|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van't Veer LJ, Dai H, van de Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li S, Rong M and Iacopetta B: DNA
hypermethylation in breast cancer and its association with
clinicopathological features. Cancer Lett. 237:272–280. 2006.
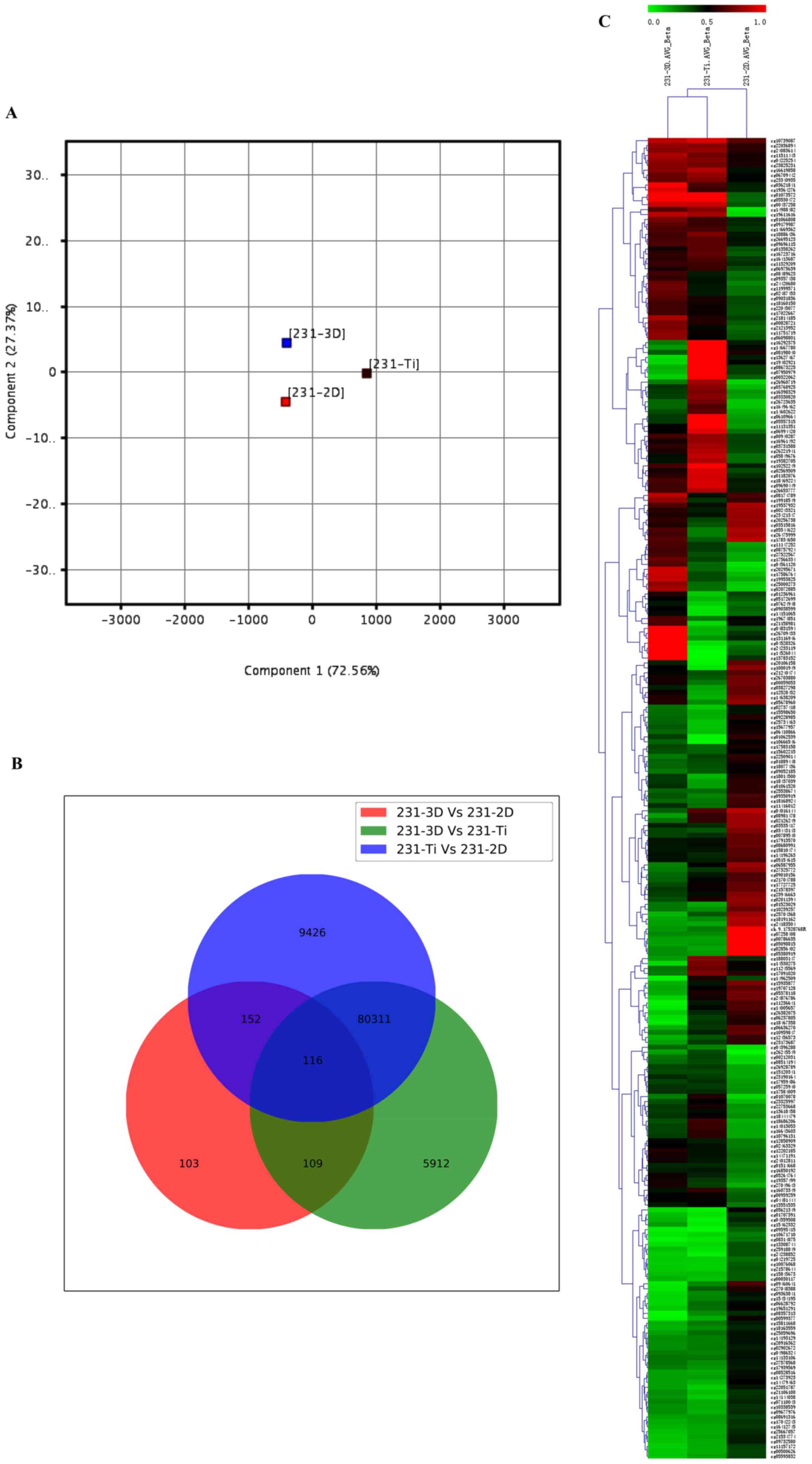
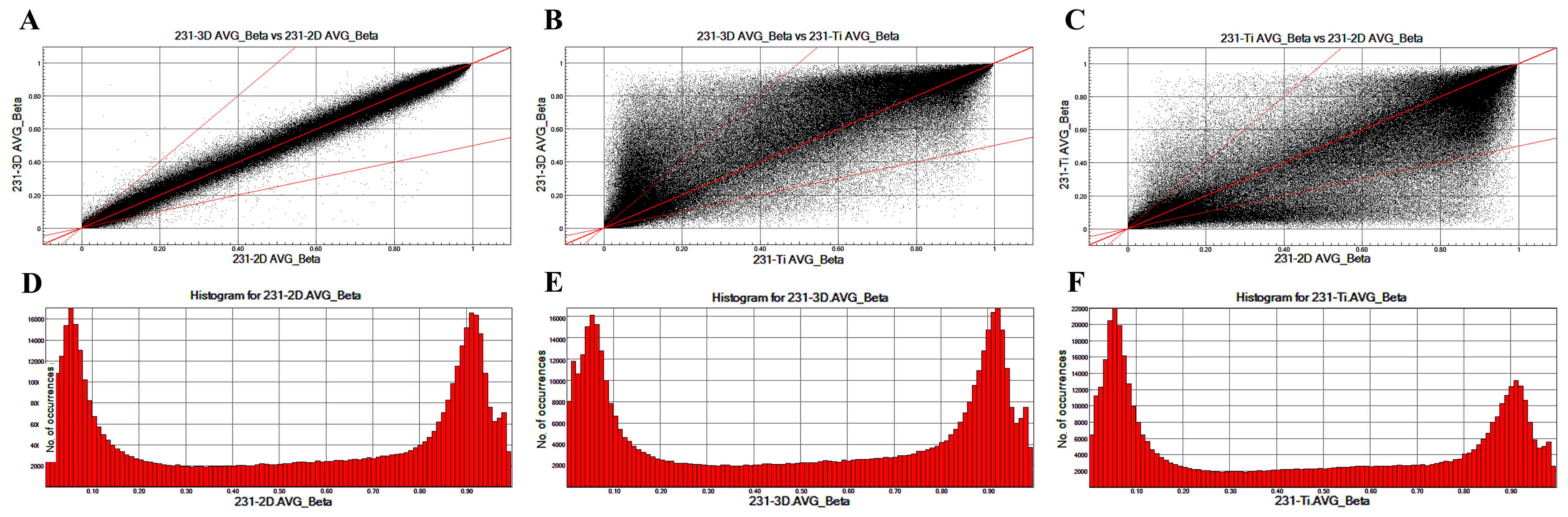
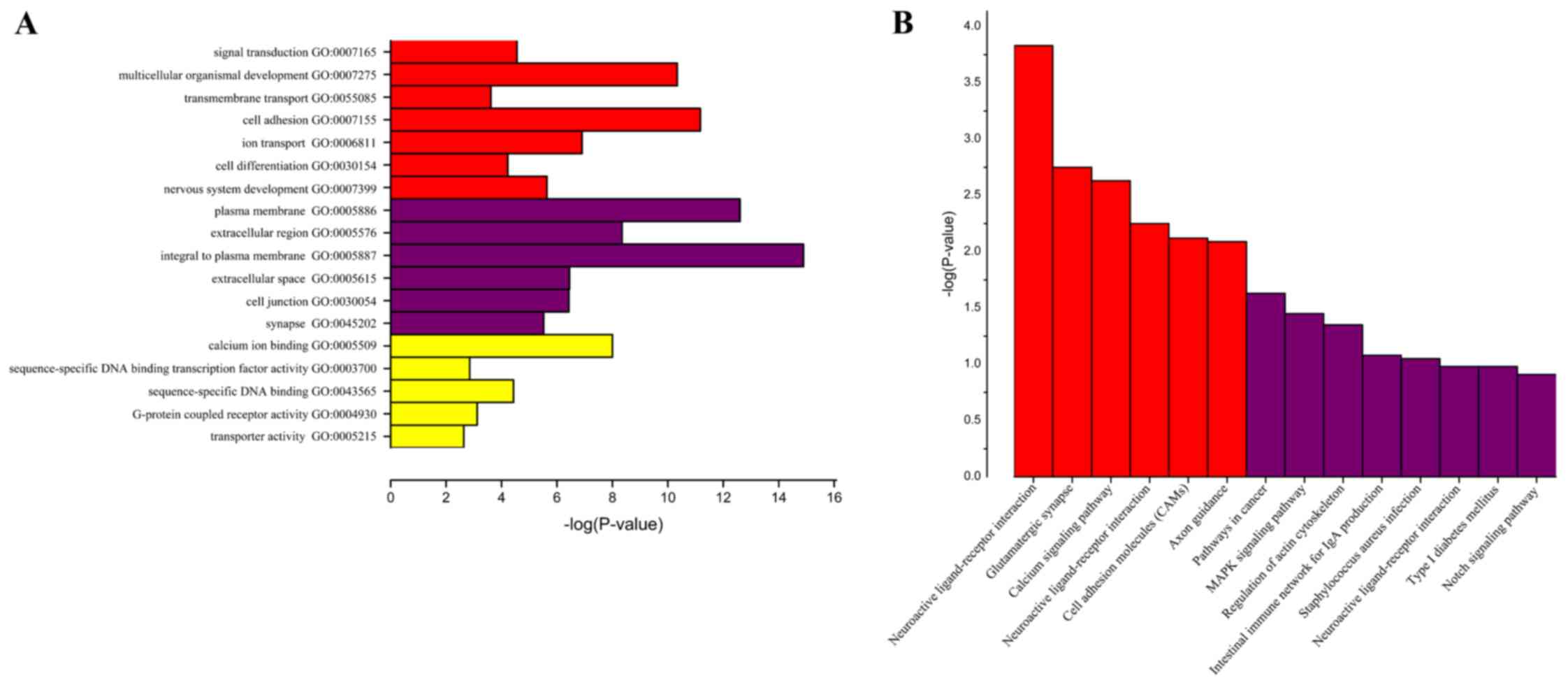
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yegnasubramanian S, Haffner MC, Zhang Y,
Gurel B, Cornish TC, Wu Z, Irizarry RA, Morgan J, Hicks J, DeWeese
TL, et al: DNA hypomethylation arises later in prostate cancer
progression than CpG island hypermethylation and contributes to
metastatic tumor heterogeneity. Cancer Res. 68:8954–8967. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ji H, Ehrlich LI, Seita J, Murakami P, Doi
A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, et al:
Comprehensive methylome map of lineage commitment from
haematopoietic progenitors. Nature. 467:338–342. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Doi A, Park IH, Wen B, Murakami P, Aryee
MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al:
Differential methylation of tissue- and cancer-specific CpG island
shores distinguishes human induced pluripotent stem cells,
embryonic stem cells and fibroblasts. Nat Genet. 41:1350–1353.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Portela A and Esteller M: Epigenetic
modifications and human disease. Nat Biotechnol. 28:1057–1068.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsai HC and Baylin SB: Cancer epigenetics:
Linking basic biology to clinical medicine. Cell Res. 21:502–517.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fackler MJ, McVeigh M, Evron E, Garrett E,
Mehrotra J, Polyak K, Sukumar S and Argani P: DNA methylation of
RASSF1A, HIN-1, RAR-beta, Cyclin D2 and Twist in in situ and
invasive lobular breast carcinoma. Int J Cancer. 107:970–975. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dimas AS, Deutsch S, Stranger BE,
Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C,
Gutierrez Arcelus M, Sekowska M, et al: Common regulatory variation
impacts gene expression in a cell type-dependent manner. Science.
325:1246–1250. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murrell A, Heeson S, Cooper WN, Douglas E,
Apostolidou S, Moore GE, Maher ER and Reik W: An association
between variants in the IGF2 gene and Beckwith-Wiedemann syndrome:
Interaction between genotype and epigenotype. Hum Mol Genet.
13:247–255. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kerkel K, Spadola A, Yuan E, Kosek J,
Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al: Genomic
surveys by methylation-sensitive SNP analysis identify
sequence-dependent allele-specific DNA methylation. Nat Genet.
40:904–908. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li XJ, Valadez AV, Zuo P and Nie Z:
Microfluidic 3D cell culture: Potential application for
tissue-based bioassays. Bioanalysis. 4:1509–1525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Baharvand H, Hashemi SM, Kazemi Ashtiani S
and Farrokhi A: Differentiation of human embryonic stem cells into
hepatocytes in 2D and 3D culture systems in vitro. Int J Dev Biol.
50:645–652. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tibbitt MW and Anseth KS: Hydrogels as
extracellular matrix mimics for 3D cell culture. Biotechnol Bioeng.
103:655–663. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu M, Huang S, Yu KJ and Clyne AM: Dextran
and polymer polyethylene glycol (PEG) coating reduce both 5 and 30
nm iron oxide nanoparticle cytotoxicity in 2D and 3D cell culture.
Int J Mol Sci. 13:5554–5570. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weigelt B, Ghajar CM and Bissell MJ: The
need for complex 3D culture models to unravel novel pathways and
identify accurate biomarkers in breast cancer. Adv Drug Deliv Rev.
69–70:42–51. 2014. View Article : Google Scholar
|
|
18
|
Hájková H, Fritz MH, Haškovec C, Schwarz
J, Šálek C, Marková J, Krejčík Z, Dostálová Merkerová M, Kostečka
A, Vostrý M, et al: CBFB-MYH11 hypomethylation signature and PBX3
differential methylation revealed by targeted bisulfite sequencing
in patients with acute myeloid leukemia. J Hematol Oncol. 7:662014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Jia X, Yuan H, Ma K, Chen Y, Jin Y,
Deng M, Pan W, Chen S, Chen Z, et al: DNA methyltransferase 1
functions through C/ebpa to maintain hematopoietic stem and
progenitor cells in zebrafish. J Hematol Oncol. 8:152015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma CH, Lv Q, Cao Y, Wang Q, Zhou XK, Ye BW
and Yi CQ: Genes relevant with osteoarthritis by comparison gene
expression profiles of synovial membrane of osteoarthritis patients
at different stages. Eur Rev Med Pharmacol Sci. 18:431–439.
2014.PubMed/NCBI
|
|
21
|
Chen H and Boutros PC: VennDiagram: A
package for the generation of highly-customizable Venn and Euler
diagrams in R. BMC Bioinformatics. 12:352011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis CM, Cler LR, Bu DW, Zöchbauer-Müller
S, Milchgrub S, Naftalis EZ, Leitch AM, Minna JD and Euhus DM:
Promoter hypermethylation in benign breast epithelium in relation
to predicted breast cancer risk. Clin Cancer Res. 11:166–172.
2005.PubMed/NCBI
|
|
24
|
Stirzaker C, Song JZ, Davidson B and Clark
SJ: Transcriptional gene silencing promotes DNA hypermethylation
through a sequential change in chromatin modifications in cancer
cells. Cancer Res. 64:3871–3877. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Abdi H and Williams LJ: Principal
component analysis. Wiley Interdiscip Rev Comput Stat. 2:433–459.
2010. View
Article : Google Scholar
|
|
26
|
Bro R and Smilde AK: Principal component
analysis. Anal Methods. 6:2812–2831. 2014. View Article : Google Scholar
|
|
27
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo- and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Farkas SA, Milutin-Gašperov N, Grce M and
Nilsson TK: Genome-wide DNA methylation assay reveals novel
candidate biomarker genes in cervical cancer. Epigenetics.
8:1213–1225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matias-Guiu X, Catasus L, Bussaglia E,
Lagarda H, Garcia A, Pons C, Muñoz J, Argüelles R, Machin P and
Prat J: Molecular pathology of endometrial hyperplasia and
carcinoma. Hum Pathol. 32:569–577. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kumari G, Singhal PK, Rao MR and
Mahalingam S: Nuclear transport of Ras-associated tumor suppressor
proteins: Different transport receptor binding specificities for
arginine-rich nuclear targeting signals. J Mol Biol. 367:1294–1311.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reardon SN, King ML, MacLean JA II, Mann
JL, DeMayo FJ, Lydon JP and Hayashi K: CDH1 is essential for
endometrial differentiation, gland development, and adult function
in the mouse uterus. Biol Reprod. 86(141): 1–10. 2012. View Article : Google Scholar
|
|
32
|
Simpson L and Parsons R: PTEN: Life as a
tumor suppressor. Exp Cell Res. 264:29–41. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maehama T and Dixon JE: PTEN: A tumour
suppressor that functions as a phospholipid phosphatase. Trends
Cell Biol. 9:125–128. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stemke-Hale K, Gonzalez-Angulo AM, Lluch
A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, et
al: An integrative genomic and proteomic analysis of PIK3CA, PTEN
and AKT mutations in breast cancer. Cancer Res. 68:6084–6091. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blyth K, Cameron ER and Neil JC: The RUNX
genes: Gain or loss of function in cancer. Nat Rev Cancer.
5:376–387. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Levanon D and Groner Y: Structure and
regulated expression of mammalian RUNX genes. Oncogene.
23:4211–4219. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito
K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al: RUNX3 is
frequently inactivated by dual mechanisms of protein
mislocalization and promoter hypermethylation in breast cancer.
Cancer Res. 66:6512–6520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Allen NP, Donninger H, Vos MD, Eckfeld K,
Hesson L, Gordon L, Birrer MJ, Latif F and Clark GJ: RASSF6 is a
novel member of the RASSF family of tumor suppressors. Oncogene.
26:6203–6211. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Djos A, Martinsson T, Kogner P and Carén
H: The RASSF gene family members RASSF5, RASSF6 and RASSF7 show
frequent DNA methylation in neuroblastoma. Mol Cancer. 11:402012.
View Article : Google Scholar : PubMed/NCBI
|