Introduction
Split-hand/split-foot malformation (SHFM) is a rare
congenital limb defect with a wide phenotypic spectrum and high
genetic heterogeneity. SHFM is primarily characterized by a deep
median cleft of the hand and/or foot due to the poor
differentiation of the apical ectodermal ridge (AER) during early
embryonic development. Typical clinical manifestations include the
following: ectrodactyly of the digits; presence of a wedge-shaped
cleft on the palm (also known as crab-claw or lobster-claw
anomaly); hypoplasia of the phalanges, metacarpals, and
metatarsals; and polydactyly or syndactyly. Absence of the radial
axis without cleft and monodactyly of the unaffected fifth digits
have also been reported (1). To
date, six different genetic loci of SHFM have been identified
(2–6). Based on the different clinical
manifestations of the affected organs, SHFM is classified as an
isolated trait, as an asymptomatic disease (OMIM: 1863600), or part
of a multiple congenital anomaly syndrome. Non-syndromic SHFM
occurs as a sporadic deformity or as part of a syndrome associated
with other limb defects, such as split-hand/foot malformation with
long bone deficiency (SHFLD; OMIM: 119100), including tibial
aplasia (7) (Table I). In general, the SHFM phenotypes
of the different affected loci show no significant differences,
making the genetic diagnosis of SHFM more challenging.
 | Table I.Human genetic mapping of SHFM and
SHFLD. |
Table I.
Human genetic mapping of SHFM and
SHFLD.
| Phenotype | Omim database
number | Location | Gene/locus | Mode of
inheritance |
|---|
| SHFM1 | OMIM 183,600 | 7q21 | DLX5, DLX6,
DSS1 | Autosomal
dominant |
| SHFM2 | OMIM 246,560 | Xq26 | FGF13 | X-linked
recessive |
| SHFM3 | OMIM 246,560 | 10q24 | HOX11,
FGF8 | Autosomal
dominant |
| SHFM4 | OMIM 605,289 | 3q27 | TP63
(p63), | Autosomal
dominant |
| SHFM5 | OMIM 606,708 | 2q31 | HOXD13 | Autosomal
dominant |
| SHFM6 | OMIM 183,600 | 12q13 | WNT10b | Autosomal
recessive |
| SHFLD1 | OMIM 119,100 | 1q42.2-q43 |
| Autosomal
dominant |
| SHFLD2 | OMIM 610685 | 6q14.1 |
| Autosomal
dominant |
| SHFLD3 | OMIM 612,576 | 17p13.3 | BHLHA9 | Autosomal
dominant |
The TP63 protein plays a significant role as
a transcription factor involved in limb, epithelial, and
craniofacial formation during the development of the mammalian
embryonic endoderm (8).
Approximately 10% of isolated SHFM4 cases are attributed to
mutations in the human TP63 gene. In addition, TP63
mutations were detected in 93% of patients with
ectrodactyly-ectodermal dysplasia-cleft (EEC) syndrome (9,10).
All clinical conditions related to TP63 mutations exhibit
features that largely overlap with those of the EEC syndrome,
thereby increasing the difficulty of diagnosis (11). In this study, we investigated the
phenotype and genetic mechanisms underlying SHFM in a Chinese
family with two members exhibiting isolated SHFM.
Subjects and methods
Clinical data of propositus and
familial members
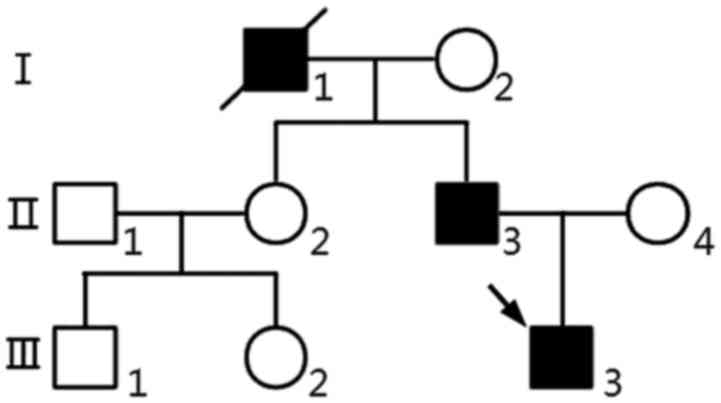
We performed whole-exome sequencing (WES) of proband
(III3) using the Complete Genomics (CG) platform. Clinical data of
propositus and familial members (Fig.
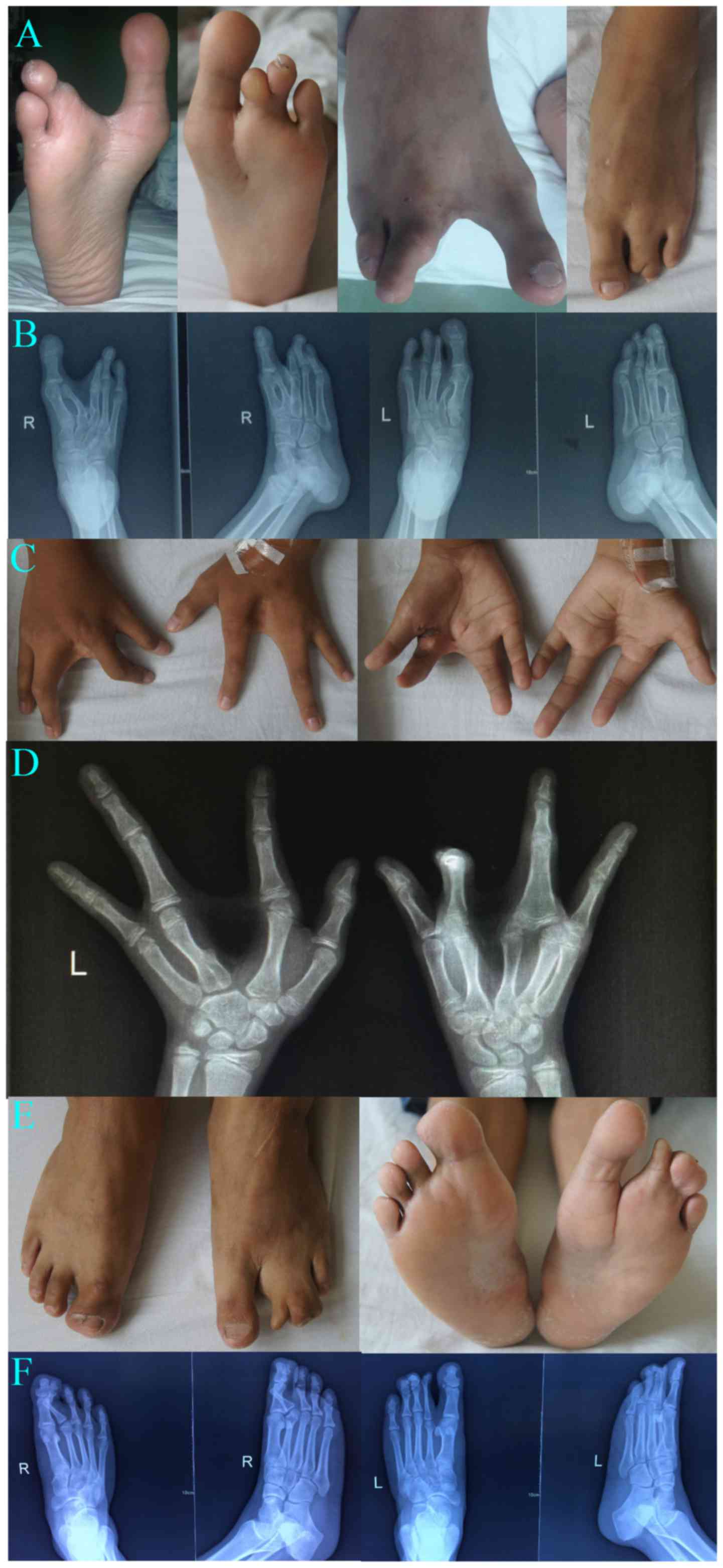
1), Clinical characteristics of the proband (III3) (Fig. 2A-D) are as follows. The proband is
a 14-year-old male who was admitted to the hospital mainly because
of deformities in both hands and feet for 12 years and
camptodactyly of the second finger of the right hand for 11 years.
Physical examination showed spinal physiological curvature without
lateral bending. He was diagnosed with SHFM with only four digits
in both hands and feet and with hands splitted into the ulnar and
radial parts. Left hand fingers and bilateral toes functioned
normally. Striated cicatricial contracture was observed in the 1st
finger web and the second finger of the right hand, with
camptodactyly of the second finger, a deep finger web between the
separated 3rd and 4th metacarpal. X-ray results showed the absence
of the 3rd phalange bilaterally and 3rd metacarpus of the left
hand. The capitate of the left hand was significantly larger than
that of the right hand, with the fourth proximal phalanx obviously
expanded and formed a joint with the third and fourth metacarpals.
Osteochondroma was present on the fourth proximal radial metacarpal
of the left hand. The second toes were absent bilaterally, and only
two sections of phalanxes of the third toes were present, which
were accompanied by blurred and narrowed related
metatarsophalangeal joint space. The second metatarsals of the left
foot were smaller, especially the distal end, and was accompanied
by deformity of the second metatarsal of the right foot.
Osteochondroma was formed on the basal and proximal part of the
second metatarsal of the right side, respectively. Pseudoarthrosis
was present in the third metatarsal and the first phalanges,
indicating deformity of both hands and feet with partial bone
dysplasia.
Clinical manifestations of (II3) are as follows
(Fig. 2D and F). The proband was
diagnosed with SHFM and had five digits in both feet. X-ray results
showed that the proximal joints of the second toe of the right side
was buckling and fused with the soft tissue of the first toe. In
addition, the first and second toes of the left foot were
separated. The proximal soft tissues of the second to fourth toes
of the left foot were fused. The second, third, and fourth distal
soft tissues of the left foot were separated. The rest of the bone
cortex was intact and showed continuous trabecular bone. There were
no obvious abnormalities in bone structure, joint relationship, and
shapes of the hand and hand joints. The two patients and their
family members did not exhibit deafness, mental retardation, and
external body malformations, such as face, palate, anadontia, and
other obvious deformities.
DNA extraction
Genomic DNA was extracted from peripheral blood
samples (QIAamp DNA Blood Mini kit; Qiagen, Hilden, Germany). The
concentration and purity of the DNA extracts were determined using
a NanoDrop 1000 instrument (Nanodrop Technologies; Thermo Fisher
Scientific, Inc., Pittsburgh, PA, USA). All procedures were
performed in accordance to the tenets of the Declaration of
Helsinki and approved by the Ethics Committee of Fujian Provincial
Hospital (Fuzhou, China). All participants and legal guardians of
the minors involved in the present study provided written informed
consent.
CG whole exome sequencing
The proband (III3) was examined via WES using the CG
platform (Complete Genomics, Inc., San Jose, CA, USA) for next
generation sequencing (NGS). First, Covaris was used to randomly
fragment the genomic DNA (gDNA). Fragments ranging from 200 to 400
bp were selected after two rounds of bead purification. Next, the
AdA 5′- and 3′-adaptors were ligated to the 5′- and 3′-ends of the
fragments, respectively, before PCR amplification. The PCR products
were then subjected to exon capture. Captured exon fragments were
purified via DynabeadsM-280 Streptavidin Bead purification and
further amplified by another round of PCR. The products were
cyclized to generate double-strand (ds) circles, which were
digested with Ecop15I. Small fragments were collected after bead
purification. Following the same procedure as in AdA adaptor
ligation, AdB adaptors were also ligated to both ends of the
purified fragments. The fragments then underwent single-strand (ss)
cyclization. The resulting ss circles were used as the final
library products for sequencing on the CG Black Bird platform.
Finally, high-throughput sequencing was performed for each captured
library to ensure that each sample meets the desired average
sequencing depth.
Bioinformatics analysis
After base calling, reads sequence of each DNA
nano-balls (DNBs) are derived. Initial mapping is conducted by an
in-house mapping tool, Teramap, developed by Complete Genomics,
Inc.. Based on the initial mapping results, the regions which are
deemed to differ from the reference genome are identified. Then
individual reads that lie in those regions are chosen to perform a
local de novo assembly. The assembly results are converted together
with the initial mapping into a BAM file which only includes mapped
reads. Next according to the initial mapping and the assembly
results, a probability statistical model is adopted to acquire
variants. Variants extracted from the model with a probability
higher than a significant threshold are reported. Finally, small
variants with high confidence are selected and annotated. A strict
data analysis quality control system (QC) is built throughout the
whole analysis (12–14).
Sanger DNA sequencing
Variants were confirmed using Sanger DNA sequencing
in this family (I2, II1-4 and III1-3). Primers for amplification of
the target sequences were designed using Premier 5 software and
synthesized by Thermo Fisher Scientific, Inc. (Shanghai, China).
The TP63 gene sequence was obtained from GenBank
(NM_003722.4), and the length of the target sequence was 226 bp and
the primers were synthesized by Thermo Fisher Scientific, Inc. The
following primer sequences were used for amplification:
TP63-F: 5′-GACATGCCCCATCCAGATCAA-3′ and TP63-R:
5′-AGGTGGGTCTCAAACAAAAATGC-3′. PCR products were purified using the
Omega E.Z.N.A.™ Gel Extraction kit (Omega Bio-tek, Norcross, GA,
USA) according to the manufacturer's instructions. Sanger
sequencing was performed using the BigDye Terminator v1.1 kit
following the manufacturer's instructions and on a 3730xl DNA
Analyzer (Thermo Fisher Scientific Inc.).
Results
Statistics of whole exome
sequencing
WES of one DNA sample produced an average of
503,436,416 DNBs using the CG platform. Duplicate DNBs, DNBs with
too many good reference mappings, and DNBs with no consistent mate
mappings were filtered out prior to variant calling. After
filtering, a total of 475,501,541 DNBs were used as input for local
de novo assembly and variant detection. In this study,
58.97-Mb target regions were captured, and an average of 11.56 GB
of mapped bases were generated per individual. On average, 99.53%
of the target bases were sequenced with at least 1X coverage per
sample, and 97.34% of the bases were sequenced with at least 10X
coverage per sample.
A total of 43,846 SNPs were identified in all
individuals. Furthermore, 97.37% of all variants were represented
in dbSNP, while 95.64% were annotated in the 1000 Genomes Project
database. We identified 755 novel SNPs with a
transition-to-transversion ratio of 2.61. Of all SNPs, 10,280 were
synonymous mutations, while 9,442 were missense mutations. A total
of 34 SNPs were stop-loss, 66 stop-gain, and 16 start-loss
mutations. In addition, 65 SNPs were located in splice sites.
A total of 3,591 indels were identified in all
samples. Of these, 80.76% were represented in dbSNP and 66.33% were
annotated in the 1000 Genome Project database. The analysis
identified 608 novel indels. Of all the indels, 178 were
frameshift, three were stop-loss, three were start-loss, and 44
were splice-site mutations. A total of 1,702 point mutations had
maf ≤1%, which included 1,056 amino acid substitutions and splice
mutations and 107 indel mutations.
Determination of suspected pathogenic
mutations in the propositus
The DNBs of each sample were compared with the
reference human genome sequence (GRCh37/HG19). Based on the
candidate genes listed in Table I,
we performed direct screening to identify putative mutation sites.
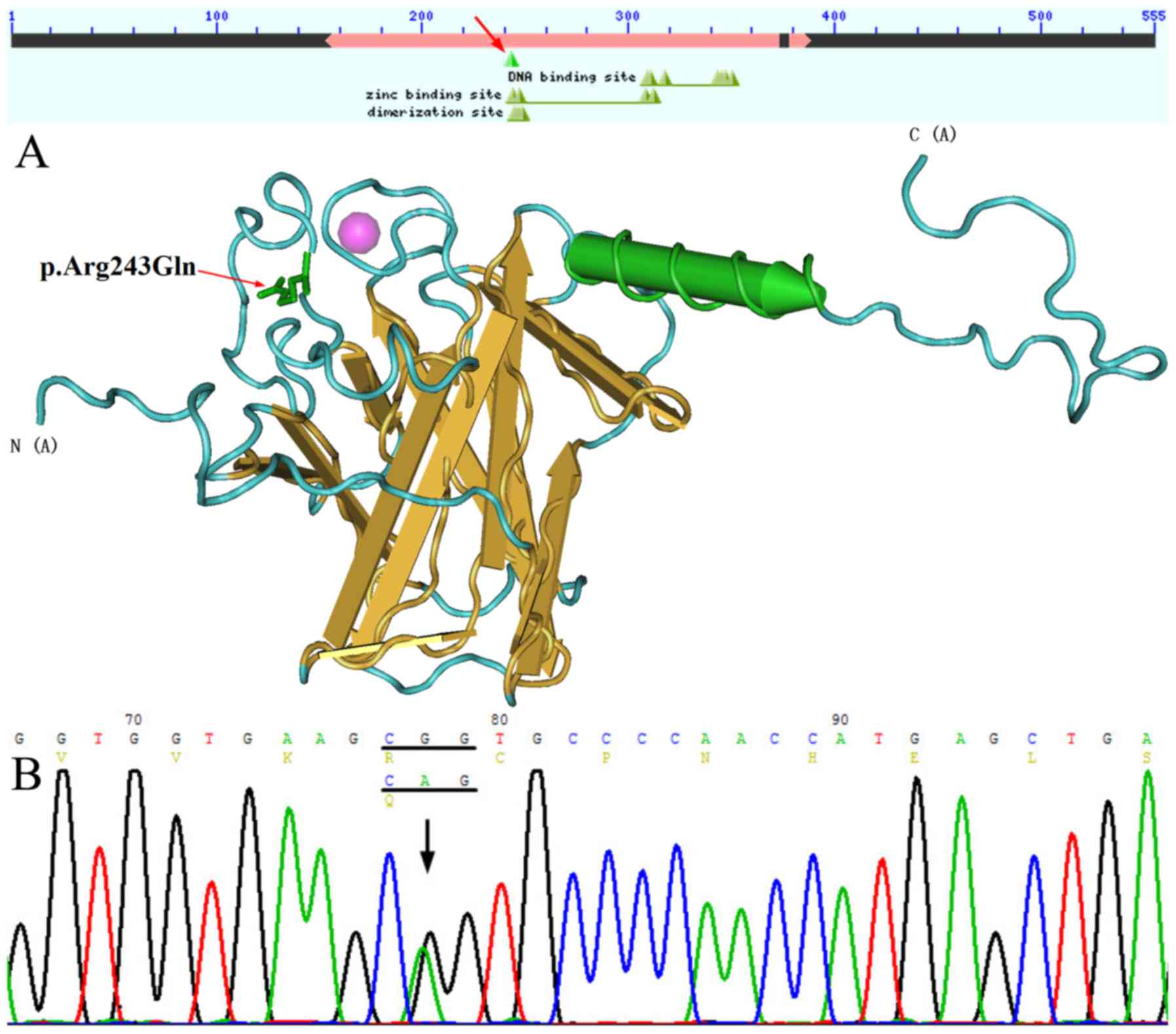
The c.728G>A (p.Arg243Gln) (rs121908836) mutation located within
TP63 was detected in the proband (III3) and his father
(II3), who was diagnosed with SHFM. A CGG-to-CAG mutation
(c.728G>A) in exon 5 predicted an arginine-to-glutamine
substitution at amino acid position 243 (R243Q), which is located
in the zinc binding site and dimerization site of TP63
(15,16) (Fig.
3). This missense variant was predicted to be pathogenic by
PolyPhen-2 and was not detected in other family members. In
addition, a synonymous variant, c.1059C>T
(p.His353His)/(rs1051886), was identified in the candidate gene
WNT10b of the proband.
Discussion
SHFM disorders are highly heterogeneous and exhibit
variable clinical manifestations, which are attributed to multiple
loci and various inheritance modes. TP63 is an important
functional gene that is involved in tissue development and
apoptosis. Mutations in human TP63 can lead to developmental
disorders, including the EEC syndrome, ankyloblepharon-ectodermal
defects-cleft lip/palate syndrome (AEC), limb-mammary syndrome
(LMS), acro-derma-to-ungual-lacrimal-tooth syndrome (ADULT),
Rapp-Hodgkin syndrome (RHS), SHFM4, and nonsyndromic cleft lip
(NSCL) (17). Similarly, previous
studies have reported that EEC syndrome in a small number of
families were caused by a p.Arg243Gln mutation in TP63
(5,9). We first discovered the isolated SHFM4
genetic phenotype corresponding to this point mutation, which is
characterized by congenital ectrodactyly, syndactyly, without
prosopo-cleft, and ectodermal dysplasia.
The majority of EEC syndrome cases are caused by a
missense mutation in the core DNA binding domain (DBD) of
TP63, thereby disrupting its ability to translate proteins
normally. On the other hand, only 10% of the nonsyndromic SHFM
cases are attributed to TP63 mutations. Mutations in the
domain of the TP63 gene can lead to isolated/nonsyndromic
SHFM but also result in syndrome SHFM, which indicated that
morphological classification may not be reliable for accurate
diagnosis (1). The mutation
spectrum of the EEC syndrome reflects a specific pathogenetic
mechanism. Several studies have provided strong evidence that
codons 204, 279, 280, and 304, could result in both EEC and SHFM
(5,18). Mutations in codon 204 are
consistent with our current results showing the effects of the
R243Q mutation (rs121908836, NM_003722.4). The above findings
demonstrate the partial overlap between the mutational spectra of
EEC and SHFM. ADULT and EEC are generally caused by missense
mutations in the DBD, whereas AEC and LM syndromes are caused by
missense mutations in the SAM region and in other nearby regions.
SHFM is caused by mutations in various regions of TP63.
Different mutations have been shown to result in different effects,
indicating that TP63 is involved in various developmental
functions (8,9). So far, p.Arg97Cys, p.Lys233Glu, and
p.Arg319Cys mutations in TP63 were identified in isolated
SHFM4 patients (19,20). The p.Arg97Cys and p.Lys233Glu
mutations occur in the N-terminal transactivation domain (TA)
domain, whereas p.Arg319Cys mutations are found in DBD loci. The
Human Gene Mutation Database (HGMD) reported that missense variants
in position 243 (R243Q, R243W, and R243L) and in nearby residues
(V241M, H247Y, H247D, and H247R) are associated with
TP63-related disorders, thereby supporting the functional
importance of this TP63 region. The most common phenotypes
of patients harboring mutations in position R243 include hair,
lacrimal duct, and nail defects, which can be observed in the EEC3
syndrome (604292) (21).
All amino acids of p53 that directly bind DNA are
conserved in both TP63 and p73. The transcription factors
TP63 and p73 belong to the p53 family and have been
predicted to perform similar functions. The human TP63 gene
is located in the 3q27-3q29 chromosomal region, which encodes
specific domains (22,23), including the TA domain, DBD, and
oligomerization domain (OD). Unlike p53, TP63 contains a
unique N-terminal SAM (sterile alpha motif) region, which is also
found in many signaling proteins involved in cell development and
differentiation (24,25).
P73 and TP63 encode various homologs
which differ based on the carboxyl termini. In particular, the α,
β, and γ isoforms exhibit diverse biological characteristics
(22). Under the influence of the
P1 and P2 promoters, P73 and TP63 respectively
transcribe two isomers, namely, Tap73/63, which contains the TA
region, and the N-terminal truncated isomer ∆Np73/63. These two
proteins have opposing biological characteristics; Tap73/63
promotes p53 function, whereas ∆Np73/63 antagonizes p53 function
(26). The zinc binding site and
dimerization site are located in the TA region. The TAp63 subtype
can induce developmental cell apoptosis. p53, pTP63, and p73
are involved in similar but distinct physiological processes.
P53-knockout mice may exhibit high frequency of spontaneous
tumors during development. The active p53 protein does not
participate in physiological apoptosis during the entire embryonic
development process, whereas TP63- and P73-knockout
mice exhibit specific limb and epithelial developmental
malformations without inducing the formation of spontaneous tumors
(27).
∆Np63 subtypes are primarily expressed in late
embryos and during postnatal epidermal development. The gene
knockout mouse model demonstrated that the ∆Np63 protein is
essential for maintaining the integrity of the epidermal basal
layer, final differentiation of keratinocytes, and initial
stratification of the epithelium during embryonic development
(28,29). These processes can be induced by
activation of the ∆Np63 gene during epidermal differentiation. The
∆Np63 mutation not only determines ectodermal fate, but also
influences ectodermal embryonic stem cell proliferation and
epidermal formation, which are important in maintaining the
proliferation potential of epidermal stem cells in the mature
epithelium (30). In
TP63-deficient mice, multiple layers of regenerated
epithelial stem cells can be inactivated and can undergo asymmetric
division (31). Knockout mice
overexpress ∆Np63α in the skin, which leads to characteristic
changes, such as delayed wound healing, reduced skin thickness,
decreased subcutaneous adipose tissue, hair loss, reduced cell
proliferation, and accelerated skin aging, some of which can be
ameliorated by Sirt1 regulation (32).
The p.R243Q mutation in the TA region affects the
binding of zinc ions, which can lead to incorrect folding of the
protein and impair tap63 function. In turn, reduced tap63 function
leads to ∆Np63 overexpression, which inhibits apoptosis by
downregulating the expression of pro-apoptotic genes (33) and ultimately affects the
development of the epidermis. In addition, SHFM can produce the
ectrodactyly phenotype and mainly effects the development of
central rays of the autopod. The most common cause of SHFM is
interference in the AER signaling pathway. Reduced AER signaling
promotes AER cell death or inhibits cell proliferation. As a
result, the activity of central AER cannot be maintained, which
directly causes distal limb defects. AER abnormalities occur during
limb development (34,35).
A few studies have reported the role of TP63
in SHFM, and some case reports have implicated mutations in the
zinc binding site of TP63 in SHFM. In the present study, we
demonstrated that the R243Q mutation in the TP63 gene
produces a new phenotype called SHFM4, thereby demonstrating the
mutational overlap between EEC and SHFM4. The genetic and clinical
heterogeneity of SHFM significantly increases the difficulty of
genetic counseling. Therefore, identifying the genetic alterations
that are responsible for SHFM in individual patients is of
practical importance.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the
Financial Scheme for Young Talents Training Program of Fujian
Health industry (grant no. 2015-ZQN-ZD-7), Fujian Provincial
Natural Science Fund Project (grant no. 2016J01501) and Fujian
Provincial Health and Family Planning Youth Research Program, China
(grant no. 2016-1-84).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL conceived and designed the study and drafted the
manuscript. XY, XL, YZ and GL performed the data collection,
statistical analyses and drafted the manuscript. The final version
of the manuscript was read and approved by all authors.
Ethics approval and consent to
participate
All procedures were performed in accordance to the
tenets of the Declaration of Helsinki and the study was approved by
the Ethics Committee of Fujian Provincial Hospital (Fuzhou, China).
All participants and legal guardians of the minors involved in the
present study provided written informed consent.
Consent for publication
Written informed consent was obtained for the
publication of the participants data and clinical images.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Basel D, Kilpatrick MW and Tsipouras P:
The expanding panorama of split hand foot malformation. Am J Med
Genet A. 140:1359–1365. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Duijf PH, van Bokhoven H and Brunner HG:
Pathogenesis of split-hand/split-foot malformation. Hum Mol Genet.
12:R51–R60. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Amalnath SD, Gopalakrishnan M and Dutta
TK: Split-hand/feet malformation in three tamilian families and
review of the reports from India. Indian J Hum Genet. 20:92–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Klar AJ: Split hand/foot malformation
genetics supports the chromosome 7 copy segregation mechanism for
human limb development. Philos Trans R Soc Lond B Biol Sci.
371:201504152016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Celli J, Duijf P, Hamel BC, Bamshad M,
Kramer B, Smits AP, Newbury-Ecob R, Hennekam RC, Van Buggenhout G,
van Haeringen A, et al: Heterozygous germline mutations in the p53
homolog p63 are the cause of EEC syndrome. Cell. 99:143–153. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khan S, Basit S, Zimri FK, Ali N, Ali G,
Ansar M and Ahmad W: A novel homozygous missense mutation in WNT10B
in familial split-hand/foot malformation. Clin Genet. 82:48–55.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Armour CM, Bulman DE, Jarinova O, Rogers
RC, Clarkson KB, DuPont BR, Dwivedi A, Bartel FO, McDonell L,
Schwartz CE, et al: 17p13.3 microduplications are associated with
split-hand/foot malformation and long-bone deficiency (SHFLD). Eur
J Hum Genet. 19:1144–1151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Van Bokhoven H, Melino G, Candi E and
Declercq W: p63, a story of mice and men. J Invest Dermatol.
131:1196–1207. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Bokhoven H, Hamel BC, Bamshad M,
Sangiorgi E, Gurrieri F, Duijf PH, Vanmolkot KR, van Beusekom E,
van Beersum SE, Celli J, et al: p63 Gene mutations in EEC syndrome,
limb-mammary syndrome and isolated split hand-split foot
malformation suggest a genotype-phenotype correlation. Am J Hum
Genet. 69:481–492. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sowińska-Seidler A, Socha M and Jamsheer
A: Split-hand/foot malformation-molecular cause and implications in
genetic counseling. J Appl Genet. 55:105–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Celik TH, Buyukcam A, Simsek-Kiper PO,
Utine GE, Ersoy-Evans S, Korkmaz A, Yntema HG, Bodugroglu K and
Yurdakok M: A newborn with overlapping features of AEC and EEC
syndromes. Am J Med Genet A. 155A:3100–3103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Robinson JT, Thorvaldsdóttir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Thorvaldsdóttir H, Robinson JT and Mesirov
JP: Integrative Genomics Viewer (IGV): High-performance genomics
data visualization and exploration. Brief Bioinform. 14:178–192.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lam HY, Clark MJ, Chen R, Chen R,
Natsoulis G, O'Huallachain M, Dewey FE, Habegger L, Ashley EA,
Gerstein MB, et al: Performance comparison of whole-genome
sequencing platforms. Nat Biotechnol. 30:78–82. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Addess KJ, Chen J, Geer LY, He J,
He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, et al: MMDB:
annotating protein sequences with Entrez's 3D-structure database.
Nucleic Acids Res. 35:(Database issue). D298–D300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Var view Protein 3D, . https://www.ncbi.nlm.nih.gov/Structure/cblast/cblast.cgi?client=snp&master_gi=169234657&neighbor_gi=212374861
|
|
17
|
Rinne T, Brunner HG and van Bokhoven H:
p63-associated disorders. Cell Cycle. 6:262–268. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berdón-Zapata V, Granillo-Alvarez M,
Valdés-Flores M, García-Ortiz JE, Kofman-Alfaro S and Zenteno JC:
p63 gene analysis in Mexican patients with syndromic and
non-syndromic ectrodactyly. J Orthop Res. 22:1–5. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zenteno JC, Berdón-Zapata V, Kofman-Alfaro
S and Mutchinick OM: Isolated ectrodactyly caused by a heterozygous
missense mutation in the transactivation domain of TP63. Am J Med
Genet A. 134A:74–76. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ianakiev P, Kilpatrick MW, Toudjarska I,
Basel D, Beighton P and Tsipouras P: Split-hand/split-foot
malformation is caused by mutations in the p63 gene on 3q27. Am J
Hum Genet. 67:59–66. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rinne T, Hamel B, van Bokhoven H and
Brunner HG: Pattern of p63 mutations and their phenotypes-update.
Am J Med Genet A. 140:1396–1406. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mangiulli M, Valletti A, Caratozzolo MF,
Tullo A, Sbisà E, Pesole G and D'Erchia AM: Identification and
functional characterization of two new transcriptional variants of
the human p63 gene. Nucleic Acids Res. 37:6092–6104. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gonfloni S, Caputo V and Iannizzotto V:
P63 in health and cancer. Int J Dev Biol. 59:87–93. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Thanos CD and Bowie JU: p53 Family members
p63 and p73 are SAM domain-containing proteins. Protein Sci.
8:1708–1710. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Enthart A, Klein C, Dehner A, Coles M,
Gemmecker G, Kessler H and Hagn F: Solution structure and binding
specificity of the p63 DNA binding domain. Sci Rep. 6:267072016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stiewe T, Zimmermann S, Frilling A, Esche
H and Pützer BM: Transactivation-deficient DeltaTA-p73 acts as an
oncogene. Cancer Res. 62:3598–3602. 2002.PubMed/NCBI
|
|
27
|
Yang A, Schweitzer R, Sun D, Kaghad M,
Walker N, Bronson RT, Tabin C, Sharpe A, Caput D, Crum C, et al:
p63 is essential for regenerative proliferation in limb,
craniofacial and epithelial development. Nature. 398:714–718. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vincek V, Knowles J, Li J and Nassiri M:
Expression of p63 mRNA isoforms in normal human tissue. Anticancer
Res. 23:3945–3948. 2003.PubMed/NCBI
|
|
29
|
Koster MI, Dai D, Marinari B, Sano Y,
Costanzo A, Karin M and Roop DR: p63 induces key target genes
required for epidermal morphogenesis. Proc Natl Acad Sci USA.
104:3255–3260. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koster MI and Roop DR: Mechanisms
regulating epithelial stratification. Annu Rev Cell Dev Biol.
23:93–113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koster MI, Dai D and Roop DR: Conflicting
roles for p63 in skin development and carcinogenesis. Cell Cycle.
6:269–273. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sommer M, Poliak N, Upadhyay S, Ratovitski
E, Nelkin BD, Donehower LA and Sidransky D: DeltaNp63alpha
overexpression induces downregulation of Sirt1 and an accelerated
aging phenotype in the mouse. Cell Cycle. 5:2005–2011. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan W and Chen X: GPX2, a direct target of
p63, inhibits oxidative stress-induced apoptosis in a p53-dependent
manner. J Biol Chem. 281:7856–7862. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Restelli M, Lopardo T, Lo Iacono N,
Garaffo G, Conte D, Rustighi A, Napoli M, Del Sal G, Perez-Morga D,
Costanzo A, et al: DLX5, FGF8 and the Pin1 isomerase control ΔNp63α
protein stability during limb development: A regulatory loop at the
basis of the SHFM and EEC congenital malformations. Hum Mol Genet.
23:3830–3842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lo Iacono N, Mantero S, Chiarelli A,
Garcia E, Mills AA, Morasso MI, Costanzo A, Levi G, Guerrini L and
Merlo GR: Regulation of Dlx5 and Dlx6 gene expression by p63 is
involved in EEC and SHFM congenital limb defects. Development.
135:1377–1388. 2008. View Article : Google Scholar : PubMed/NCBI
|