Introduction
Renal cell carcinoma (RCC) is a common human
urologic cancer that accounts for approximately 3% of all
malignancies (1). The incidence
and death rates of RCC have been steadily increased in recently
years (2). RCC can be
histologically classified into four subtypes, namely, clear cell
RCC (ccRCC), papillary RCC, chromophobe RCC and oncocytoma
(3). ccRCC, the most common
subtype of RCC, accounts for 70–80% of RCC cases (4). Several risk factors, including
dietary habit, physical activity and occupational exposure to
specific carcinogens, have been identified to play important roles
in ccRCC pathogenesis and progression (5). Despite considerable advancements in
cancer diagnosis and therapy, the curative effects on patients at
advanced stages remain unsatisfactory with a 5-year survival rate
of only 5–10% (6). Surgical
resection offers no therapeutic benefit for patients diagnosed with
advanced stages of ccRCC (7). In
addition, ccRCC is resistant to standard chemotherapy and
radiotherapy (8). Therefore, the
mechanisms underlying the formation and progression of ccRCC should
be elucidated, and novel therapeutic methods should be developed
for the clinical management of patients with this malignancy.
microRNAs (miRNAs) are a large group of highly
conserved, short and non-coding RNAs with lengths of 19–23
nucleotides (9). miRNAs have
emerged as novel gene regulators that control gene expression by
specifically binding to the 3′-untranslated regions (3′-UTRs) of
their target genes in a base-pairing manner, thereby repressing
translation and/or inducing mRNA degradation (10,11).
It is estimated that at least one thirds of human genes are
regulated by miRNAs (12). MiRNAs
have been acknowledged to be aberrantly expressed in various types
of human cancer, including ccRCC (13). The dysregulation of miRNAs in ccRCC
is associated with clinicopathological characteristics and
prognosis. For example, miR-181a is overexpressed in ccRCC, and
this upregulation is strongly associated with tumour size,
tumour/node/metastasis (TNM) staging and Fuhrman grade (14). Furthermore, abnormally expressed
miRNAs contribute to the tumourigenesis and tumour development of
ccRCC by controlling numerous crucial cellular processes (15–17).
Therefore, miRNAs may be potential targets of therapeutic
intervention for patients with ccRCC.
miR-599 plays critical roles in different types of
human cancers, such as breast cancer (18), hepatocellular carcinoma (19) and glioma (20). However, the expression pattern,
biological function and molecular mechanism of miR-599 in ccRCC
remain unknown. Thus, this study aimed to detect the expression
level of miR-599 in ccRCC, examine its effect on ccRCC progression
and further explore the possible mechanisms underlying the tumour
suppressive roles of miR-599 in ccRCC.
Materials and methods
Clinical samples
Twenty-one paired ccRCC tissues and normal adjacent
tissues (NATs) were obtained from patients (17 males, 4 females;
age range, 42–75 years old; mean age, 59 years old) who underwent
nephrectomy in Yidu Central Hospital of Weifang between August 2014
and March 2016. None of these patients were treated with
chemotherapy or radiotherapy prior to nephrectomy. All of the
tissue specimens were immediately frozen and stored in liquid
nitrogen until further RNA isolation. This study was approved by
the Ethics Committee of Yidu Central Hospital of Weifang, and
written informed consent was collected from all patients before
they participated in this research.
Cell culture and transfection
Two human ccRCC cell lines, A498 (21–23)
and 786-O and one normal renal cell line (HK-2) were purchased from
American Type Culture Collection (ATCC, Manassas, VA, USA). All
ccRCC cell lines were cultured in Dulbecco's Modified Eagle's
Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), and
1% penicillin/streptomycin (all from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). HK-2 cells were grown at
keratinocyte-SFM (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with bovine pituitary extract and human recombinant
epidermal growth factor (all from Gibco; Thermo Fisher Scientific,
Inc.). All these cell lines were maintained at 37°C in a humidified
incubator containing 5% carbon dioxide (CO2).
miR-599 mimics, negative control miRNA mimics
(miR-NC), small interfering RNA (siRNA) against the expression of
HMGA2 (si-HMGA2) and negative control siRNA (si-NC) were designed
and synthesised by Shanghai GenePharma Co., Ltd. (Shanghai, China).
HMGA2 overexpression plasmid (pCMV-HMGA2) and empty pCMV plasmid
were acquired from OriGene Technologies, Inc., (Beijing, China).
Cells were inoculated into six-well plates and transfected with
miRNA mimics, siRNA or plasmid by using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol. Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and western blot analysis were
performed to evaluate the transfection efficacy.
RT-qPCR
Total RNA was isolated from tissue samples or cells
by using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) in accordance with the manufacturer's protocol. The
concentration and purity of total RNA were determined using a
NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Inc.).
The cDNA of miR-599 was synthesised by using a TaqMan
MicroRNA reverse transcription kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Quantitative PCR (qPCR) was conducted to
detect miR-599 expression by using a TaqMan MicroRNA PCR kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with U6 snRNA
as an internal control. To quantify the mRNA expression of HMGA2,
we conducted reverse transcription with a PrimeScript RT reagent
kit (Takara Bio, Inc., Otsu, Japan) followed by qPCR with a SYBR
Premix Ex Taq™ kit (Takara Bio, Inc.). GAPDH was employed as
an internal control for the mRNA level of HMGA2. Relative gene
expression was analysed through the 2−∆∆Ct method
(24).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) was applied to evaluate cell proliferation of
ccRCC. Transfected cells were harvested at 24 h post-transfection.
In each well of a 96-well plate, 3,000 transfected cells were
plated and incubated at 37°C in a humidified incubator with 5%
CO2 for 0, 24, 48 and 72 h. At every time point, CCK-8
assay was performed in accordance with the manufacturer's protocol.
Briefly, 10 µl CCK-8 reagent was added into each well, and then
incubated at 37°C for another 2 h. The absorbance value of each
well was detected at a wavelength of 450 nm with the ELISA
microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Transwell invasion assay
Cellular invasion ability was examined by using a
Boyden chamber containing 24-well Transwell plates (Corning Costar,
Corning, NY, USA) with 8 µm pore membranes. After transfection for
48 h, cells were collected, suspended in FBS-free DMEM and seeded
in the upper chamber of the insert, which was pre-coated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). At 24 h
post-incubation, the remaining cells at the upper side of the 8 µm
pore membranes were wiped away with cotton swabs. The invasive
cells that had invaded the bottom of the inserts were fixed in 4%
cold paraformaldehyde and stained with 0.5% crystal violet. The
stained cells were photographed and counted under an inverted
microscope (Olympus Corporation, Tokyo, Japan) at ×200
magnification.
miR-599 target prediction and
luciferase reporter assay
TargetScan (http://www.targetscan.org/) and microRNA (www.microrna.org) algorithms were used to predict the
putative targets of miR-599. HMGA2 was predicted as a potential
target of miR-599. Luciferase reporter assay was performed to
further determine whether miR-599 could directly bind to the 3′-UTR
of HMGA2. A total of 6×104 cells were seeded in
triplicates in 24-well plates. Luciferase reporter plasmids,
namely, pGL3-HMGA2-3′-UTR wild-type (Wt) and pGL3-HMGA2-3′-UTR
mutant (Mut), were designed and synthesised by Shanghai GenePharma
Co., Ltd. When the cell density reached 70–80% confluence, miR-599
mimics or miR-NC was transfected into cells with pGL3-HMGA2-3′-UTR
Wt or pGL3-HMGA2-3′-UTR Mut by using Lipofectamine 2000. After 48 h
of incubation, a dual luciferase reporter assay system (Promega
Corp., Madison, Wisconsin, USA) was adopted to measure the
luciferase activity in accordance with the manufacturer's
procedure. The firefly luciferase activity was normalised to
Renilla luciferase activity.
Western blot analysis
Total protein was extracted from tissue samples or
cells in an ice bath by using RIPA lysis buffer (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The protein concentration
was detected by using a BCA protein assay kit (Bio-Rad
Laboratories, Inc.). Equal amounts of proteins were subjected to
10% sodium dodecyl sulphate polyacrylamide gel electrophoresis and
transferred to a polyvinylidene difluoride membrane (EMD Millipore,
Billerica, MA, USA), which was then blocked with 5% skimmed milk
dissolved in TBS containing 0.1% Tween-20 (TBST). Afterwards, the
membranes were incubated with primary antibodies against HMGA2
(1:1,000 dilution, ab184616; Abcam, Cambridge, UK) or GAPDH
(1:1,000 dilution, sc-47724; Santa Cruz Biotechnology, Inc.)
overnight at 4°C, rinsed with TBST, further incubated with the
corresponding horseradish peroxidase-conjugated secondary antibody
(1:5,000 dilution, sc-2005; Santa Cruz Biotechnology, Inc.) at room
temperature for 2 h and washed with TBST. The protein signals were
then visualised with an ECL detection kit (GE Healthcare Life
Sciences, Chalfont, UK) and analysed with Image J 1.41 (National
Institutes of Health, Bethesda, MD, USA). GAPDH was used as an
internal control.
Statistical analysis
Data were expressed as mean ± standard deviation and
analysed using SPSS v21.0 (SPSS Inc., Chicago, IL, USA). Each assay
was repeated at least thrice. Differences between two groups were
compared using t test, whereas differences between multiple groups
were compared through one-way ANOVA. Student-Newman-Keuls test was
used as a post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-599 is downregulated in ccRCC
tissues and cell lines
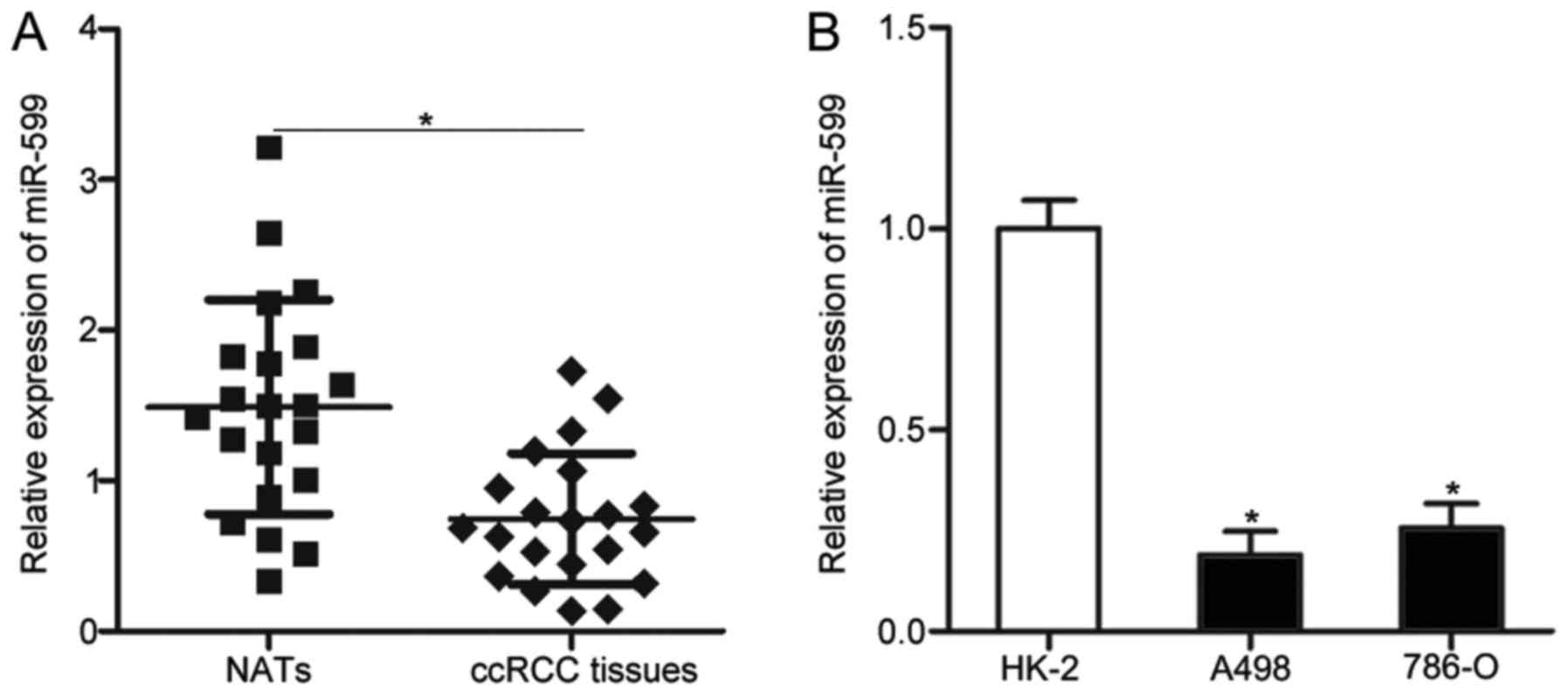
miR-599 expression levels were detected in 21 paired
ccRCC tissues and NATs. The results of RT-qPCR revealed that
miR-599 was underexpressed in ccRCC tissues compared with that in
NATs (P<0.05; Fig. 1A). The
expression level of miR-599 was further examined in two ccRCC cell
lines (A498 and 786-O) and one normal renal cell line (HK-2) by
conducting RT-qPCR. As shown in Fig.
1B, miR-599 expression level decreased in the A498 and 786-O
cell lines compared with HK-2 (P<0.05). These results suggest
that the downregulation of miR-599 may be correlated with ccRCC
progression.
miR-599 attenuates the proliferation
and invasion of ccRCC cells
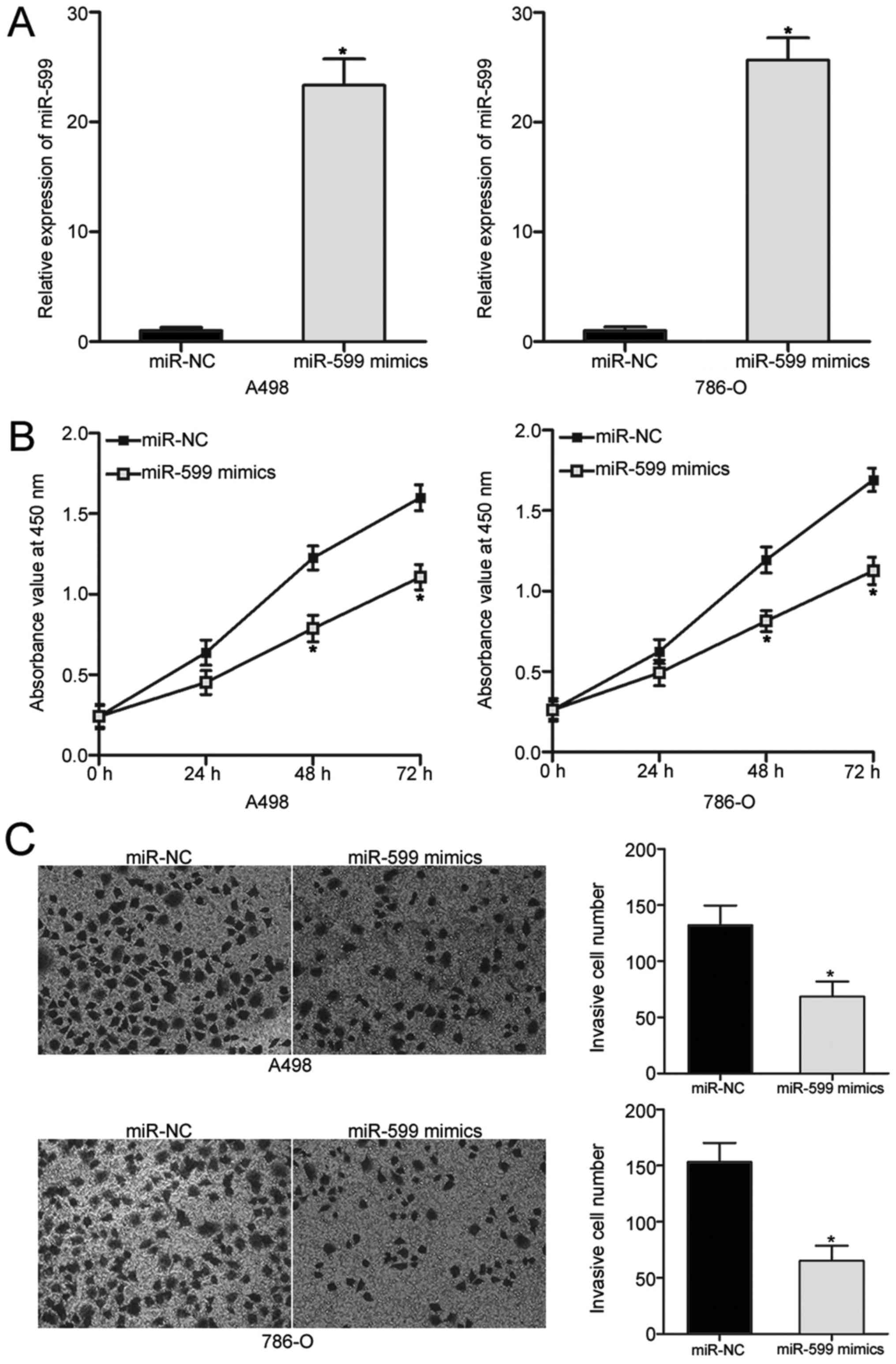
To elucidate the effects of miR-599 in ccRCC
development, we overexpressed miR-599 in A498 and 786-O cells.
RT-qPCR analysis confirmed that miR-599 was markedly upregulated in
A498 and 786-O cells transfected with miR-599 mimics (P<0.05;
Fig. 2A). Then, CCK-8 assays were
performed to determine the effects of miR-599 overexpression on the
proliferation of A498 and 786-O cells. Ectopic expression of
miR-599 significantly restricted the proliferation of A498 and
786-O cells compared with that in the cells transfected with miR-NC
(P<0.05; Fig. 2B).
Additionally, Transwell invasion assays were conducted to
investigate the invasion abilities of A498 and 786-O cells
transfected with miR-599 mimics or miR-NC. Upregulation of miR-599
resulted in the reduced invasion capabilities of A498 and 786-O
cells (P<0.05; Fig. 2C). These
data suggest that miR-599 may play tumour suppressive roles in
ccRCC.
HMGA2 is a direct target of miR-599 in
ccRCC
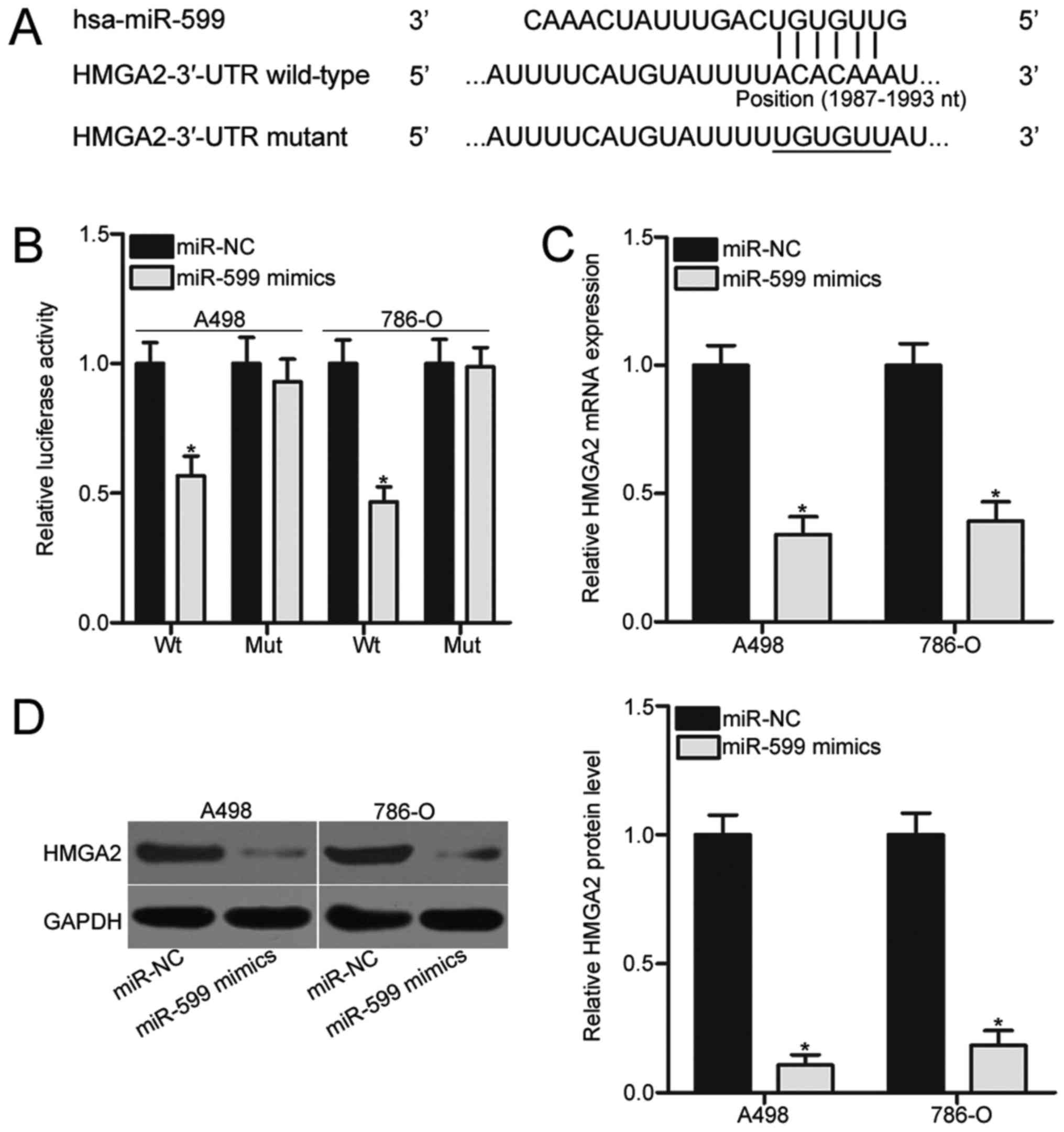
Bioinformatics analysis was performed to predict the
putative targets of miR-599 and to investigate the molecular
mechanism by which miR-599 affected the aggressive phenotype of
ccRCC cells. Among the candidates, HMGA2 was selected for further
confirmation (Fig. 3A) because it
is aberrantly highly expressed in ccRCC and significantly
associated with ccRCC progression (25–28).
To identify whether HMGA2 is a direct target of miR-599, we
subjected A498 and 786-O cells cotransfected with miR-599 mimics or
miR-NC and pGL3-HMGA2-3′-UTR Wt or pGL3-HMGA2-3′-UTR Mut to
luciferase reporter assays. As shown in Fig. 3B, enforced expression of miR-599
significantly decreased the luciferase activities of the wild-type
3′-UTR reporter plasmid in A498 and 786-O cells (P<0.05).
However, this suppressive effect was abrogated in the mutant 3′-UTR
reporter plasmid, in which the binding sequences mutated. RT-qPCR
and western blot analysis were performed to detect the mRNA and
protein expression of HMGA2 in A498 and 786-O cells transfected
with miR-599 mimics or miR-NC and to evaluate the association
between miR-599 and HMGA2 in ccRCC. The results indicated that
restored miR-599 expression in the two ccRCC cell lines resulted in
significantly reduced HMGA2 expression at both mRNA (Fig. 3C, P<0.05) and protein
(P<0.05; Fig. 3D) levels.
Therefore, HMGA2 is a direct target gene of miR-599 in ccRCC.
HMGA2 knockdown inhibits cell
proliferation and invasion in ccRCC
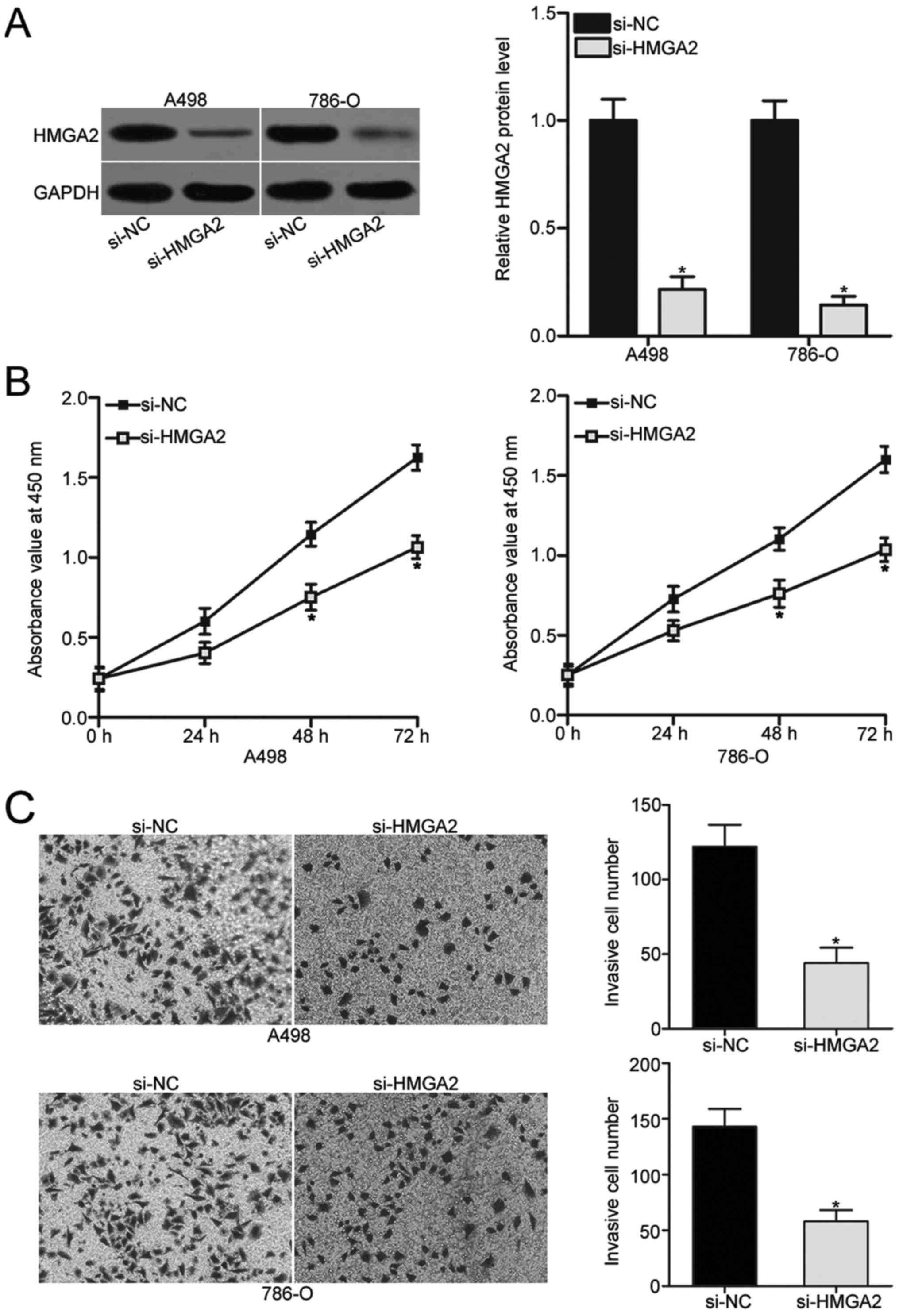
HMGA2 was validated as a direct target of miR-599 in
ccRCC. As such, we hypothesised that the tumour suppressive roles
of miR-599 in ccRCC might be exhibited by HMGA2 knockdown. To
confirm this hypothesis, si-HMGA2 was transfected into A498 and
786-O cells to knock down endogenous HMGA2 expression. Western blot
analysis revealed that HMGA2 was obviously downregulated in A498
and 786-O cells following transfection with si-HMGA2 (P<0.05;
Fig. 4A). Subsequent CCK-8 and
Transwell invasion assays demonstrated that the inhibition of HMGA2
prohibited the proliferation (P<0.05; Fig. 4B) and invasion (P<0.05; Fig. 4C) of A498 and 786-O cells. This
phenomenon resembled the inhibitory effects of miR-599
overexpression on ccRCC cells, further suggesting that HMGA2 is a
direct downstream target of miR-599 in ccRCC.
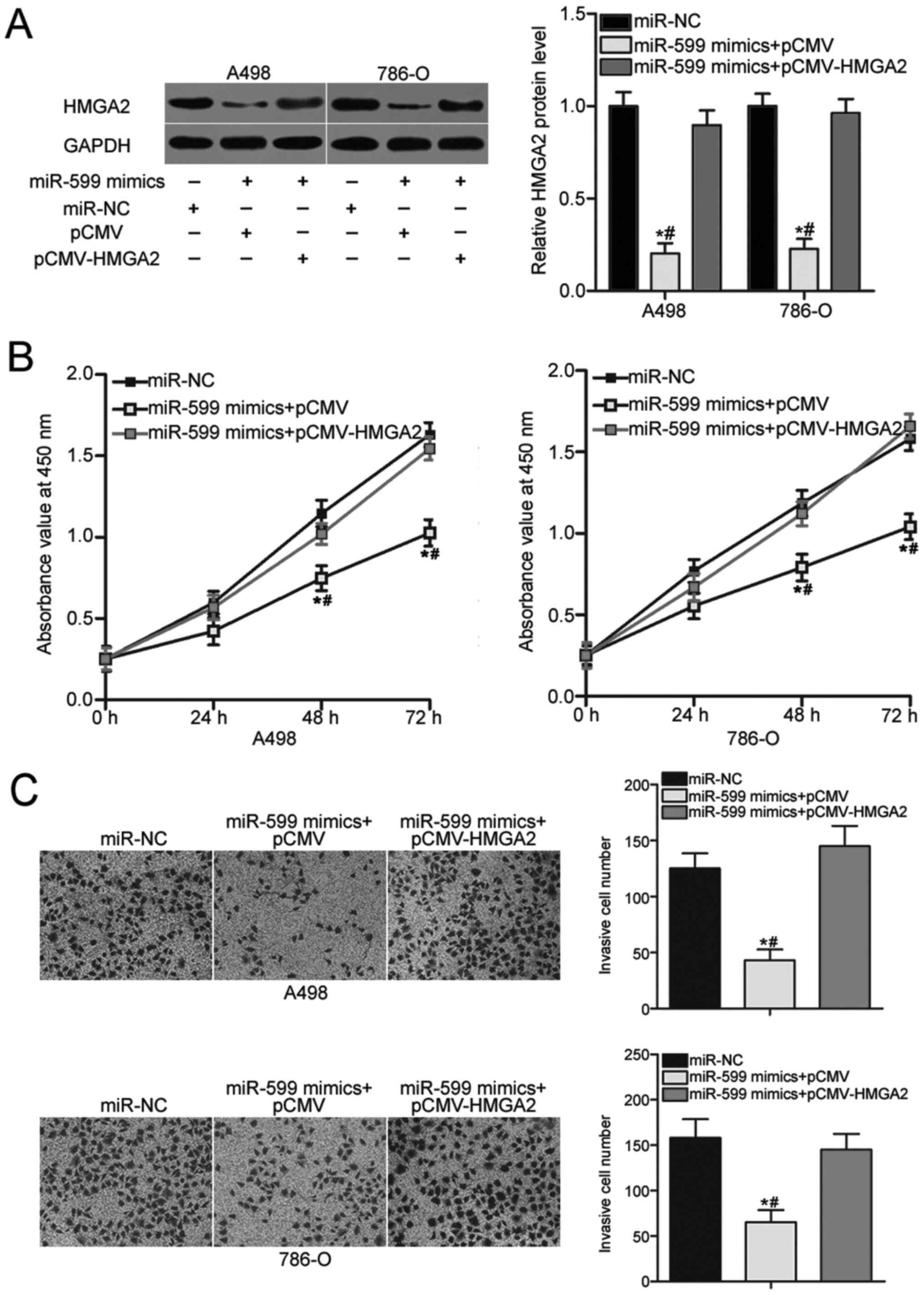
Restored expression of HMGA2 reverses
the suppressive effects induced by miR-599 overexpression in ccRCC
cells
A rescue experiment was performed to evaluate
whether HMGA2 is responsible for the inhibitory effects of miR-599
on ccRCC cells. A498 and 786-O cells were cotransfected with
miR-599 mimics and pCMV-HMGA2 or empty pCMV plasmid. Western blot
analysis was conducted to examine the transfection efficiency, and
the results confirmed that the downregulation of HMGA2 caused by
miR-599 overexpression was recovered after the A498 and 786-O cells
were cotransfected with pCMV-HMGA2 (P<0.05; Fig. 5A). In addition, CCK-8 and Transwell
invasion assays revealed that recovered HMGA2 expression eliminated
the inhibitory effects on cell proliferation (Fig. 5B, P<0.05) and invasion (Fig. 5C, P<0.05) induced by miR-599
overexpression in A498 and 786-O cells. Therefore, miR-599 inhibits
cell proliferation and invasion of ccRCC partly by inhibiting the
HMGA2 expression.
Discussion
Dysregulation of miRNAs is involved in the
occurrence and development of ccRCC through their participation in
many important biological processes, such as cell proliferation,
cycle, apoptosis, angiogenesis, migration, invasion and metastasis
(29–31). Thus, an in-depth investigation on
miRNAs and their biological roles in ccRCC may provide useful
insights into the identification of novel therapeutic methods for
patients with this disease. In this study, miR-599 expression was
significantly reduced in ccRCC tissues and cell lines. Ectopic
expression of miR-599 suppressed the proliferation and invasion of
ccRCC cells. HMGA2 was identified as a direct target gene of
miR-599 in ccRCC, and HMGA2 knockdown could mimic the suppressive
effects of miR-599 overexpression on ccRCC cells. Moreover,
restored HMGA2 expression rescued the inhibitory properties of
ccRCC cells caused by miR-599 overexpression. These results suggest
that miR-599 might be a novel therapeutic agent for ccRCC.
miR-599 has been reported to be aberrantly expressed
in many types of human malignancies. For example, miR-599
expression is reduced in gastric cancer tissues and cell lines. The
downregulation of miR-599 was associated with poor prognostic
features, including lymph node metastasis and advanced TNM stage.
The 5-year overall survival of patients with gastric cancer and a
low miR-599 expression is shorter than that of patients with high
miR-599 levels (32). miR-599 is
also observed to be underexpressed in breast cancer (18), hepatocellular carcinoma (19) and glioma (20). Nevertheless, the miR-599 expression
is relatively higher in non-small cell lung cancer tissues than in
normal lung tissues (33). These
conflicting studies suggest that the expression pattern of miR-599
exhibits tissue specificity and possibly represents a biomarker for
the diagnosis of specific cancer types.
The dysregulation of miR-599 has been implicated in
the carcinogenesis and progression of multiple types of human
cancer. For instance, miR-599 overexpression suppresses cell
metastasis and epithelial-mesenchymal transition in gastric cancer
(32). Wang et al (18) reported that restored expression of
miR-599 reduces cell proliferation, colony formation and metastasis
in vitro and decreases tumour growth in vivo. Tian
et al (19) revealed that
enforced expression of miR-599 restricts cell growth and motility
of hepatocellular carcinoma in vitro. Zhang et al
(20) demonstrated that ectopic
expression of miR-599 prohibits the migration and invasion of
glioma cells. However, miR-599 serves as an oncogene in non-small
cell lung cancer by promoting cell migration and invasion (33). These findings suggested that
miR-599 may be a promising therapeutic target for patients with
these human cancers.
Several miR-599 targets, including EIF5A2 (32) in gastric cancer, BRD4 (18) in breast cancer, MYC (19) in hepatocellular carcinoma,
periostin (20) in glioma and
STAB2 (33) in non-small cell lung
cancer, have been validated. HMGA2, a membrane of high-mobility
group A proteins, was identified as a novel target of miR-599 in
ccRCC. Aberrantly overexpressed HMGA2 has been reported in various
types of human cancer, such as breast cancer (34), thyroid cancer (35), colorectal cancer (36) and glioblastoma (37). HMGA2 is highly expressed in ccRCC
at mRNA and protein levels (25).
The upregulation of HMGA2 is strongly correlated with tumour size,
lymph node metastasis and Fuhrman grade. In addition, the prognosis
of patients with ccRCC with high HMGA2 levels is poorer than that
of patients with low HMGA2 levels. Furthermore, HMGA2 expression
level is an independent prognostic factor for patients with ccRCC
(26). Besides, HMGA2 deregulation
affected the onset and development of ccRCC by regulating cell
proliferation, invasion and epithelial-mesenchymal transition
(26,28). These findings suggested that
targeting HMGA2 may show potential for the treatment of ccRCC in
the future.
In conclusion, miR-599 was significantly
downregulated in ccRCC tissues and cell lines. In vitro
functional experiments demonstrated that miR-599 inhibited the
proliferation and invasion of ccRCC cells. Mechanistically, HMGA2
was identified as a direct target gene of miR-599 in ccRCC.
Therefore, the miR-599/HMGA2 pathway may provide a novel
therapeutic target for the treatment of patients with ccRCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL designed this research. HaZ and HuZ performed the
experiments. XX analyzed the data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Yidu Central Hospital of Weifang and was performed in accordance
with the Declaration of Helsinki. Written informed consent was
collected from all patients prior to their participation.
Consent for publication
Written informed consent was obtained from all
participants for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rasmussen F: Metastatic renal cell cancer.
Cancer Imaging. 13:374–380. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheville JC, Lohse CM, Zincke H, Weaver AL
and Blute ML: Comparisons of outcome and prognostic features among
histologic subtypes of renal cell carcinoma. Am J Surg Pathol.
27:612–624. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Z, Zhang J, Zhang Z, Feng Z, Wei J,
Lu J, Fang Y, Liang Y, Cen J, Pan Y, et al: The putative tumor
suppressor microRNA-30a-5p modulates clear cell renal cell
carcinoma aggressiveness through repression of ZEB2. Cell Death
Dis. 8:e28592017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chow WH and Devesa SS: Contemporary
epidemiology of renal cell cancer. Cancer J. 14:288–301. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hadoux J, Vignot S and De La Motte Rouge
T: Renal cell carcinoma: Focus on safety and efficacy of
temsirolimus. Clin Med Insights Oncol. 4:143–154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu TY, Li J, Wen XH, Zhang H and Gui Q:
The efficacy of open nephron-sparing surgery in the treatment of
complex renal cell carcinoma. Eur Rev Med Pharmacol Sci.
20:3959–3964. 2016.PubMed/NCBI
|
|
8
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lui PY, Jin DY and Stevenson NJ: MicroRNA:
Master controllers of intracellular signaling pathways. Cell Mol
Life Sci. 72:3531–3542. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jonas S and Izaurralde E: Towards a
molecular understanding of microRNA-mediated gene silencing. Nat
Rev Genet. 16:421–433. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Giannakakis A, Coukos G, Hatzigeorgiou A,
Sandaltzopoulos R and Zhang L: miRNA genetic alterations in human
cancers. Expert Opin Biol Ther. 7:1375–1386. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang R, Ma Y, Yu D, Zhao J and Ma P:
miR-377 functions as a tumor suppressor in human clear cell renal
cell carcinoma by targeting ETS1. Biomed Pharmacother. 70:64–71.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lei Z, Ma X, Li H, Zhang Y, Gao Y, Fan Y,
Li X, Chen L, Xie Y, Chen J, et al: Up-regulation of miR-181a in
clear cell renal cell carcinoma is associated with lower KLF6
expression, enhanced cell proliferation, accelerated cell cycle
transition, and diminished apoptosis. Urol Oncol. Oct 20–2017.(Epub
ahead of print). PubMed/NCBI
|
|
15
|
Cao J, Liu J, Xu R, Zhu X, Liu L and Zhao
X: MicroRNA-21 stimulates epithelial-to-mesenchymal transition and
tumorigenesis in clear cell renal cells. Mol Med Rep. 13:75–82.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakata W, Uemura M, Sato M, Fujita K,
Jingushi K, Ueda Y, Kitae K, Tsujikawa K and Nonomura N: Expression
of miR-27a-3p is an independent predictive factor for recurrence in
clear cell renal cell carcinoma. Oncotarget. 6:21645–21654. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao H, Zeng J, Li H, Chen K, Yu G, Hu J,
Tang K, Zhou H, Huang Q, Li A, Li Y, et al: MiR-1 downregulation
correlates with poor survival in clear cell renal cell carcinoma
where it interferes with cell cycle regulation and metastasis.
Oncotarget. 6:13201–13215. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang Y, Sui Y, Zhu Q and Sui X:
Hsa-miR-599 suppresses the migration and invasion by targeting BRD4
in breast cancer. Oncol Lett. 14:3455–3462. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tian J, Hu X, Gao W, Zhang J, Chen M,
Zhang X, Ma J and Yuan H: Identification a novel tumor-suppressive
hsa-miR-599 regulates cells proliferation, migration and invasion
by targeting oncogenic MYC in hepatocellular carcinoma. Am J Transl
Res. 8:2575–2584. 2016.PubMed/NCBI
|
|
20
|
Zhang T, Ma G, Zhang Y, Huo H and Zhao Y:
miR-599 inhibits proliferation and invasion of glioma by targeting
periostin. Biotechnol Lett. 39:1325–1333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang D, Zhu C, Zhang Y, Zheng Y, Ma F, Su
L and Shao G: MicroRNA-30e-3p inhibits cell invasion and migration
in clear cell renal cell carcinoma by targeting Snail1. Oncol Lett.
13:2053–2058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fan W, Huang J, Xiao H and Liang Z:
MicroRNA-22 is downregulated in clear cell renal cell carcinoma,
and inhibits cell growth, migration and invasion by targeting PTEN.
Mol Med Rep. 13:4800–4806. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma X, Shen D, Li H, Zhang Y, Lv X, Huang
Q, Gao Y, Li X, Gu L, Xiu S, et al: MicroRNA-185 inhibits cell
proliferation and induces cell apoptosis by targeting VEGFA
directly in von Hippel-Lindau-inactivated clear cell renal cell
carcinoma. Urol Oncol. 33:169.e1–11. 2015. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Fu QZ, Pu L, Meng QG, Liu XF, Dong
SF, Yang JX and Lv GY: HMGA2 expression in renal carcinoma and its
clinical significance. J Med Biochem. 34:338–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Na N, Si T, Huang Z, Miao B, Hong L, Li H
and Qiu J and Qiu J: High expression of HMGA2 predicts poor
survival in patients with clear cell renal cell carcinoma. Onco
Targets Ther. 9:7199–7205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Fu QZ, Pu L, Song LL, Wang YY, Liu
J, Wang ZL and Wang ZM: Effect of RNA interference of the
expression of HMGA2 on the proliferation and invasion ability of
ACHN renal cell carcinoma cells. Mol Med Rep. 16:5107–5112. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kou B, Liu W, Tang X and Kou Q: HMGA2
facilitates epithelial-mesenchymal transition in renal cell
carcinoma by regulating the TGF-β/Smad2 signaling pathway. Oncol
Rep. 39:101–108. 2018.PubMed/NCBI
|
|
29
|
Liu LJ, Yu JJ and Xu XL: MicroRNA-93
inhibits apoptosis and promotes proliferation, invasion and
migration of renal cell carcinoma ACHN cells via the TGF-β/Smad
signaling pathway by targeting RUNX3. Am J Transl Res. 9:3499–3513.
2017.PubMed/NCBI
|
|
30
|
He YH, Chen C and Shi Z: The biological
roles and clinical implications of microRNAs in clear cell renal
cell carcinoma. J Cell Physiol. Dec 7–2017.(Epub ahead of
print).
|
|
31
|
Xiao W, Lou N, Ruan H, Bao L, Xiong Z,
Yuan C, Tong J, Xu G, Zhou Y, Qu Y, et al: Mir-144-3p promotes cell
proliferation, metastasis, sunitinib resistance in clear cell renal
cell carcinoma by downregulating ARID1A. Cell Physiol Biochem.
43:2420–2433. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X, Jin Y, Zhang H, Huang X, Zhang Y
and Zhu J: MicroRNA-599 inhibits metastasis and
epithelial-mesenchymal transition via targeting EIF5A2 in gastric
cancer. Biomed Pharmacother. 97:473–480. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian W, Wang G, Liu Y, Huang Z, Zhang C,
Ning K, Yu C, Shen Y, Wang M, Li Y, et al: The miR-599 promotes
non-small cell lung cancer cell invasion via SATB2. Biochem Biophys
Res Commun. 485:35–40. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun M, Song CX, Huang H, Frankenberger CA,
Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C and Rosner
MR: HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer
growth and metastasis. Proc Natl Acad Sci USA. 110:pp. 9920–9925.
2013; View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Belge G, Meyer A, Klemke M, Burchardt K,
Stern C, Wosniok W, Loeschke S and Bullerdiek J: Upregulation of
HMGA2 in thyroid carcinomas: A novel molecular marker to
distinguish between benign and malignant follicular neoplasias.
Genes Chromosomes Cancer. 47:56–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Esmailzadeh S, Mansoori B, Mohammadi A,
Shanehbandi D and Baradaran B: siRNA-mediated silencing of HMGA2
induces apoptosis and cell cycle arrest in human colorectal
carcinoma. J Gastrointest Cancer. 48:156–163. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kaur H, Ali SZ, Huey L, Hütt-Cabezas M,
Taylor I, Mao XG, Weingart M, Chu Q, Rodriguez FJ, Eberhart CG and
Raabe EH: The transcriptional modulator HMGA2 promotes stemness and
tumorigenicity in glioblastoma. Cancer Lett. 377:55–64. 2016.
View Article : Google Scholar :
|