Introduction
Breast cancer (BC) is the leading cause of
cancer-related mortality in women between the ages of 20 and 59
years (1,2), and affects 12% of women worldwide
(3). A number of risk factors have
been associated with the occurrence of BC in women, including
obesity, early menarche, late menopause, hormone replacement
therapy, late first-full pregnancy and diets rich in high-fat foods
or red meats (4). Therefore, it is
necessary to investigate effective diagnostic methods for BC
(5).
MicroRNAs (miRNAs) are a class of small non-coding
RNAs that provide a novel perspective to predict and screen for
cancer, and previous studies have reported that miRNA expression
levels may be aberrant in tumors (6–8).
Associations between the clinicopathological features of BC and
miRNA expression levels have been reported previously (9,10).
miR-142-3p was demonstrated to directly regulate its target genes
in a several types of cancer, including non-small cell lung cancer,
colon cancer and hepatic cancer (11–13).
miR-142 expression levels were elevated in human BC stem cells
(BCSCs) compared with non-tumorigenic BC cells (14). In addition, miR-142-5p expression
levels were reported to be markedly higher in the lymph node-cancer
group compared with expression in the non-lymph node-cancer group,
which were detected by miRNA microarray analysis (15). Upregulation of miR-142-5p was
identified in atherosclerotic plaques, and regulated oxidized
low-density lipoprotein-induced apoptosis in macrophages (16). miR-142-5p was involved in squamous
lung cancer via regulation of cell cycle related genes (17). miR-142-5p contributes to
Hashimoto's thyroiditis by targeting CLDN1 (18) and promotes the development of
colorectal cancer by targeting SDHB (19). However, the function of miR-142-5p
in BC has not yet been reported. Consequently, the present study
aimed to investigate a target gene of miR-142-5p in BC cells and to
provide a novel potential target for the treatment of BC.
Materials and methods
Study population
BC tissues and the adjacent healthy tissues were
obtained from 60 female patients who underwent surgery in Renmin
Hospital of Wuhan University (Wuhan, China) between March, 2013 and
July, 2016. Clinicopathological characteristics of patients' age,
tumor size and metastasis were collected and are presented in
Table I. There were 40 patients
aged <50 years and 20 patients aged ≥50 years. Written informed
consent was obtained from all the patients prior to the start of
the study; the present study was approved by the ethics committee
of Renmin Hospital of Wuhan University.
 | Table I.Association between miR-142-5p or PTEN
and patients clinicopathological characteristics. |
Table I.
Association between miR-142-5p or PTEN
and patients clinicopathological characteristics.
| Factor | n |
miR-142-5pa | P-value | PTENa | P-value |
|---|
| Age (years) |
|
| 0.92 |
| 0.73 |
|
<50 | 40 | 2.33±0.59 |
| 0.69±0.21 |
|
| ≥50 | 20 | 2.42±0.45 |
| 0.58±0.13 |
|
| Tumor size (cm) |
|
| 0.04b |
| 0.01b |
| ≥5 | 18 | 2.67±0.28 |
| 0.37±0.22 |
|
|
<5 | 42 | 2.09±0.14 |
| 0.96±0.12 |
|
| Metastasis |
|
| 0.04b |
| 0.01b |
| No | 32 | 2.25±0.21 |
| 0.96±0.24 |
|
|
Yes | 28 | 2.53±0.12 |
| 0.32±0.11 |
|
Cell culture
Normal breast epithelial cell line MCF-10A and BC
cell lines SK-BR-3 and MDA-MB-231 were purchased from the American
Type Culture Collection (Manassas, VA, USA) and cultured in
Dulbecco's modified Eagle's medium (DMEM)/Ham's F12 medium
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), penicillin (100 U/ml) and
streptomycin (100 µg/ml) in an incubator at 37°C with humidified
atmosphere of 5% CO2. MDA-MB-231 cells with the highest
miR-142-5p expression levels were selected for the remaining in
vitro experiments as these cells exhibited the highest
expression levels of miR-142-5p compared with other cell lines.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from BC tissues (1
cm3) and cell cultures (1×106 cells/well)
using a miRNeasy kit (Qiagen Inc., Valencia, CA, USA) according to
the manufacturer's protocol. Reverse transcription of miR142-5p and
phosphatase and tensin homolog (PTEN) to cDNA was performed using
miRNA cDNA Synthesis Kit (Takara bio, Inc., Otsu, Japan) and First
Strand cDNA Synthesis kit (Takara, Dalian, China), respectively.
Expression levels of miR-142-5p and PTEN were detected with a
TaqMan miRNA assay kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.) on an ABI 7500 thermocycler (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Thermocycling conditions were as following: 95°C for 30
sec, followed by 35 cycles of 95°C for 10 sec and 60°C for 25 sec.
Relative expression levels of miRNA-142-5p were normalized to U6,
and relative PTEN expression levels were normalized to GAPDH.
Primers were as follows: PTEN, forward
5′-TGGATTCGACTTAGACTTGACCT-3′, reverse
5′-GGTGGGTTATGGTCTTCAAAAGG-3′; GAPDH, forward
5′-ACAAGATGGTGAAGGTCGGTGTGA-3′, reverse
5′-AGCTTCCCATTCTCAGCCTTGACT-3′; miR-142-5p, forward
5′-AACTCCAGCTGGTCCTTAG-3′, reverse 5′-TCTTGAACCCTCATCCTGT-3′; and
U6, forward 5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse
5′-CGCTTCACGAATTTGCGT-3′. The expression levels were compared with
the 2−ΔΔCq method (20).
Plasmid transfection
MDA-MB-231 cells (1×105 cells/well) were
seeded in 24-well plates and transfected with 30 µM miR-142-5p
inhibitor or miR-negative control (NC) inhibitor using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) and
incubated at 37°C for 48 h according to the manufacturer's
protocol. MDA-MB-231 cells were randomly divided into three groups,
including the Control group (untreated cells), the miR-142-5p
inhibitor (5′-AGUAGUGCUUUCUACUUUAUG-3′; Guangzhou RiboBio Co.,
Ltd.) group and the miR-NC inhibitor (5′-CAGUACUUUUGUGUAGUACAA-3′;
Guangzhou RiboBio Co., Ltd.) group. Cells were collected at 48 h
after plasmid transfection for the following experiments.
Cell viability analysis
An MTT assay was conducted to evaluate cell
viability. MDA-MB-231 cells from each of the three groups were
seeded (3×104 cells/well) in 96-well plates and
incubated for 12, 24 and 48 h. Following the addition of MTT (5
mg/ml) into each well, cells were incubated for 1.5 h at 37°C.
Subsequently, the supernatant was discarded, 200 µl
dimethylsulfoxide was added to dissolve the formazan crystals and
the optical density was evaluated by reading the absorbance at 450
nm of each well with a spectrophotometer.
Cell apoptosis analysis
MDA-MB-231 cells (2×105 cells/well) were
seeded in 12-well plates and cultured for 48 h in an incubator at
37°C in a humidified atmosphere of 5% CO2. The cells
were collected by centrifugation at of 23,200 × g at 4°C for 5 min,
washed with cold PBS and fixed in ice-cold 70% ethanol overnight at
−20°C. Cells were subsequently stained with annexin V-fluorescein
isothiocyanate and propidium iodide (Roche Diagnostics, Basel,
Switzerland) for 15 min at room temperature in the dark. Cells were
collected and the percentage of cells with apoptotic nuclei (early
and late apoptosis) was calculated using a flow cytometer (Beckman
Coulter, Inc., Miami, FL, USA) and analyzed by Cell Quest software
version FCS2.0 (BD Biosciences, Franklin Lakes, NJ, USA).
Western blotting
Total protein was extracted from MDA-MB-231 cell
(1×105 cells/plate of 6-well plates) using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology, Shanghai, China). Protein concentration was
determined by a bicinchoninic acid kit (Beyotime Institute of
Biotechnology). Proteins (15 µg/lane) were separated by 10%
SDS-PAGE and transferred onto polyvinylidene difluoride membranes.
The membranes were blocked with 5% non-fat milk at room temperature
for 2 h and incubated with the following primary antibodies
overnight at 4°C: PTEN (1:1,000; cat. no. 9552; Cell Signaling
Technology, Inc., Danvers, MA, USA); phosphorylated
(p)-phosphoinositide 3-kinase (PI3K; 1:1,000; cat. no. 4228; Cell
Signaling Technology, Inc.); p-RACα serine/threonine-protein kinase
(AKT; 1:1,000; cat. no. D9E; Cell Signaling Technology, Inc.); and
GAPDH (1:1,000; cat. no. FL-355; Santa Cruz Biotechnology, Inc.,
Dallas, TX USA). Subsequently, membranes were incubated with
anti-rabbit horseradish peroxidase-conjugated secondary antibody
(1:1,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 2 h
at room temperature. Following washing with TBS + 0.1% Tween-20,
blots were visualized with an Enhanced Chemiluminescence kit
(Beyotime Institute of Biotechnology). Experiments were repeated in
triplicate. Protein expression levels were normalised to GAPDH, and
Quantity One version 4.6.2 (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) was used for densitometric analysis.
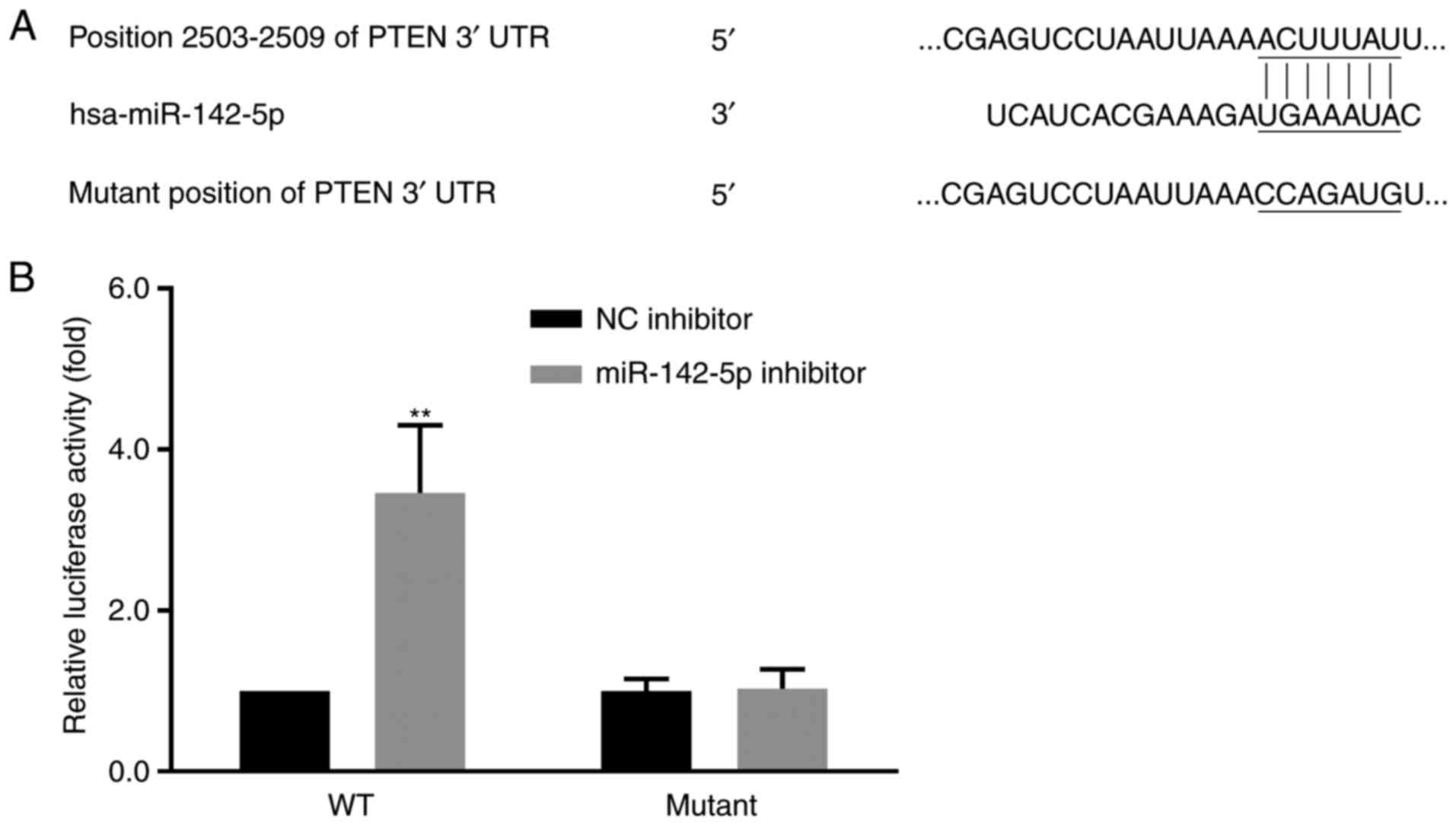
Luciferase activity assay
To verify whether PTEN was a direct target of
miR-142-5p, a position of the PTEN 3′UTR containing a putative
miR-142-5p target site was cloned into the luciferase open reading
frame. The plasmids of PTEN were cloned into reporter pLuc control
vector (Promega Corporation, Madison, WI, USA). MDA-MB-231 cells
(1×104 cells/plate) were seeded in 48-well plates and
incubated for 24 h in an incubator at 37°C in a humidified
atmosphere of 5% CO2, followed by co-transfection with
miR-142-5p inhibitor (20 nM) or miR-NC inhibitor (20 nM) and either
PTEN wild-type (WT)-3′ untranslated region (UTR; 1 mg, pLuc-PTEN-WT
3′UTR) or PTEN mutant (Mut)-3′UTR (1 mg, pLuc-PTEN-Mut 3′UTR) by
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
After 48 h transfection, the luciferase activity was examined by
Dual Luciferase Reporter Assay system (Promega Corporation,
Madison, WI, USA), and the results were normalized to the Renilla
luciferase activity.
Statistical analysis
Data were analyzed by software of SPSS version 21.0
(IBM Corp., Armonk, NY, USA) and expressed as the mean ± standard
error of mean. Data between two groups were analyzed by t-test and
data among three groups were analyzed by one-way analysis of
variance followed with Student-Newman-Keuls test. P<0.05 was
considered to indicate a statistically significance difference.
Results
Differing miR-142-5p expression levels
between tumoral tissues and adjacent healthy tissues
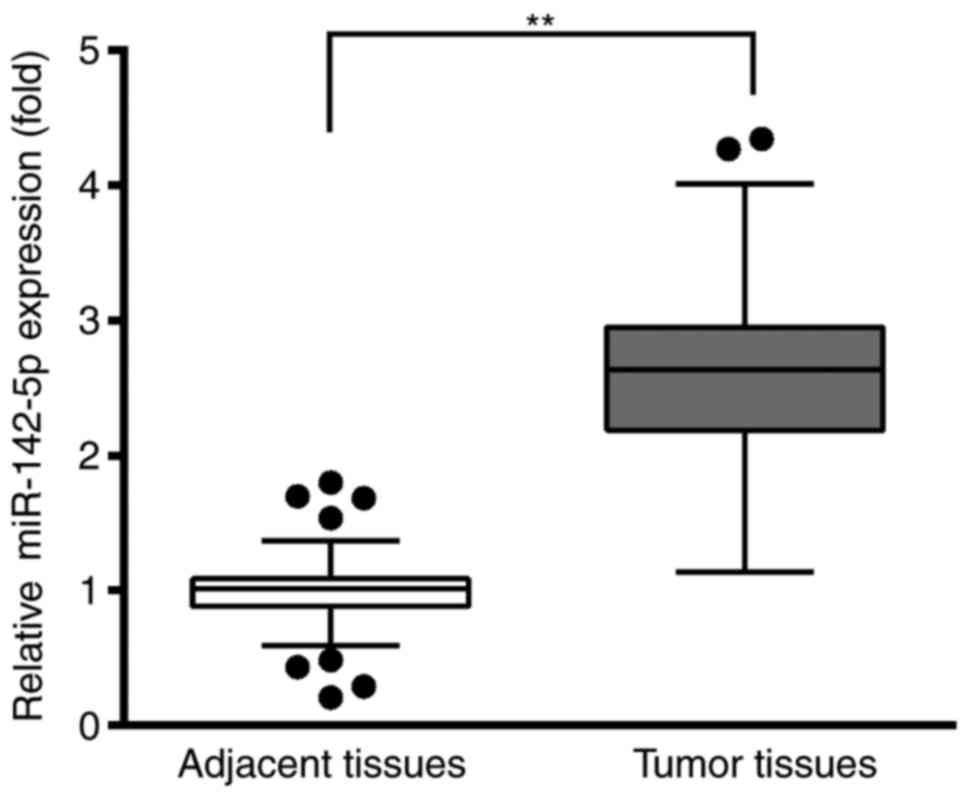
RT-qPCR was used to analyze the relative expression
levels of miR-142-5p in tumoral tissues and adjacent healthy
tissues. The results demonstrated that miR-142-5p expression levels
were significantly higher in the BC tissues compared with the
adjacent healthy tissues (Fig. 1).
This result suggested that miR-142-5p may be an oncogene during the
development of BC.
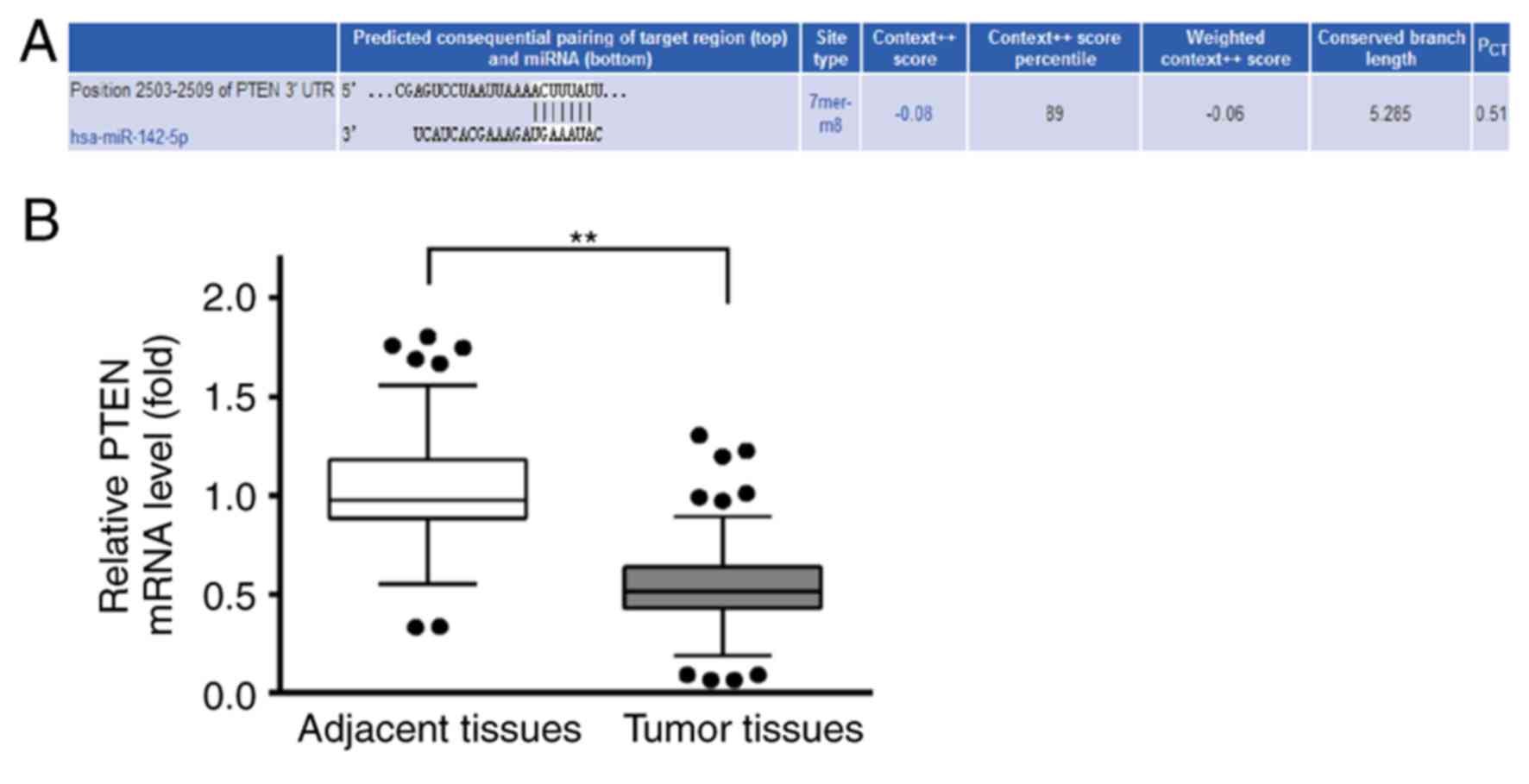
PTEN may be a target of miR-142
Potential targets of miR-142-5p were predicted by
TargetScan (http://www.targetscan.org/vert_71), and the analysis
revealed three target sequences of miR-142-5p in positions
2,189–2,195, 2,427–2,433 and 2,503–2,509 within the 3′UTR of PTEN
(data not shown). The target site at position 2,189–2,195 scored 59
(the lowest), the target sequence at position 2,427–2,433 scored 88
and at position 2,503–2,509 scored 89, which was highest (Fig. 2A). Therefore, position 2,503–2,509
of 3′UTR of PTEN was used for further analysis. RT-qPCR was used to
examine the relative expression levels of PTEN in BC tissues and
the adjacent healthy tissues. The results demonstrated that PTEN
mRNA expression levels were lower in BC tissues compared with
adjacent healthy tissues (Fig.
2B). This result suggested that PTEN may be a tumor suppressor
gene during the development of BC.
Association between miR-142-5p or PTEN
expression and patient clinicopathological characteristics
The characteristics of 60 patients with BC including
age, tumor size and metastasis were presented in Table I. Gene expression levels of
miR-142-5p and PTEN in BC tissues were investigated using RT-qPCR;
results were presented as fold change compared with the adjacent
healthy tissues. No significant associations were identified
between miR-142-5p or PTEN expression level and patient age.
Conversely, compared with tumor size <5 cm and patients with no
metastasis, miR-142-5p expression levels were significantly higher
(P=0.04), whereas PTEN expression levels were significantly lower
(P=0.01) in patients with tumors ≥5 cm and who exhibited
metastasis.
Relative expression levels of
miR-142-5p in MCF-10A, SK-BR-3 and MDA-MB-231 cell lines
RT-qPCR analysis was performed to investigate the
variations in miR-142-5p expression levels among three different
cell lines. The results indicated that miR-142-5p expression levels
were the lowest in MCF-10A normal breast epithelial cells, no
significant difference was observed between MCF-10A and SK-BR-3,
whereas the highest expression level was detected in MDA-MB-231 BC
cells (Fig. 3A). Therefore,
MDA-MB-231 cells were selected for further analysis in the present
study. The MDA-MB-231 cells were divided into three groups,
including the Control group, the miR-NC inhibitor group and the
miR-142-5p inhibitor group. The successful transfection of
miR-142-5p inhibitor into MDA-MB-231 cells was identified by the
reduced miR-142-5p expression level in the miR-142-5p
inhibitor-treated group compared with the other two groups
(Fig. 3B).
In addition, the effects of miR-142-5p inhibition on
PTEN expression were investigated by RT-qPCR. In MDA-MB-231 cells
transfected with miR-142-5p inhibitor, the mRNA expression levels
of PTEN were significantly increased compared with Control group
and miR-NC group (Fig. 3C). These
results suggested a potential the interaction between miR-142-5p
and PTEN.
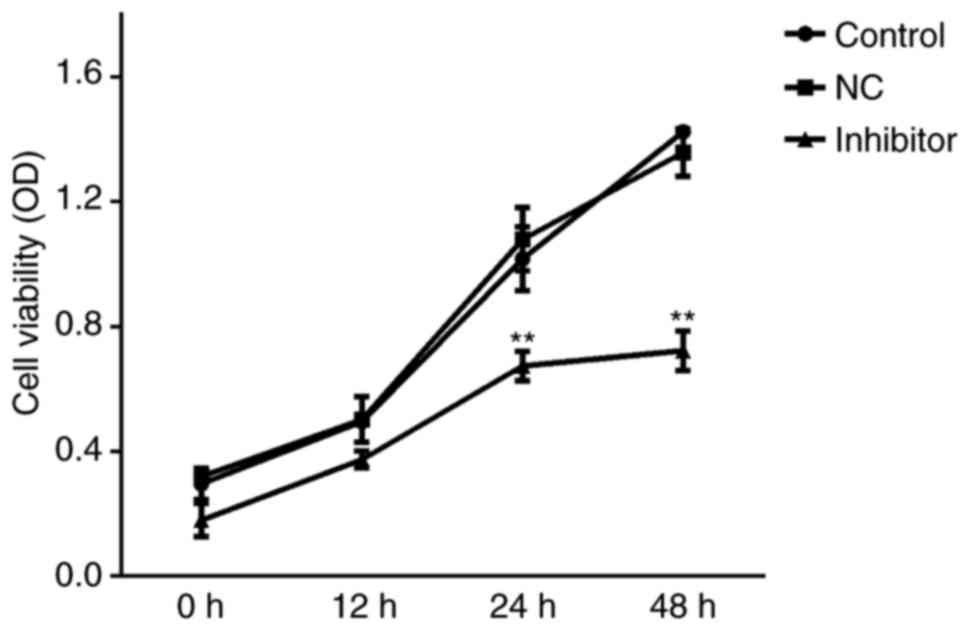
Effects of miR-142-5p inhibition on
MDA-MB-231 cell viability
MTT assays were conducted to examine the effects of
miR-142-5p inhibition on cell viability. No significant differences
in viability were identified between Control and miR-NC
inhibitor-treated groups; whereas the inhibition of miR-142-5p
significantly reduced MDA-MB-231 cell viability at 24 and 48 h
following transfection compared with Control and miR-NC inhibitor
groups (Fig. 4).
Effects of miR-142-5p inhibition on
MDA-MB-231 cell apoptosis
Flow cytometry results revealed no significant
differences between Control and miR-NC inhibitor groups, and that
the inhibition of miR-142-5p significantly increased MDA-MB-231
cell apoptosis (Fig. 5).
Effects of miR-142-5p inhibition on
PTEN, p-PI3K and p-AKT protein expression levels
The protein expression levels of PTEN, p-PI3K and
p-AKT were examined by western blot analysis, which revealed no
significant difference between the Control and miR-NC inhibitor
groups; the inhibition of miR-142-5p significantly increased the
expression of PTEN and reduced the expression of p-PI3K and p-AKT
in MDA-MB-231 cells compared with the Control and miR-NC
inhibitor-treated groups (Fig.
6).
Interaction between miR-142-5p and
PTEN
To verify whether PTEN was a direct target of
miR-142-5p, a portion of the PTEN 3′UTR containing a putative
miR-142-5p target site was cloned into the luciferase open reading
frame. Co-transfection of the miR-142-5p inhibitor and the
PTEN-3′UTR-WT vector into MDA-MB-231 cells (pLuc-PTEN-WT 3′UTR)
exhibited a significant increase in the luciferase activity
compared with miR-NC inhibitor. However, no significant difference
was identified in luciferase activity in cells co-transfected with
the miR-142-5p inhibitor and the mutant PTEN 3′UTR vector
(pLuc-PTEN-Mut 3′UTR); similar levels of luciferase activity were
observed in the cells harboring the mutant 3′UTR and WT-NC control
cells (Fig. 7).
Discussion
BC affects 12% women worldwide (3), thus it is urgent to investigate
current diagnostic methods (5).
miR-142 was upregulated in BCSCs compared with in non-tumorigenic
BC cells (14). Additionally,
miR-142-5p expression levels were reported to be higher in patients
with BC with lymph node metastasis compared with patients without
lymph node metastasis (15). The
present study aimed to investigate a target of miR-142-5p in BC
cells and provide a novel target for the treatment of BC.
RT-qPCR revealed that miR-142-5p expression levels
were significantly higher in BC tissues compared with adjacent
healthy tissues. These data suggested a potential oncogenic role of
miR-142-5p in BC, which was consistent with previous studies of BC
(14,15). Subsequently, TargetScan was
employed to predict the mRNA targets of miR-142-5p, which revealed
that the target sequence of miR-142-5p was present in the 3′UTR of
PTEN. In addition, RT-qPCR demonstrated that the expression levels
of PTEN were lower in BC tissues compared with adjacent healthy
tissues, which was in accordance with a previous report that PTEN
gene expression levels were lower in 93 human BC tissues compared
with in healthy breast tissues (21). These results suggested that the
overexpression of miR-142-5p may inhibit PTEN mRNA expression in BC
tissues.
The PTEN protein is a protein tyrosine phosphatase
that is encoded by the PTEN gene (22) and may be mutated in human brain,
breast and prostate cancers (23).
Subsequently, the association of miR-142-5p and PTEN expression
with clinicopathological characteristics in patient with BC were
investigated. No significant association between miR-142-5p or PTEN
expression levels and patient age was observed in present study;
however, miR-142-5p and PTEN expression levels were positively and
negatively associated, respectively, with patient tumor size and
metastasis. The results of the present study suggested that
miR-142-5p may have negatively affected the expression levels of
PTEN in BC tissues. Additionally, in vitro experiments were
conducted to investigate the effect of miR-142-5p expression in
human BC cell lines. The MDA-MB-231 BC cell line exhibited a high
expression level of miR-142-5p, which was reduced upon transfection
with the miR-142-5p inhibitor, whereas mRNA expression levels of
PTEN increased. These data further suggested an interaction and a
negative association between miR-142-5p and PTEN expression.
Results from the present study demonstrated that
inhibition of miR-142-5p significantly reduced MDA-MB-231 cell
viability and increased apoptosis. These data were in line with
previous studies that have reported an upregulation of miR-142-5p
expression levels in breast cancer patients with lymph node
metastasis compared with those without lymph node metastasis
(15), atherosclerotic plaques
(16), Hashimoto's thyroiditis
(18) and in the serum of patients
with colorectal cancer (19),
which was associated with the promotion of cell proliferation and
colony formation, and with the inhibition of apoptosis in
colorectal cancer cell lines by targeting succinate dehydrogenase
(ubiquinone) iron-sulfur subunit B (19). miR-142-5p was also reported to be
elevated in human renal cell carcinoma tissues, and was
demonstrated to induce growth and migration of renal cell carcinoma
cells by targeting BTG anti-proliferation factor 3 (24); however, the mechanisms responsible
for the alterations in BC required further investigation.
Therefore, western blotting was performed in the present study to
analyze protein expression levels of PTEN; in cells transfected
with the miR-142-5p inhibitor, a significant increase in the
expression of PTEN was observed. These results suggested that the
expression of PTEN may be affected by miR-142-5p. The present study
also aimed to determine the molecules that may be regulated and
influenced by PTEN by using western blot analysis.
The PI3K signaling pathway is an important kinase
cascade that regulates cell proliferation, migration, apoptosis and
angiogenesis through its downstream effector AKT; dysregulation of
this pathway may be caused by mutations or altered expression
levels of an upstream regulator of AKT activity such as PTEN
(25,26). Previous studies have also reported
that mutations in PI3Kα were common in BC and the mutation
frequency ranged between 27 and 36% (27). Additionally, PTEN was reported to
be a regulator of PI3K cytoplasmic signaling (28) and an inhibitor of the
growth-promoting PI3K/AKT/mechanistic target of rapamycin (mTOR)
signaling pathway (29).
Furthermore, a lack of PTEN protein expression was reported to be
associated with overactivation of the PI3K/AKT/mTOR signaling
pathway (30). In the present
study, western blotting revealed that the inhibition of miR-142-5p
significantly reduced the expression of p-PI3K and p-AKT in
MDA-MB-231 cells, which agreed with the above studies that further
inferred that PI3K or AKT expression was attributed to PTEN.
Results from the luciferase activity assay
demonstrated that PTEN was a direct target of miR-142-5p, confirmed
by higher luciferase activity after miR-142-5p inhibitor
administration in WT group than in the mutant group. Additionally,
similar levels of luciferase activity were observed in the cells
harboring the WT 3′UTR between the miR-142-5p inhibitor group and
miR-NC group in the present study. In a recent study, miR-142-5p
was found to target 3′UTR of PTEN in cutaneous squamous cell
carcinoma by Bai et al (31), which was in line with the findings
of the present study.
In conclusion, miR-142-5p may be a possible target
for treating BC in the future; the PTEN/p-PI3K/p-AKT signaling
pathway was associated with the effects of miR-142-5p in MDA-MB-231
cells. However, the effects of miR-142-5p mimics on BC were not
analyzed in the present study and should be investigated in the
future; the effects on PI3K or AKT expression attributed to PTEN,
can only be inferred, as no direct evidence has been provided. It
is necessary for this to be explored in future studies.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WX designed and carried out the experiments and
analyzed the data. WW designed the experiments, analyzed the data
and prepared the manuscript. Both authors have seen and approved
the manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of Renmin Hospital of Wuhan University (Wuhan,
China).
Consent for publication
Written informed consent was obtained from all
patients prior to the start of the study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Siegel R, Bandi P and Jemal A:
Breast cancer statistics, 2011. CA Cancer J Clin. 61:409–418. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McGuire A, Brown JA, Malone C, McLaughlin
R and Kerin MJ: Effects of age on the detection and management of
breast cancer. Cancers (Basel). 7:908–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Veronesi U, Boyle P, Goldhirsch A,
Orecchia R and Viale G: Breast cancer. Lancet. 365:1727–1741. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang BN, Cao XC, Chen JY, Chen J, Fu L,
Hu XC, Jiang ZF, Li HY, Liao N, Liu DG, et al: Guidelines on the
diagnosis and treatment of breast cancer (2011 edition). Gland
Surg. 1:39–61. 2011.
|
|
6
|
Farazi TA, Hoell JI, Morozov P and Tuschl
T: MicroRNAs in human cancer. Adv Exp Med Biol. 774:1–20. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nohata N, Hanazawa T, Enokida H and Seki
N: microRNA-1/133a and microRNA-206/133b clusters: Dysregulation
and functional roles in human cancers. Oncotarget. 3:9–21. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Melo SA and Esteller M: Dysregulation of
microRNAs in cancer: Playing with fire. FEBS Lett. 585:2087–2099.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lerebours F, Cizeron-Clairac G, Susini A,
Vacher S, Mouret-Fourme E, Belichard C, Brain E, Alberini JL,
Spyratos F, Lidereau R and Bieche I: miRNA expression profiling of
inflammatory breast cancer identifies a 5-miRNA signature
predictive of breast tumor aggressiveness. Int J Cancer.
133:1614–1623. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu
H, Liu MF and Wang ED: MicroRNA-155 functions as an OncomiR in
breast cancer by targeting the suppressor of cytokine signaling 1
gene. Cancer Res. 70:3119–3127. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J
and Zhang HT: MiR-142-3p represses TGF-β-induced growth inhibition
through repression of TGFβR1 in non-small cell lung cancer. FASEB
J. 28:2696–2704. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen WW, Zeng Z, Zhu WX and Fu GH:
MiR-142-3p functions as a tumor suppressor by targeting CD133,
ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl).
91:989–1000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chai S, Tong M, Ng KY, Kwan PS, Chan YP,
Fung TM, Lee TK, Wong N, Xie D, Yuan YF, et al: Regulatory role of
miR-142-3p on the functional hepatic cancer stem cell marker CD133.
Oncotarget. 5:5725–5735. 2001.
|
|
14
|
Isobe T, Hisamori S, Hogan DJ, Zabala M,
Hendrickson DG, Dalerba P, Cai S, Scheeren F, Kuo AH, Sikandar SS,
et al: miR-142 regulates the tumorigenicity of human breast cancer
stem cells through the canonical WNT signaling pathway. eLife.
3:e019772014. View Article : Google Scholar :
|
|
15
|
Wang B, Li J, Sun M, Sun L and Zhang X:
miRNA expression in breast cancer varies with lymph node metastasis
and other clinicopathologic features. IUBMB Life. 66:371–377. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu R, Bi C, Song J, Wang L, Ge C, Liu X
and Zhang M: Upregulation of miR-142-5p in atherosclerotic plaques
and regulation of oxidized low-density lipoprotein-induced
apoptosis in macrophages. Mol Med Rep. 11:3229–3234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su YH, Zhou Z, Yang KP, Wang XG, Zhu Y and
Fa XE: MIR-142-5p and miR-9 may be involved in squamous lung cancer
by regulating cell cycle related genes. Eur Rev Med Pharmacol Sci.
17:3213–3220. 2013.PubMed/NCBI
|
|
18
|
Zhu J, Zhang Y, Zhang W, Zhang W, Fan L,
Wang L, Liu Y, Liu S, Guo Y, Wang Y, et al: MicroRNA-142-5p
contributes to Hashimoto's thyroiditis by targeting CLDN1. J Transl
Med. 14:1662016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu S, Xiao Z, Ai F, Liu F, Chen X, Cao K,
Ren W, Zhang X, Shu P and Zhang D: miR-142-5p promotes development
of colorectal cancer through targeting SDHB and facilitating
generation of aerobic glycolysis. Biomed Pharmacother.
92:1119–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Liu M, Yang H, Wang J, Liu H and
Li X, Li J, Xu J and Li X: PIK3CA mutations are a predictor of
docetaxel plus epirubicin neoadjuvant chemotherapy clinical
efficacy in breast cancer. Neoplasma. 61:461–467. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song MS, Salmena L and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor. Nat Rev Mol
Cell Biol. 13:283–296. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Yen C, Liaw D, Podsypanina K, Bose
S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al:
PTEN, a putative protein tyrosine phosphatase gene mutated in human
brain, breast, and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu L, Liu S, Duan Q, Chen L, Wu T, Qian
H, Yang S, Xin D, He Z and Guo Y: MicroRNA-142-5p promoted cell
growth and migration in renal cell carcinoma by targeting BTG3. Am
J Transl Res. 9:2394–2402. 2017.PubMed/NCBI
|
|
25
|
Stemke-Hale K, Gonzalez-Angulo AM, Lluch
A, Neve RM, Kuo WL, Davies M, Carey M, Hu Z, Guan Y, Sahin A, et
al: An integrative genomic and proteomic analysis of PIK3CA, PTEN,
and AKT mutations in breast cancer. Cancer Res. 68:6084–6091. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nagata Y, Lan KH, Zhou X, Tan M, Esteva
FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, et al: PTEN
activation contributes to tumor inhibition by trastuzumab, and loss
of PTEN predicts trastuzumab resistance in patients. Cancer Cell.
6:117–127. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Board RE, Thelwell NJ, Ravetto PF, Little
S, Ranson M, Dive C, Hughes A and Whitcombe D: Multiplexed assays
for detection of mutations in PIK3CA. Clin Chem. 54:757–760. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Milella M, Falcone I, Conciatori F, Incani
Cesta U, Del Curatolo A, Inzerilli N, Nuzzo CM, Vaccaro V, Vari S,
Cognetti F and Ciuffreda L: PTEN: Multiple functions in human
malignant tumors. Front Oncol. 5:242015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baselga J: Targeting the
phosphoinositide-3 (PI3) kinase pathway in breast cancer.
Oncologist. 16 Suppl 1:S12–S19. 2011. View Article : Google Scholar
|
|
30
|
Carracedo A and Pandolfi PP: The PTEN-PI3K
pathway: Of feedbacks and cross-talks. Oncogene. 27:5527–5541.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bai X, Zhou Y, Chen P, Yang M and Xu J:
MicroRNA-142-5p induces cancer stem cell-like properties of
cutaneous squamous cell carcinoma via inhibiting PTEN. J Cell
Biochem. 119:2179–2188. 2018. View Article : Google Scholar : PubMed/NCBI
|