Introduction
Cleft lip with or without cleft palate (CL/P) is one
of the most prevalent developmental deformities world-wide, with a
prevalence ranging from approximately 1/300 to 1/2,500 of live
births (1,2). CL/P results from a complex etiology
due to multiple genetic and environmental risk factors (3). Environmental factors, such as
smoking, alcohol, caffeine, infections, corticosteroids, retinoic
acid, dioxin, as well as other occupational pollutants, can
seriously affect development of the fetus during pregnancy to
eventually result in conditions such as CL/P (4).
In specific,
2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), a highly toxic
halogenated environmental contaminant that produces adverse
biological effects, including developmental toxicity and
teratogenesis (5), has been shown
to result in the formation of a cleft palate within mice (6). Recent findings have established that
epigenetic modifications are involved in murine palatogenesis
induced by TCDD, with DNA methylation being considered as playing a
key role in cleft palate formation (7). The significance of DNA methylation
has been revealed from findings demonstrating that global DNA
methylation status was substantially enhanced in fetal mice
following maternal exposure to TCDD, and this factor was thought to
be a causative factor in palate malformation (8). However, the underlying molecular
mechanisms of DNA methylation caused by TCDD in mice require
further elucidation.
DNA methylation is among the best studied epigenetic
modifications and is closely related to disease and homeostatic
imbalances in mammals, through its capacity of participating in
proper embryonic development, gene expression regulation and even
tumorigenesis (9, 10). The dynamic patterns of DNA methylation are
catalyzed by DNA methyltransferases (DNMTs) and then read and
interpreted by specific proteins, called methyl-CpG binding domain
proteins (MBDs) (11). As it has
been reported that congenital cleft palate patients usually show
distinct methylation profiles (12), we hypothesized that DNMTs and MBDs,
which greatly affect DNA methylation patterns, would participate in
cleft palate formation induced by TCDD. It is well established that
DNA methyltransferases are crucial for establishing DNA methylation
patterns in mammals, and are thus responsible for maintenance of
methylation (DNMT1) and de novo methylation (DNMT3a, 3b) (13, 14).
The methyl-CpG binding domain proteins, especially MeCP2, MBD2,
MBD3, are critical players in determining the transcriptional state
of epigenosomes, and therefore play an important role as
facilitators of epigenetic-based repression (15). Results from previous studies have
demonstrated that cigarette smoke and alcohol cause changes in the
expression of DNA methyltranferases and methyl-CpG binding domain
proteins in murine embryonic cells (16, 17). In this study, we
utilized the mouse cleft palate model as accomplished by maternal
exposure to TCDD, to elucidate the effects of epigenetic
regulation, including DNMT1, DNMT3a, DNMT3b, MeCP2, MBD2 and MBD3
expression in vivo, and investigated their relationship and
participation in TCDD-induced cleft palate.
Materials and methods
Animals
The present study was approved by the Transformation
Center of Harbin Medical University (Harbin, China). C57BL/6J mice
(8–10 weeks of age) consisting of 30 female experimental mice and
15 male mice used to impregnate the females were purchased from the
Institute of Laboratory Animals of the Chinese Academy of Medical
Sciences. The mice were housed under controlled laboratory
conditions of 22–24°C, 55±5% humidity, under a 12/12 h light/dark
cycle, with standard laboratory chow and water ad libitum.
After being acclimated to the laboratory conditions, two female
mice were paired with one male overnight. Presence of vaginal plugs
on the following morning was interpreted as a successful mating and
was designated as embryonic day 0.5 (E0.5).
Drug administrations
At E10.5, pregnant mice were randomly divided into
two groups: i) TCDD group-consisted of gavage with a single dose of
64 µg/kg TCDD; and ii) control group-mice that received an equal
volume of corn oil. Following treatment, pregnant female mice were
caged and fed separately. At E13.5, E14.5, E15.5 or E17.5, mice
were euthanized using CO2. A total of three pregnant
mice were euthanized at each time-point, and the total number of
fetal mice present was recorded. Fetal palate tissue was quickly
isolated from each embryo and stored at −80°C.
Histological staining of sections
For histological analysis, the heads of the fetal
mice were fixed in 4% paraformaldehyde (PFA), dehydrated in a
graded series of ethanol, embedded in paraffin and then sectioned
at 10 µm in the coronal plane. To assess the general morphology,
deparaffinized sections were subjected to hematoxylin (Solarbio,
Beijing, China) and eosin (Sinopharm Chemical Reagent Co., Ltd.,
Beijing, China) staining using established procedures.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from palatal shelves using a
RNeasy Mini kit (BioTeke, Beijing, China) according to the
manufacturer's instructions. The Super M-MLV RT kit (BioTeke) was
used to synthesize first-standed cDNA. Each q-PCR reaction system
was performed in a total volume of 20 µl: cDNA template 1 µl,
forward and reverse primers (10 µm) each 0.5 µl, SYBR-Green
Mastermix 10 µl and ddH2O 8 µl. The amplification
protocol consisted of 40 cycles of denaturation for 10 sec at 95°C,
annealing for 20 sec at 60°C, and elongation for 30 sec at
72°C.
A SYBR-Green fluorescence quantification system
(Bioneer, Daejeon, Korea) was utilized to perform the Real-time PCR
analysis. β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) was used as an internal control and gene-specific mRNA
expression was normalized to β-actin expression by using the
2−∆∆Ct method. Primer sequences are shown in Table I.
 | Table I.Mouse primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Mouse primers used for reverse
transcription-quantitative polymerase chain reaction.
| Primers | Forward (5′-3′) | Reverse (5′-3′) |
|---|
| Dnmt1 |
TGGAGCCCAGCAAAGAGTA |
TAATGGTAGAAGGAGGAACAGTG |
| Dnmt3a |
AACGGAAACGGGATGAGTG |
TCGTCGGCTGCTTTGGTAG |
| Dnmt3b |
CGAAGACGCACAACCAATG |
ACAGAGCCCACCCTCAAAG |
| Mecp2 |
AAGCCTCTGAGACCCTATCC |
GCAGTGGAGACAACCCTT |
| Mbd2 |
CAGCTCCATTGCCTGCATAG |
GGAATCAGACGGAAAGAAAA |
| Mbd3 |
AGATGAATAAGAGTCGCCAG |
AGGCACTCAATCCACTTAGC |
| β-actin |
CTGTGCCCATCTACGAGGGCTAT |
TTTGATGTCACGCACGATTTCC |
Western blotting
Proteins extracts of the TCDD and control groups
from E13.5 to E15.5 were performed using standard protocols and
antibodies recognizing DNMT1, DNMT3a, DNMT3b, MeCP2, MBD2 and MBD3
(all from Bioss, Beijing, China). All data were normalized to the
expression of β-actin (Santa Cruz Biotechnology, Inc.). The
intensity of the bands was analyzed by densitometry.
Statistical analysis
Data from E13.5 to E15.5 in the two groups were
presented as the mean ± SD and analyzed using SPSS 17.0 (SPSS,
Inc., Chicago, IL, USA). Independent t-tests were used to establish
statistically significant differences between two groups. A
P<0.05 was required for results to be considered statistically
significant. All experiments were performed at least in
triplicate.
Results
TCDD treatments at E10.5 induced cleft
palate in mice
In fetuses excised from pregnant females who
received TCDD at E10.5, the number of fetuses with CP only, without
other obvious orofacial defects were: 14/14 at E13.5, 16/16 at
E14.5, 13/14 at E15.5, and 13/13 at E17.5. In contrast, no cleft
palates were observed in fetal mice within the control group.
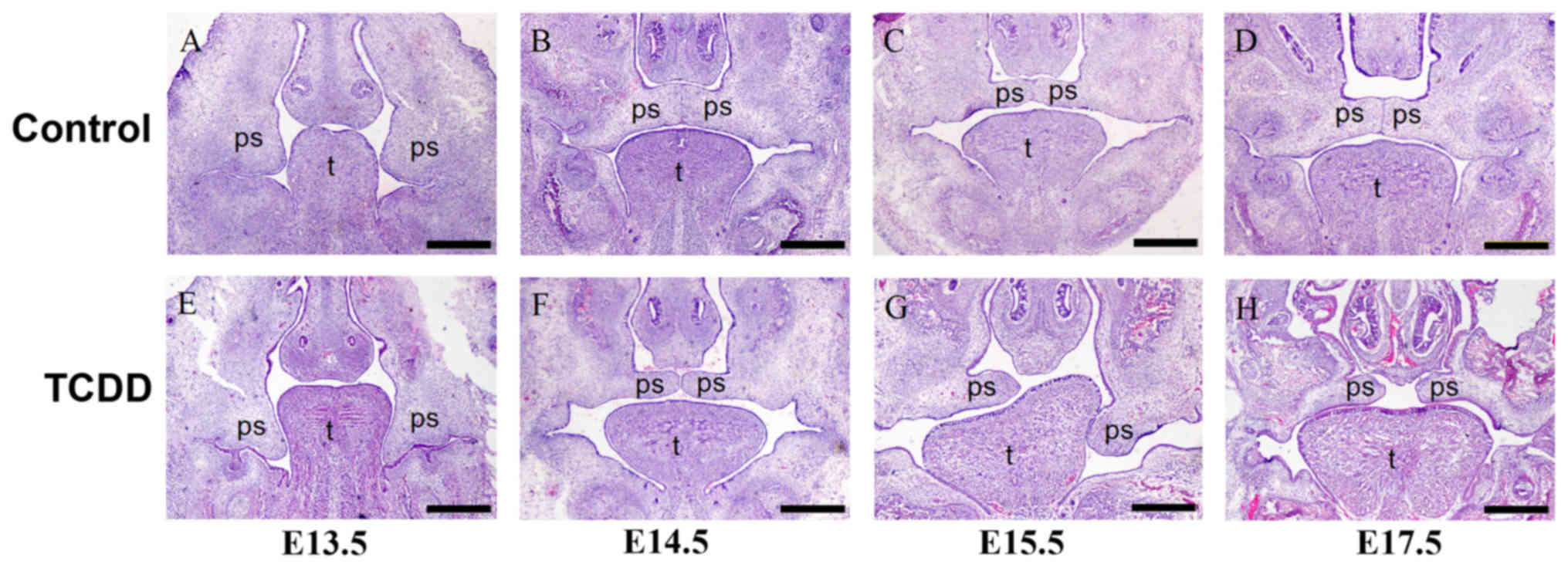
The morphological features of palatal development
from E13.5 to E17.5 in fetuses from pregnant females treated with
TCDD were examined under a stereomicroscope (Fig. 1). H&E staining revealed that at
E13.5, bilateral palate shelves of TCDD mice were observed to
project downward on each side of the tongue and failed to elevate,
which was similar to that observed in controls. At E14.5, palates
of the controls were in the fusion stage, while TCDD-treated
embryos showed bilateral palate shelves that were not fused. At
E15.5 and E17.5, fetuses in the control group demonstrated a
complete palate; while the absence of a successful adherence in
palate shelves within the TCDD group resulted in a cleft
palate.
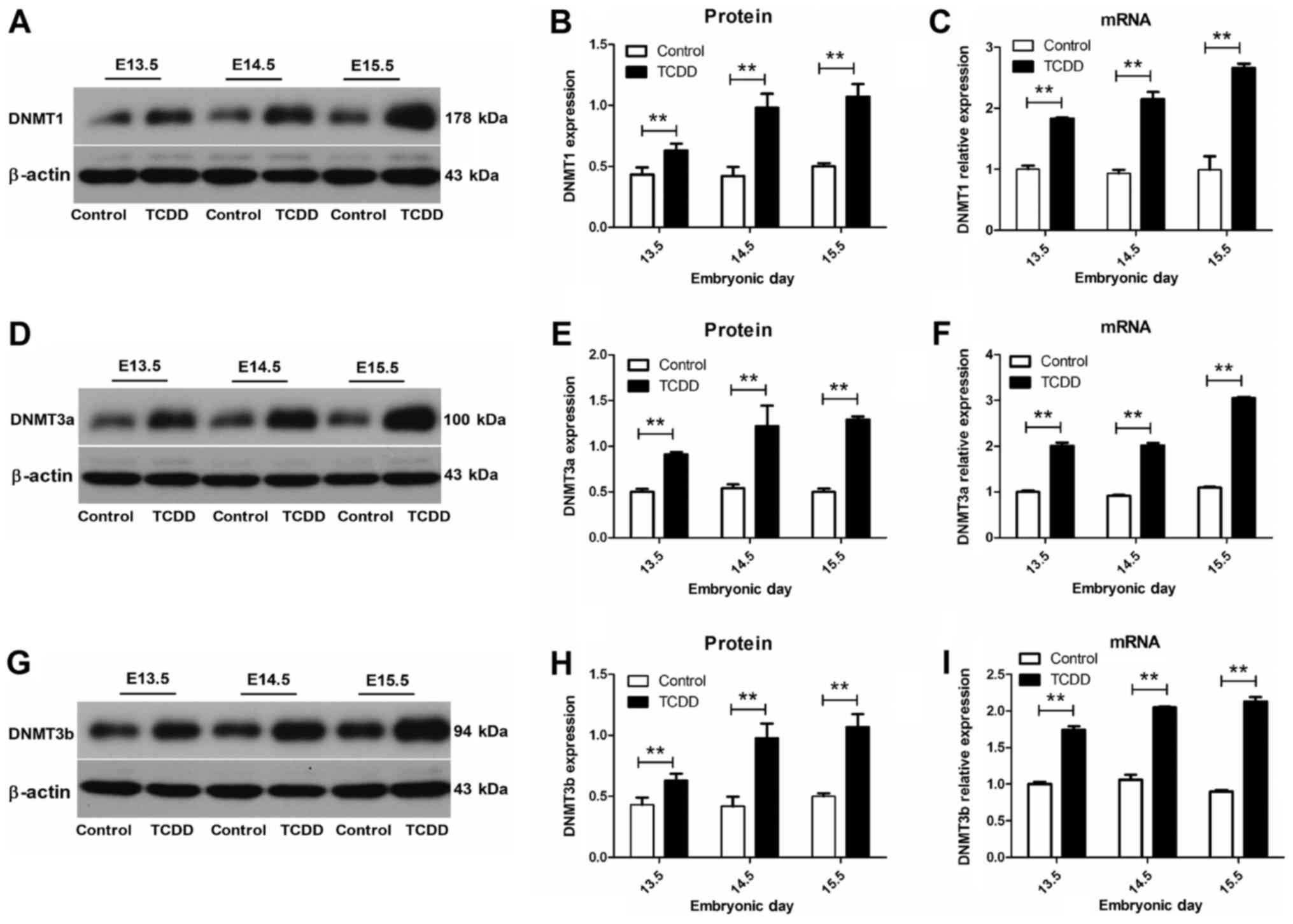
TCDD activated the expression of DNMTs
in palatal shelves
To identify which DNA methyltransferase was
regulated, we performed real-time quantitative PCR and western blot
analysis for DNMT1, DNMT3a and DNMT3b. β-actin served as an
internal reference and relative transcript and protein levels are
shown in Fig. 2. Our results
revealed that mRNA levels of DNMT1, DNMT3a and DNMT3b were
significantly increased in the TCDD, as compared with that of the
control group at E13.5 to E15.5 (P<0.01). Within palatal
shelves, a strong expression in DNMTs was obtained in the TCDD
group from E13.5 to E15.5, while a relatively weak expression was
present in the control group.
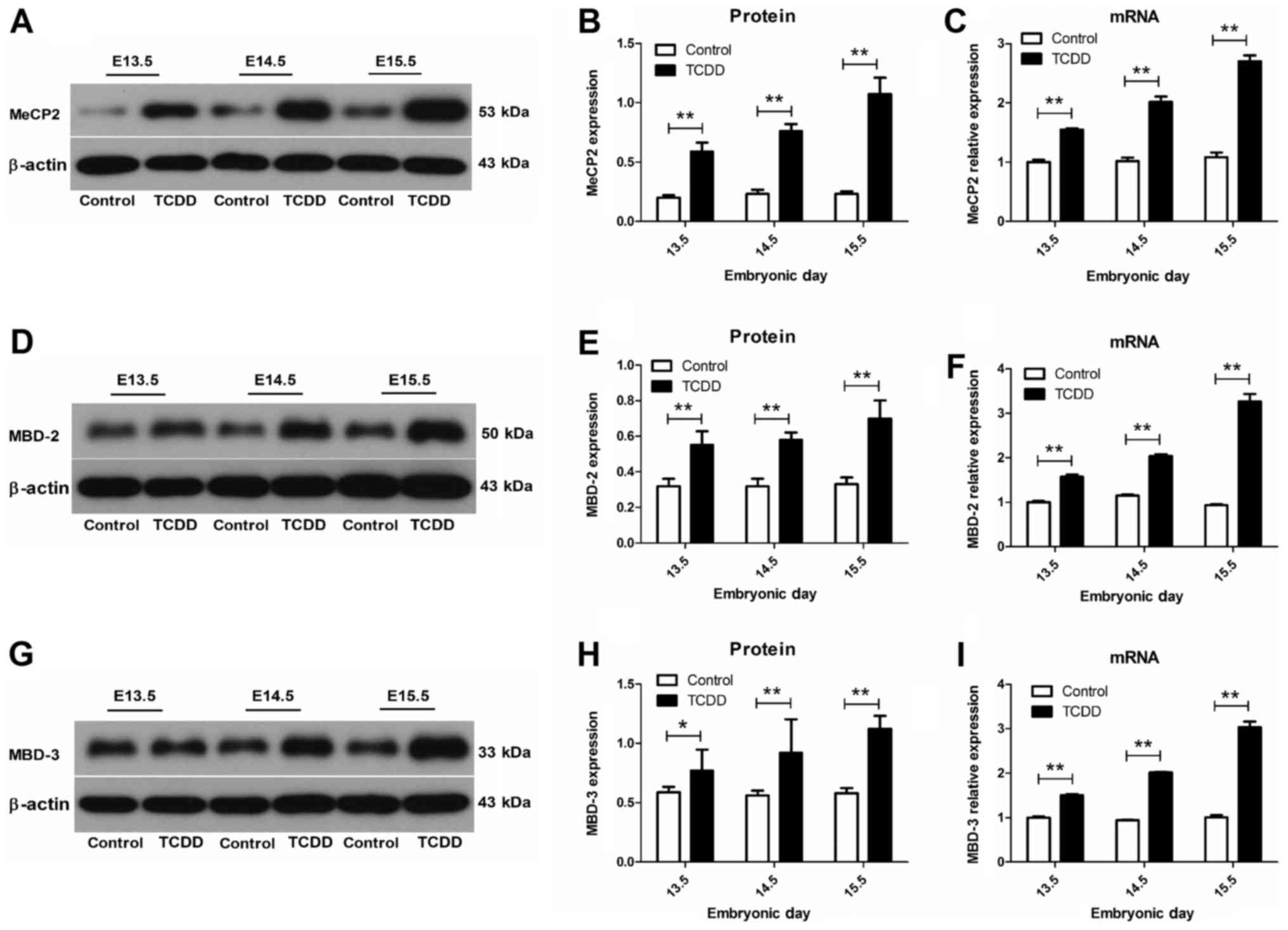
MeCP2, MBD2 and MBD3 are upregulated
by TCDD in mice palates
The mRNA expression of methyl-CpG binding domain
proteins was detected using real-time quantitative PCR. The mRNA
levels of MeCP2, MBD2 and MBD3 in the TCDD group at E13.5 to E15.5
were significantly increased (P<0.05), compared with those of
controls (Fig. 3). Protein
expressions of MeCP2, MBD2 and MBD3 were identified using western
blotting. Results from protein imprinting showed that protein
expression levels of the TCDD group were significantly greater than
in controls from E13.5 to E15.5 (P<0.05), which was in
accordance with the trend observed for mRNA.
Temporal accumulations of DNMTs and
MBDs in mouse embryonic palates
The intensity of the bands resulting from western
blot analysis was analyzed by densitometry. At E13.5, positive
staining was sparsely present in palatal shelves. At E14.5, DNMTs
and MBDs were strongly expressed in palatal shelves, with the
strongest expression of DNMTs and MBDs being detected at E15.5
within the TCDD group.
Discussion
CL/P is a common birth defect notable for its
significant morbidity and complex etiology. The complex etiology
reflects the interaction of multiple genetic, epigenetic and
environmental factors. One potential basis for this interaction
involves that of environmental factors which can then affect gene
expression via alterations in DNA methylation epigenetic
modifications. DNA methylation is well known as an epigenetic
marker involved in numerous biological processes which can induce
gene expression changes without altering DNA sequences (18). Although Kuriyama et al
(19) have reported that DNA
methylation may be crucial for murine palatogenesis, the precise
molecular underpinnings directing epigenetic alterations that
result in cleft palate remain obscure.
In eukaryotic cells, the establishment and
maintenance of methylation patterns depends on the methylation
modification system, which involves two important proteins, DNMTs
and MBDs. Associated with this process are three enzymatically
active DNA methaltransferases (DNMT1, DNMT3a, DNMT3b). DNMT1 is a
maintenance methyltransferase that ensures inheritance of
methylation patterns during cell division by preferentially
methylating hemimethylated CG dimucleotides. DNMT3a and DNMT3b are
responsible for de novo methylation, establishing DNA
methylation patterns in germs cells. MeCP2, MBD2 and MBD3 are
members of the MBD family of proteins that are associated with
transcriptional activation or repression, regulation of chromatin
structure, pluripotency, development and differentiation (20). Mukhopadhyay et al (16) demonstrated that upon exposure to 80
µg/ml cigarette smoke extract for 24 h, first branchial
arch-derived (1-BA) cells, which contribute to cleft palate
formation, exhibited a decline in global DNA methylation and
triggered a proteasomal-mediated degradation of DNMTs (DNMT1,
DNMT3a), MeCP2 and MBD3. However, as this study was performed using
an in vitro preparation, the question of whether such
changes in gene expression of DNMTs and MBDs would be present in
vivo has yet to be determined.
TCDD is a teratogenic reagent shown to be effective
in inducing cleft palate in mice and therefore has been widely used
as a model for the study of palate formation. Gestational exposure
to TCDD can be considered as an environmental factor which
influences the molecular development of palate tissue. Accordingly,
we utilized this cleft palate model and directed our attention at
examining potential in vivo changes that might be present in
the expressions of DNMTs and MBDs in this model.
In this study, exposure of pregnant C57BL/6J mice to
TCDD was effective in producing cleft palate in the fetuses. With
use of a TCDD dose of 64 µg/kg as administered at E10.5, we
obtained a 98.24% incidence of cleft palate. No other apparent
external anomalies or complications were observed in the
craniofacial tissue of these TCDD exposed litters. The
morphological outcomes in terms of palatal development at E13.5,
E14.5, E15.5 fetuses were evaluated with use of a stereomicroscope.
H&E staining of embryo head sections at E14.5 and E15.5 showed
a failure of palatal fusion in the TCDD group resulting in a cleft
defect. By contrast, palatal shelve fusion was complete in the
control group.
As it has been reported that abnormal methylation
status may be a probable reason for cleft palate (12,19,21),
we next examined DNMT gene expression patterns in these mice. Our
findings reveal that developmental exposure to TCDD significantly
affects DNMT gene expression patterns. Results obtained from q-PCR
analysis showed that Dnmt1, Dnmt3a and Dnmt3b were overexpressed at
E13.5 in the TCDD group, and increased further at later stages of
development. This trend was consistent with results obtained from
western blotting. Overall, these changes are in accordance with
recent studies by Wang et al (8). One slight difference between our
current results and that of Wang et al was the observation
of a decline in the expression of Dnmt3b in their group, a
difference which may be attributable to their use of a lower dose
of 28 µg/kg TCDD as administered via oral gavage in the mice.
However, the increased expression of DNMTs was clearly suggestive
of stimulation by TCDD in maintenance (DNMT1) and de novo
(DNMT3a and DNMT3b) DNMT activities. The fact that DNA
methyltransferase activity is considered to be increased by folic
acid (22), and folic
acid is regarded as the most effective treatment for fetal
congenital malformations, including neural tube defects and CL/P
(22,23), the significance of these increased
expressions of DNMTs can be appreciated. Interestingly, the lack of
folic acid intake in the peri-conceptional period has been
identified as one of the high risk factors for CL/P (24). Accordingly, we hypothesized that
the aberrant increase of DNMTs may consume a large amount of folic
acid and result in a folate deficiency within the body.
Coinciding with the changes in the expression of
DNMT genes, we also observed a significant increase in MBDs during
palatal development. Specifically, we observed that TCDD produced a
significant over expression in both mRNA and proteins levels of the
three methyl-CpG binding domain proteins, MeCP2, MBD2 and MBD3, in
the palatal shelves of mice fetuses. These findings differ from
those of Aluru et al (14)
who reported a degradation in the expressions of MeCP2, MBD2 and
MBD3 in zebrafish, the difference is likely due to the different
species of mice and zebrafish. With regard to MBDs, Zhang et
al (25) reported that MeCP2
was indispensable for normal neurological functioning and over
expression of MeCP2 in mice resulted in neurobehavioural disorders,
dendritic abnormalities and synaptic defects. Hendrich et al
(26) showed that MBD2
and MBD3 played selective roles with regard to their function as
transcriptional repressors. While Mbd3−/− mice died
during early embryogenesis, Mbd2−/− mice were viable and
fertile, suggesting that MBD2 and MBD3 exert distinct but
interacting effects in mouse development. The results presented
here are in accord with the changes in DNMTs expression levels
observed, and it seems likely that the combined actions of DNMTs
and MBDs may ultimately contribute to the aberrant methylation
status of mice as induced by TCDD. The differential recruitment of
DNA methyltransferases and methyl CpG-binding domain proteins are
considered as factors which initiate the repression and silencing
of Oct4 via hypomethylation of the Oct4 promoter (27). As based upon a review of a large
genome-wide association study of datasets, Oct4 was deemed to
represent a loci harbor gene associated with CL/P (28).
In conclusion, the data presented here, which show
increased recruitment of DNMTs and MBDs, and TCDD effects on DNMTs
and MBDs expression as demonstrated in vivo in this mouse
model of cleft palate, suggest that both the establishment and
maintenance of methylation patterns may play an important role in
murine palatogenesis.
Acknowledgements
This study was revised for submission in English by
the ED-IT Editorial Service.
Glossary
Abbreviations
Abbreviations:
|
CL/P
|
cleft lip with or without cleft
palate
|
|
TCDD
|
2,3,7,8-tetrachlorodibenzo-p-dioxin
|
|
DNMT
|
DNA methyltransferase
|
|
MBD
|
methyl-CpG binding domain protein
|
References
|
1
|
Murray JC: Gene/environment causes of
cleft lip and/or palate. Clin Genet. 61:248–256. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stanier P and Moore GE: Genetics of cleft
lip and palate: Syndromic genes contribute to the incidence of
non-syndromic clefts. Hum Mol Genet. 1:R73–R81. 2004. View Article : Google Scholar
|
|
3
|
Adeyemo WL and Butali A: Genetics and
genomics etiology of nonsyndromic orofacial clefts. Mol Genet
Genomic Med. 5:3–7. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Honein MA, Rasmussen SA, Reefhuis J,
Romitti PA, Lammer EJ, Sun L and Correa A: Maternal smoking and
environmental tobacco smoke exposure and the risk of orofacial
clefts. Epidemiology. 18:226–233. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Abbott BD: Review of the interaction
between TCDD and glucocorticoids in embryonic palate. Toxicology.
105:365–373. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamada T, Hirata A, Sasabe E, Yoshimura T,
Ohno S, Kitamura N and Yamamoto T: TCDD disrupts posterior
palatogenesis and causes cleft palate. J Craniomaxillofac Surg.
42:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yuan X, Qiu L, Pu Y, Liu C, Zhang X, Wang
C, Pu W and Fu Y: Histone acetylation is involved in TCDD-induced
cleft palate formation in fetal mice. Mol Med Rep. 14:1139–1145.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang C, Yuan XG, Liu CP, Zhai SN, Zhang DW
and Fu YX: Preliminary research on DNA methylation changes during
murine palatogenesis induced by TCDD. J Craniomaxillofac Surg.
45:678–684. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jones PA and Laird PW: Cancer epigenetics
comes of age. Nat Genet. 21:163–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith ZD and Meissner A: DNA methylation:
Roles in mammalian development. Nat Rev Genet. 14:204–220. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gigek CO, Chen ES and Smith MA:
Methyl-CpG-binding protein (MBD) family: Epigenomic read-outs
functions and roles in tumorigenesis and psychiatric diseases. J
Cell Biochem. 117:29–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sharp GC, Ho K, Davies A, Stergiakouli E,
Humphries K, McArdle W, Sandy J, Davey Smith G, Lewis SJ and Relton
CL: Distinct DNA methylation profiles in subtypes of orofacial
cleft. Clin Epigenetics. 9:632017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jurkowska RZ, Jurkowski TP and Jeltsch A:
Structure and function of mammalian DNA methyltransferases.
Chembiochem. 12:206–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aluru N, Kuo E, Helfrich LW, Karchner SI,
Linney EA, Pais JE and Franks DG: Developmental exposure to
2,3,7,8-tetrachlorodibenzo-p-dioxin alters DNA methyltransferase
(dnmt) expression in zebrafish (Danio rerio). Toxicol Appl
Pharmacol. 284:142–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Du Q, Luu PL, Stirzaker C and Clark SJ:
Methyl-CpG-binding domain proteins: Readers of the epigenome.
Epigenomics. 7:1051–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mukhopadhyay P, Rezzoug F, Kaikaus J,
Greene RM and Pisano MM: Alcohol modulates expression of DNA
methyltranferases and methyl CpG-/CpG domain-binding proteins in
murine embryonic fibroblasts. Reprod Toxicol. 37:40–48. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mukhopadhyay P, Greene RM and Pisano MM:
Cigarette smoke induces proteasomal-mediated degradation of DNA
methyltransferases and methyl CpG-/CpG domain-binding proteins in
embryonic orofacial cells. Reprod Toxicol. 58:140–148. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seelan RS, Mukhopadhyay P, Warner DR, Webb
CL, Pisano M and Greene RM: Epigenetic regulation of Sox4 during
palate development. Epigenomics. 5:131–146. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kuriyama M, Udagawa A, Yoshimoto S,
Ichinose M, Sato K, Yamazaki K, Matsuno Y, Shiota K and Mori C: DNA
methylation changes during cleft palate formation induced by
retinoic acid in mice. Cleft Palate Craniofac J. 45:545–551. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Menafra R and Stunnenberg HG: MBD2 and
MBD3: Elusive functions and mechanisms. Front Genet. 5:4282014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alvizi L, Ke X, Brito LA, Seselgyte R,
Moore GE, Stanier P and Passos-Bueno MR: Differential methylation
is associated with non-syndromic cleft lip and palate and
contributes to penetrance effects. Sci Rep. 7:24412017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Luo S, Zhang X, Yu M, Yan H, Liu H, Wilson
JX and Huang G: Folic acid acts through DNA methyltransferases to
induce the differentiation of neural stem cells into neurons. Cell
Biochem Biophys. 66:559–566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wehby GL and Murray JC: Folic acid and
orofacial clefts: A review of the evidence. Oral Dis. 16:11–19.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Figueiredo RF, Figueiredo N, Feguri A,
Bieski I, Mello R, Espinosa M and Damazo AS: The role of the folic
acid to the prevention of orofacial cleft: An epidemiological
study. Oral Dis. 21:240–247. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang D, Yu B, Liu J, Jiang W, Xie T,
Zhang R, Tong D, Qiu Z and Yao H: Altered visual cortical
processing in a mouse model of MECP2 duplication syndrome. Sci Rep.
7:64682017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hendrich B, Guy J, Ramsahoye B, Wilson VA
and Bird A: Closely related proteins MBD2 and MBD3 play distinctive
but interacting roles in mouse development. Genes Dev. 15:710–723.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gu P, Xu X, Le Menuet D, Chung AC and
Cooney AJ: Differential recruitment of methyl CpG-binding domain
factors and DNA methyltransferases by the orphan receptor germ cell
nuclear factor initiates the repression and silencing of Oct4. Stem
Cells. 29:1041–1051. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dunkhase E, Ludwig KU, Knapp M, Skibola
CF, Figueiredo JC, Hosking FJ, Ellinghaus E, Landi MT, Ma H,
Nakagawa H, et al: Nonsyndromic cleft lip with or without cleft
palate and cancer: Evaluation of a possible common genetic
background through the analysis of GWAS data. Genom Data. 10:22–29.
2016. View Article : Google Scholar : PubMed/NCBI
|