increase in
miR-125a-3p may lead to a decrease in the expression level of
ERBB2 and LIFR genes at the implantation stage. It is
possible that a reduced expression level of these genes at the
implantation stage may negatively affect the implantation process.
Ninio-Many et al revealed that increase in miR-125a-3p
expression prevents the expression of the Fyn gene which is
part of the Scr kinase family, which may lead to reduced
cell adhesion, proliferation, migration and viability (26). Along with the decrease of
Fyn gene expression, the expression levels of focal adhesion
kinase (FAK), paxillin and V-Akt murine thymoma viral
oncogene (Akt) genes located downstream of Fyn were
also decreased. Subsequently, this limits the viability and
mobility of cells and blocks the cell cycle (26). The present study identified that
increased miR-125a-3p expression may lead to a miscarriage by
reducing the expression levels of FAK, paxillin and
Akt genes during adhesion, proliferation and migration
processes which are required for placenta and embryo development.
Zhang et al observed that miR-125a-3p expression has been
increased and expression levels of metastasis-associated gene 1
(MTA1), which has a critical role in chromatin remodeling
and histone deacetylase complexes in cells, has been reduced in the
non-small cell lung cancer (27).
Increased expression of miR-125a in breast cancer cells suppresses
the growth of cells by reducing the expression of HuR protein which
is a stress-induced RNA binding protein (28). miR-125a binds to the
3′-untranslated region (UTR) of the gene encoding the transcription
factor estrogen-related receptor a in squamous cell carcinoma of
the mouth and reduces the expression of this gene, and thereby
suppressing the cell proliferation and increasing apoptosis
(29). Considering these previous
findings, cell migration, proliferation and cell adhesion are
suppressed, and apoptosis is increased due to the increased
expression of miR-125a-3p in the cell. The absence of migration and
adhesion, and excessive apoptosis during the development of the
embryo in the cellular dimensions may lead to EPL. According to the
Target Scan database, vascular endothelial growth factors (VEGFs)
are target genes of miR-125-3p, which are important for placenta
development. Choi et al investigated the correlation between
recurrent miscarriages and decrease in the expression level of
VEGFs, which are effective in angiogenesis (30). The present study determined that
miR-125a expression increased in the villous trophoblast, which may
lead to reduced VEGFs expression levels and lead to EPL by
adversely affecting angiogenesis.
The present study identified miR-575 expression to
be increased in tissue samples. Gu et al determined that
expression level of miR-575 was increased in maternal serums in
neural tube defects (NTDs) (31).
It was previously observed that miRNA expression levels were
severely reduced in serum within 24 h after birth. Therefore, it is
possible that these miRNAs associated with the pregnancy may be
used for the early diagnosis of NTD fetuses (31). Nardelli et al determined
that miR-575 was expressed only in pregnant obese patients
(32). Expression of this miRNA
was demonstrated to be effective in the adipocytokine and
mitogen-activated protein kinase signaling pathways which have an
important role in placental development, leading to a decrease in
the expression of adiponectin receptor 2 and solute carrier family
2 member 1 proteins and epigenetic regulation of placental
metabolism (32). Based on the
findings obtained in the current study, this may contribute to EPL
as degradation of the adipocytokine and mitogen-activated protein
kinase signal pathways disrupts the placental development and
function, as a result of the increased expression of miR-575. It
was previously identified that the increased expression of miR-575
in breast cancer cells may lead to the expression of
phosphatidylinositol specific phospholipase C X domain containing 1
to reduce and thus to suppress cell proliferation (33). The present study demonstrated that
increased expression of this miRNA may lead to EPL by having
diverse effects on important processes, such as cell growth and
proliferation in the development of placenta and embryo.
Previous studies on miR-3663-3p have revealed that
its expression was elevated in particular types of cancer, such as
primary cutaneous melanoma (34),
nasopharyngeal carcinoma (35) and
prostate cancer (36). The present
study determined that the expression in the miRNA scans of the
tissue samples obtained from the EPL cases was increased when
compared with normal tissues. However, as is no specific research
on this miRNA, only target genes and their associated functions
could be obtained following bioinformatics analysis. The genes
targeted by miR-3663-3p were found to be effective in
differentiation processes, cell division, apoptosis and cell
proliferation (37–39). Pathway analysis for predicted
target genes of the validated miRNAs in tissue samples
(miR-125a-3p, miR-3663-3p and miR-575), including epidermal growth
factor receptor (EGFR), T cell receptor, ErbB, Wnt, mitogen
activated protein kinase (MAPK), interleukin (IL)-2,-3,-5 and −7,
revealed signaling pathways involved may be apoptosis, oxidative
stress, focal adhesion and janus kinase-signal transducer and
activator of transcription (JAK-STAT) pathways (Table V) which have previously been
identified to be associated with miscarriage pathogenesis (2,3,24,40–43).
Dysregulated miRNAs obtained from
maternal plasma samples
Previous studies on maternal plasma miRNAs use
biomarkers for reproductive diseases, specifically for EPL and
URSA, are limited (44–46). To the best of our knowledge the
present study is the first to investigate circulating miRNAs in EPL
patients. The comparison of EPL and control group samples using
microarray and RT-qPCR techniques revealed that miR-135a, miR-122
and let-7c were significantly dysregulated in maternal plasma
samples.
Expression level of miR-135a was determined to be
higher in the maternal plasma samples. Petracco et al
detected increased expression of miR-135a in endometriosis
endometrium tissue samples during the menstrual cycle proliferation
and luteal phases and reduced in the ovulation phase (47). It is possible that binding of
miR-135a to the 3′-UTR region of HOXA10 gene may be
increased along with the reduced expression of HOXA10, and
may lead to EPL by adversely affecting the uterus development and
embryo implantation. In a previous study conducted on lung cancer,
it was determined that miR-135a acted as a tumor suppressor
(47). It is possible that binding
of miR-135a to the 3′-UTR region of HOXA10 gene may be
increased along with the reduced expression of HOXA10, and
may lead to EPL by adversely affecting the uterus development and
embryo implantation. In a previous study conducted on lung cancer,
it was determined that miR-135a acted as a tumor suppressor
(48). It was also determined that
miR-135a reduced the expression of Kruppel-like factor 8
(KLF8) which is a transcription factor, and suppressed the
epithelial mesenchymal transition molecular mechanism of cancer
cells, inhibiting cell migration and invasion (48). It believed that the increase in
miR-135a expression may reduce the expression of KLF8,
leading to the prevention of migration and invasion processes,
which are required during the embryo development, and may lead to
EPL. Navarro et al detected that the increased levels of
miRNA-135a in patients with classical Hodgkin's lymphoma may lead
to increased apoptosis and suppression of cell proliferation, and
that miR-135a may directly suppresses janus kinase 2 which is a
cytoplasmic tyrosine kinase, followed by the suppression of the B
cell lymphoma extra-large (Bcl-xL) gene which is an anti-apoptotic
gene (49). These events lead to
an increase in apoptosis and suppression in proliferation (49). The present study revealed that the
expression of miR-135a increases significantly in the maternal
plasma samples, leading to EPL by increasing apoptosis and
inhibiting proliferation in the villous cells. In order for this
miRNA to be used as a biomarker to identify EPL cases, the effects
of the miR-135a on the genes that affect EPL require further
investigation and treatment procedures for the disease must be
specified according to the obtained results as a further step by
investigating the targets of this miRNA. In this context, the
present study may be used as the basis for biomarker research.
The present study identified that miR-122 expression
levels were increased in the maternal plasma samples obtained from
patients with EPL. The invasion of maternal decidua to placenta
trophoblasts is the most important stage in the normal
physiological course of a normal pregnancy, and apoptosis has a
critical role in normal placental development (50). MiR-122 may be involved in apoptosis
in cancer cells (51). A different
previous study demonstrated that miR-122 increased the expression
of oncogenes cyclin G1 (CCNG1) (52) and reduced the expression of a
disintegrin and ADAM metallopeptidase domain 17, EGFR and
FAK activation levels (53). Based on the data obtained from
miR-122-associated cancer studies, it is possible that in the
present study, the increased maternal plasma levels of this miRNA
may have adversely affected the development of the embryo by
reducing the expression of genes that are effective during
migration, adhesion, invasion and apoptosis of placenta trophoblast
cells and embryo cells in important stages as preimplantation and
implantation in embryo development. Lasabová et al
investigated the expression of miR-122, which was associated with
apoptosis, and demonstrated an increased expression in the case of
preeclampsia compared with normal placental samples (54). Therefore, considering the findings
of the present study it is possible that the increased miR-122
expression in maternal plasma samples may lead to an inadequate
level of apoptosis in the villous trophoblast cells that normally
form the placenta during the developmental stage, which may to lead
to EPL by affecting the development of placenta.
According to the present study, the expression level
of let-7c, a member of the let-7 family was reduced in the maternal
plasma samples from EPL patients compared with the control group.
Previous studies have revealed that the let-7 family is associated
with the cardiovascular system (55,56).
The important genes targeted by the let-7 family, include toll-like
receptor 4 (TLR4), oxidized low density lipoprotein receptor
1, Bcl-×l and argonaute-1 (AGO1) (56–58).
It was previously determined that these genes are associated with
the immune system, increased apoptosis, anti-apoptosis and miRNA
biogenesis respectively. The genes targeted by the let-7 family
have been identified as effective in embryonic stem cell and
cardiovascular differentiation (59). Zhang et al determined the
hypoxia-inducible factor 1α (HIF1α)/AGO1/VEGF, which are the
factors related with let-7, have a role in the hypoxia-induced
angiogenesis signaling pathway (51). HIF1α is a key transcription
factor, which upregulates the expression levels of let-7, and let-7
targets AGO1, leading to the translational suppression of
VEGF mRNA which has an important role in angiogenesis
process (56). The anti-apoptosis
protein Bcl-×1 which is affected by let-7c contributes to the
apoptosis of epithelial cells and its expression is suppressed by
let-7, leading to an increase in apoptosis. Reduced let-7c
expression may lead to an increased expression of anti-apoptotic
proteins such as Bcl-×1 and prevent apoptosis during development,
which in turn may lead to miscarriage by adversely affecting the
placental and embryonic development (60). According to the TargetScan
database; Kruppel-like factor 9 (KLF9); CAMP responsive
element binding protein 1 (CREB-1) and phosphatase and
tensin homolog (PTEN) are let-7c target genes. KLF9 is a
zinc-finger transcription factor that has a role in the regulation
of estrogen and progesterone by modulating the activity of the
progesterone receptor. Regulation of the KLF9 expression in
the endometrium epithelial cells is important for embryo
implantation in the menstrual cycle (57); therefore, it is believed that
reduced expression of let-7c may lead to an increased expression of
these gene and hormonal disturbances in the menstrual cycle. To the
best of our knowledge, no research has been performed in this
regard; however, the findings of the current study suggest that the
decrease in the expression of let-7c may be associated with the
role of KLF9 in endometrium implantation. Ventura et
al demonstrated that CREB-1 and PTEN were
targeted by miR-17 and miR-19b which are expressed in the placenta
during the early weeks of pregnancy, and that these miRNAs were
associated with EPL, as they act as regulatory factors during
placental development (4). A
previous study revealed that expression of the PTEN gene in
the villous trophoblast cells in the early stages of the placental
formation occurred; however, this expression reduces with time
(58). It was previously suggested
that the increased expression of this gene in villous trophoblast
cells may be associated with miscarriage (58). Several pathways have been
identified from the pathway analysis for the predicted target genes
of validated miRNAs in tissue samples preformed in the present
study. The most important pathways which were investigated
associated with miscarriage pathogenesis, included Wnt, IL-1,
regulation of toll-like receptor and MAPK signaling as well as
apoptosis, focal adhesion and JAK-STAT pathways (Table VI) (2,3,24,41–43).
The primary aim of the present study was to
demonstrate the significant difference in the expression of miRNAs
between normal subjects and those with EPL by analyzing villous
tissue and maternal blood samples. Subsequently, bioinformatics
analysis of dysregulated miRNAs was also performed to detect
targeted genes and their associated pathways. A total of 4 miRNAs
in the tissue samples and 3 miRNAs in the maternal plasma had
significant differences in expression according to the microarray
results. The validation experiments have confirmed 6 miRNAs to be
valid. It is possible to use the 3 miRNAs that have been detected
in the maternal plasma as biomarkers for non-invasive prenatal
diagnosis for this group of patients. It was previously determined
that the genes targeted by the miRNAs detected in the villous
tissue and maternal plasma samples contribute to in
differentiation, development, cell division, proliferation,
implantation and angiogenesis. Additionally, these processes are
effective in embryo development; therefore, they are believed to be
effective in the biological progression of the disease. Pathway
analysis of the genes targeted by the validated miRNAs revealed
that the associated pathways were frequently associated with
miscarriage according to previous studies. It is possible that the
detected miRNAs in the maternal plasma and tissue are involved in
the EPL process. In order to take these findings one step further,
future studies may validate the 3 miRNAs obtained from the maternal
plasma screening in more patient samples in order for them to be
used for early diagnosis. In addition, further studies should be
designed on the target genes of the miRNAs detected in increased
number of villous and maternal plasma samples from patients with
EPL, and the associations of the miRNAs with the target mRNAs
should be experimentally verified by molecular methods including
sequencing, gene and protein array, RT-qPCR and western blot
assays.
Acknowledgements
The present study was supported by Istanbul
University Scientific Research Projects Department (grant no. 29278
and 40699). SEM Laboratory Company which is Agilent Technologies
Distributor in Turkey supported the present study financially
during the publication process. The authors would like to thank
Professor Bulent Hayri ERMIS (Department of Obstetrics and
Gynecology, Istanbul Medical Faculty, Istanbul, Turkey) who passed
away on 24th of February 2016, for his considerable contribution to
the current study. Additionally, many thanks to Mr Ismail Dolekcap
for laboratory technical assistance and Dr Halil Ibrahim Kisakesen
for bioinformatics assistance.
Glossary
Abbreviations
Abbreviations:
|
FC
|
fold-change
|
|
miRNAs
|
microRNAs
|
|
MOBG
|
molecular biology and genetics
|
|
NIPD
|
non-invasive prenatal diagnosis
|
|
URSA
|
unexplained recurrent spontaneous
abortion
|
References
|
1
|
Wang Y, Lv Y, Wang L, Gong C, Sun J, Chen
X, Chen Y, Yang L, Zhang Y, Yang X, et al: MicroRNAome in decidua:
A new approach to assess the maintenance of pregnancy. Fertil
Steril. 103:980–989 e6. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhu LJ, Chen YP, Chen BJ and Mei XH:
Changes in reactive oxygen species, superoxide dismutase, and
hypoxia-inducible factor-1α levels in missed abortion. Int J Clin
Exp Med. 7:2179–2184. 2014.PubMed/NCBI
|
|
3
|
Tang L, Gao C, Gao L, Cui Y and Liu J:
Expression profile of micro-RNAs and functional annotation analysis
of their targets in human chorionic villi from early recurrent
miscarriage. Gene. 576:366–371. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ventura W, Koide K, Hori K, Yotsumoto J,
Sekizawa A, Saito H and Okai T: Placental expression of microRNA-17
and −19b is down-regulated in early pregnancy loss. Eur J Obstet
Gynecol Reprod Biol. 169:28–32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Larsen EC, Christiansen OB, Kolte AM and
Macklon N: New insights into mechanisms behind miscarriage. BMC
Med. 11:1542013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Michels TC and Tiu AY: Second trimester
pregnancy loss. Am Fam Physician. 76:1341–1346. 2007.PubMed/NCBI
|
|
7
|
Dong F, Zhang Y, Xia F, Yang Y, Xiong S,
Jin L and Zhang J: Genome-wide miRNA profiling of villus and
decidua of recurrent spontaneous abortion patients. Reproduction.
148:33–41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rull K, Nagirnaja L and Laan M: Genetics
of recurrent miscarriage: Challenges, current knowledge, future
directions. Front Genet. 3:342012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsochandaridis M, Nasca L, Toga C and
Levy-Mozziconacci A: Circulating MicroRNAs as clinical biomarkers
in the predictions of pregnancy complications. Biomed Res Int.
2015:2949542015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo L, Ye G, Nadeem L, Fu G, Yang BB,
Honarparvar E, Dunk C, Lye S and Peng C: MicroRNA-378a-5p promotes
trophoblast cell survival, migration and invasion by targeting
Nodal. J Cell Sci. 125:3124–3132. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li Y and Kowdley KV: MicroRNAs in common
human diseases. Genomics Proteomics Bioinformatics. 10:246–253.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Z and Rana TM: Therapeutic targeting of
microRNAs: Current status and future challenges. Nat Rev Drug
Discov. 13:622–638. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gu Y, Sun J, Groome LJ and Wang Y:
Differential miRNA expression profiles between the first and third
trimester human placentas. Am J Physiol Endocrinol Metab.
304:E836–E843. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu XM, Han T, Sargent IL, Yin GW and Yao
YQ: Differential expression profile of microRNAs in human placentas
from preeclamptic pregnancies vs normal pregnancies. Am J Obstet
Gynecol. 200:661.e1–7. 2009. View Article : Google Scholar
|
|
16
|
Fu G, Brkić J, Hayder H and Peng C:
MicroRNAs in human placental development and pregnancy
complications. Int J Mol Sci. 14:5519–5544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lo YM, Corbetta N, Chamberlain PF, Rai V,
Sargent IL, Redman CW and Wainscoat JS: Presence of fetal DNA in
maternal plasma and serum. Lancet. 350:485–487. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao Z, Moley KH and Gronowski AM:
Diagnostic potential for miRNAs as biomarkers for
pregnancy-specific diseases. Clin Biochem. 46:953–960. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lanza G, Ferracin M, Gafà R, Veronese A,
Spizzo R, Pichiorri F, Liu CG, Calin GA, Croce CM and Negrini M:
mRNA/microRNA gene expression profile in microsatellite unstable
colorectal cancer. Mol Cancer. 6:542007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Siristatidis C, Vogiatzi P, Brachnis N,
Liassidou A, Iliodromiti Z, Bettocchi S and Chrelias C: Review:
MicroRNAs in assisted reproduction and their potential role in IVF
failure. In Vivo. 29:169–175. 2015.PubMed/NCBI
|
|
22
|
Sun YM, Lin KY and Chen YQ: Diverse
functions of miR-125 family in different cell contexts. J Hematol
Oncol. 6:62013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu Y, Liu CM, Qi L, He TZ, Shi-Guo L, Hao
CJ, Cui Y, Zhang N, Xia HF and Ma X: Two common SNPs in
pri-miR-125a alter the mature miRNA expression and associate with
recurrent pregnancy loss in a Han-Chinese population. RNA Biol.
85:861–872. 2011. View Article : Google Scholar
|
|
24
|
Li D and Li J: Association of
miR-34a-3p/5p, miR-141-3p/5p, and miR-24 in decidual natural killer
cells with unexplained recurrent spontaneous abortion. Med Sci
Monit. 22:922–929. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salleh N and Giribabu N: Leukemia
inhibitory factor: Roles in embryo implantation and in nonhormonal
contraception. ScientificWorldJournal. 2014:2015142014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ninio-Many L, Grossman H, Shomron N,
Chuderland D and Shalgi R: microRNA-125a-3p reduces cell
proliferation and migration by targeting Fyn. J Cell Sci.
126:2867–2876. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang H, Zhu X, Li N, Li D, Sha Z, Zheng X
and Wang H: miR-125a-3p targets MTA1 to suppress NSCLC cell
proliferation, migration, and invasion. Acta Biochim Biophys Sin
(Shanghai). 47:496–503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo X, Wu Y and Hartley RS: MicroRNA-125a
represses cell growth by targeting HuR in breast cancer. RNA Biol.
6:575–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tiwari A, Shivananda S, Gopinath KS and
Kumar A: MicroRNA-125a reduces proliferation and invasion of oral
squamous cell carcinoma cells by targeting estrogen-related
receptor α: Implications for cancer therapeutics. J Biol Chem.
289:32276–32290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Choi HK, Choi BC, Lee SH, Kim JW, Cha KY
and Baek KH: Expression of angiogenesis- and apoptosis-related
genes in chorionic villi derived from recurrent pregnancy loss
patients. Mol Reprod Dev. 66:24–31. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu H, Li H, Zhang L, Luan H, Huang T, Wang
L, Fan Y, Zhang Y, Liu X, Wang W and Yuan Z: Diagnostic role of
microRNA expression profile in the serum of pregnant women with
fetuses with neural tube defects. J Neurochem. 122:641–649. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nardelli C, Iaffaldano L, Ferrigno M,
Labruna G, Maruotti GM, Quaglia F, Capobianco V, Di Noto R, Del
Vecchio L, Martinelli P, et al: Characterization and predicted role
of the microRNA expression profile in amnion from obese pregnant
women. Int J Obes (Lond). 38:466–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fisher JN, Terao M, Fratelli M, Kurosaki
M, Paroni G, Zanetti A, Gianni M, Bolis M, Lupi M, Tsykin A, et al:
MicroRNA networks regulated by all-trans retinoic acid and
Lapatinib control the growth, survival and motility of breast
cancer cells. Oncotarget. 6:13176–13200. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sand M, Skrygan M, Sand D, Georgas D,
Gambichler T, Hahn SA, Altmeyer P and Bechara FG: Comparative
microarray analysis of microRNA expression profiles in primary
cutaneous malignant melanoma, cutaneous malignant melanoma
metastases, and benign melanocytic nevi. Cell Tissue Res.
351:85–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peng J, Feng Y, Rinaldi G, Levine P,
Easley S, Martinez E, Hashmi S, Sadeghi N, Brindley PJ, Mulvenna
JP, et al: Profiling miRNAs in nasopharyngeal carcinoma FFPE tissue
by microarray and next generation sequencing. Genom Data.
2:285–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chiyomaru T, Yamamura S, Fukuhara S,
Hidaka H, Majid S, Saini S, Arora S, Deng G, Shahryari V, Chang I,
et al: Genistein up-regulates tumor suppressor microRNA-574-3p in
prostate cancer. PLoS One. 8:e589292013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gitenay D, Lallet-Daher H and Bernard D:
Caspase-2 regulates oncogene-induced senescence. Oncotarget.
5:5845–5847. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vanbekbergen N, Hendrickx M and Leyns L:
Growth differentiation Factor 11 is an encephalic regionalizing
factor in neural differentiated mouse embryonic stem cells. BMC Res
Notes. 7:7662014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang L, Wang L and Zheng Y: Gene targeting
of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in
filopodia induction, directed migration, and proliferation in
primary mouse embryonic fibroblasts. Mol Biol Cell. 17:4675–4685.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Large MJ, Wetendorf M, Lanz RB, Hartig SM,
Creighton CJ, Mancini MA, Kovanci E, Lee KF, Threadgill DW, Lydon
JP, et al: The epidermal growth factor receptor critically
regulates endometrial function during early pregnancy. PLoS Genet.
10:e10044512014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bao SH, Shuai W, Tong J, Wang L, Chen P
and Duan T: Increased Dickkopf-1 expression in patients with
unexplained recurrent spontaneous miscarriage. Clin Exp Immunol.
172:437–443. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin YX, Cui XS, Han YJ and Kim NH: Leptin
accelerates pronuclear formation following intracytoplasmic sperm
injection of porcine oocytes: Possible role for MAP kinase
inactivation. Anim Reprod Sci. 115:137–148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xiao B, Xue X, Hu F, Sun R, Chen Q, Yang M
and Zhang W: Expression and regulatory mechanism of microRNA-155 in
the villi of patients with unexplained recurrent spontaneous
abortion patients. Zhonghua Fu Chan Ke Za Zhi. 49:130–134. 2014.(In
Chinese). PubMed/NCBI
|
|
44
|
Qin W, Tang Y, Yang N, Wei X and Wu J:
Potential role of circulating microRNAs as a biomarker for
unexplained recurrent spontaneous abortion. Fertil Steril.
105:1247–1254 e3. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luque A, Farwati A, Crovetto F, Crispi F,
Figueras F, Gratacós E and Aran JM: Usefulness of circulating
microRNAs for the prediction of early preeclampsia at
first-trimester of pregnancy. Sci Rep. 4:48822014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Al-Shorafa H and Sharif FA: MicroRNA in a
case of unexplained recurrent pregnancy loss. J Clin Case Rep.
02:2382012. View Article : Google Scholar
|
|
47
|
Petracco R, Grechukhina O, Popkhadze S,
Massasa E, Zhou Y and Taylor HS: MicroRNA 135 regulates HOXA10
expression in endometriosis. J Clin Endocrinol Metab.
96:E1925–E1933. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shi H, Ji Y, Zhang D, Liu Y and Fang P:
MiR-135a inhibits migration and invasion and regulates EMT-related
marker genes by targeting KLF8 in lung cancer cells. Biochem
Biophys Res Commun. 465:125–130. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Navarro A, Diaz T, Martinez A, Gaya A,
Pons A, Gel B, Codony C, Ferrer G, Martinez C, Montserrat E and
Monzo M: Regulation of JAK2 by miR-135a: Prognostic impact in
classic Hodgkin lymphoma. Blood. 114:2945–2951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ishihara N, Matsuo H, Murakoshi H,
Laoag-Fernandez JB, Samoto T and Maruo T: Increased apoptosis in
the syncytiotrophoblast in human term placentas complicated by
either preeclampsia or intrauterine growth retardation. Am J Obstet
Gynecol. 185:158–166. 2002. View Article : Google Scholar
|
|
51
|
Zhang ZW, Li H, Chen SS, Li Y, Cui ZY and
Ma J: MicroRNA-122 regulates caspase-8 and promotes the apoptosis
of mouse cardiomyocytes. Braz J Med Biol Res. 50:e57602017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ma L, Liu J, Shen J, Liu L, Wu J, Li W,
Luo J, Chen Q and Qian C: Expression of miR-122 mediated by
adenoviral vector induces apoptosis and cell cycle arrest of cancer
cells. Cancer Biol Ther. 9:554–561. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW,
Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al: MicroRNA-122, a
tumor suppressor microRNA that regulates intrahepatic metastasis of
hepatocellular carcinoma. Hepatology. 49:1571–1582. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lasabová Z, Vazan M, Zibolenova J and
Svecova I: Overexpression of miR-21 and miR-122 in preeclamptic
placentas. Neuro Endocrinol Lett. 36:695–699. 2015.PubMed/NCBI
|
|
55
|
Ding Z, Wang X, Schnackenberg L, Khaidakov
M, Liu S, Singla S, Dai Y and Mehta JL: Regulation of autophagy and
apoptosis in response to ox-LDL in vascular smooth muscle cells,
and the modulatory effects of the microRNA hsa-let-7g. Int J
Cardiol. 168:1378–1385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen Z, Lai TC, Jan YH, Lin FM, Wang WC,
Xiao H, Wang YT, Sun W, Cui X, Li YS, et al: Hypoxia-responsive
miRNAs target argonaute 1 to promote angiogenesis. J Clin Invest.
123:1057–1067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Du H, Sarno J and Taylor HS: HOXA10
inhibits Kruppel-like factor 9 expression in the human endometrial
epithelium. Biol Reprod. 83:205–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tokyol C, Aktepe F, Husniye Dilek F and
Yilmazer M: Comparison of placental PTEN and beta1 integrin
expression in early spontaneous abortion, early and late normal
pregnancy. Ups J Med Sci. 113:235–242. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y
and Zhou HH: Let-7 in cardiovascular diseases, heart development
and cardiovascular differentiation from stem cells. Int J Mol Sci.
14:23086–23102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Qin B, Xiao B, Liang D, Li Y, Jiang T and
Yang H: MicroRNA let-7c inhibits Bcl-×l expression and regulates
ox-LDL-induced endothelial apoptosis. BMB Rep. 45:464–469. 2012.
View Article : Google Scholar : PubMed/NCBI
|