Introduction
Epidermal growth factor receptor (EGFR), a
trans-membrane glycoprotein, may be distinguished into three parts:
an extracellular domain, a transmembrane domain and an
intracellular domain, which possesses tyrosine kinase (TK)
activity. Particular mutations of EGFR, for example EGFR exon 19
deletion and exon 21 point mutation, serve a key role in the
development of non-small cell lung cancer (NSCLC), such as
promoting cancer cell proliferation, differentiation,
revascularization and metastasis (1). EGFR TK inhibitors (TKIs), mainly
targeting EGFR TK activities, are able to effectively prolong the
survival time of patients with cancer. In particular, a number of
studies have demonstrated the efficacy of the two most widely
available TKIs, erlotinib and gefitinib, in the first-line,
maintenance and relapsed settings (2–4).
The response rate of EGFR mutation activation to
EGFR TKI treatment reaches 70%; TKI treatment may additionally
prolong the NSCLC patients' progression free survival (PFS)
(5–8). EGFR exon 19 deletion (Del 19) and
exon 21 point mutation (L858R), the main EGFR mutation activation,
primarily exist in females, never-smokers, East Asians (~50%) and
patients with lung adenocarcinoma (ADC, ~30%) (9–12).
However, activation of EGFR mutations are rare in patients with
squamous cell carcinoma (SCC) (<3%); the lack of reported
mutations may limit the use of EGFR-TKIs in lung cancer patients
with SCC (13–17). To date, the benefits of EGFR-TKIs
in EGFR-mutated patients with SCC have not been well-studied.
In the present study, the mutation rate of SCC in
all EGFR mutations was analyzed in our laboratory and the
correlation of EGFR-mutations associated with SCC under
clinicopathological parameters was conducted. A Kaplan-Meier
survival analysis performed to estimate the effect of the EGFR
mutations in the survival rates of patient with SCC. Additionally,
the potential implications of EGFR TKI treatment and the tumor
biology of EGFR mutations in patients with SCC was investigated.
The aim of the present study was to investigate the mutations of
EGFR in lung SCC; the results of the present study may contribute
to developments in treatments for lung SCC.
Materials and methods
Patients and samples
All SCC samples (tumor tissues, blood and pleural
effusion) used in the present study were collected between 2010 and
2016 from clinical data or an archived thoracic oncology tissue
repository at the Department of Thoracic Surgery of Tangdu Hospital
affiliated with The Fourth Military Medical University (Xi'an,
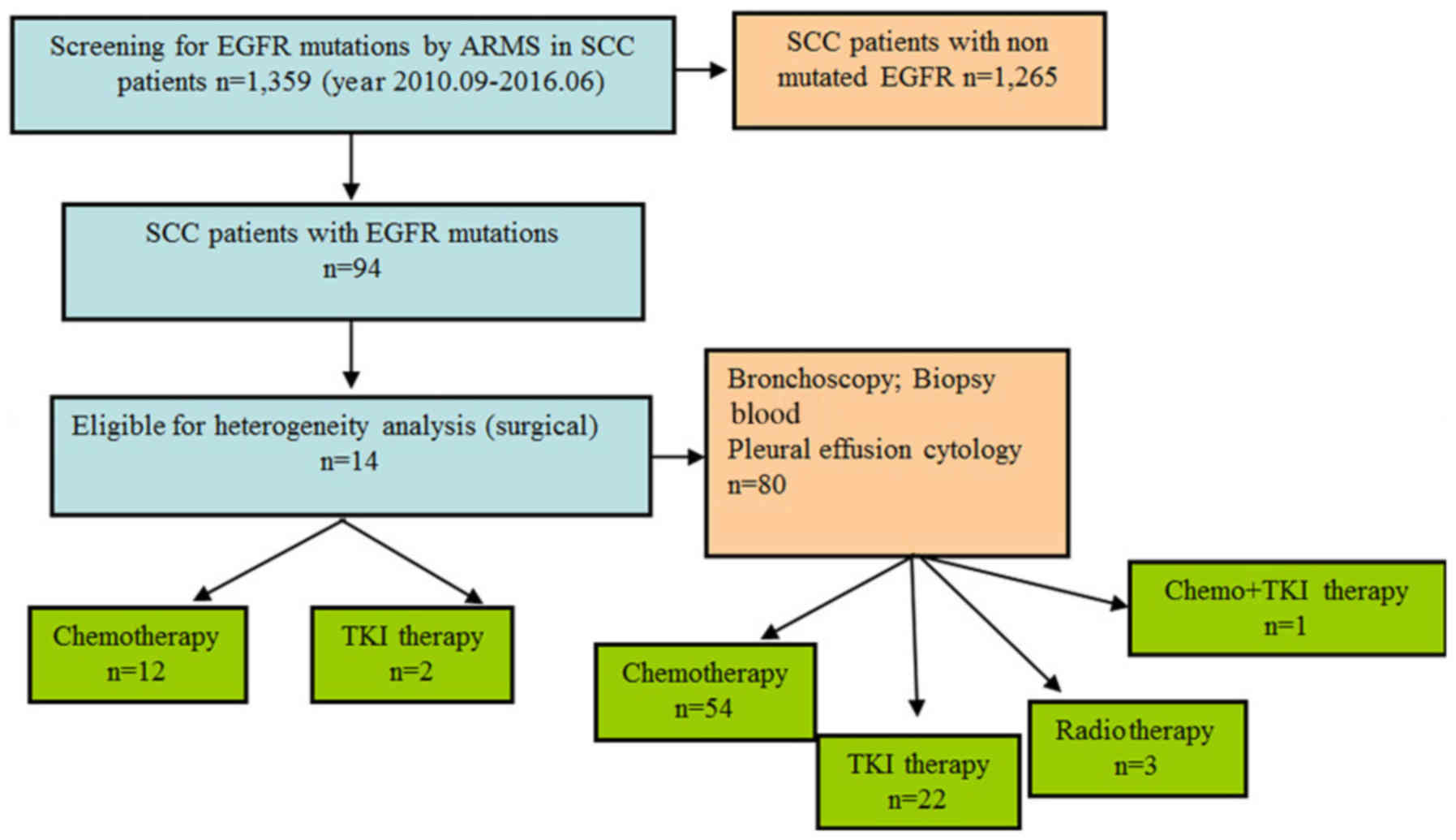
China; Fig. 1). Patients who had
received preoperative chemotherapy, radiotherapy or EGFR-targeted
therapy were excluded from the present study. Detailed
clinicopathological information, including patient ages, sex,
smoking history, tumor status, histological differentiation, nodal
status, clinical manifestation, surgical method, postoperative
treatment and follow-up information were collected and completed.
The surgery day was considered to be the starting day for
estimating postoperative survival time. The follow-up lasted until
August 13, 2016, with a median follow-up period of 39.62 months
(range, 2–63.28 months). The histological classification of the
tumors was reviewed by pathologists. And all tumors were staged
according to the pathological tumor, node, metastasis (pTNM)
classification (7th edition) of the International Union against
Cancer (18). The study protocol
was approved by the Regional Ethics Committee for Clinical Research
of The Fourth Military Medical University. All patients provided
written informed consent for use of their medical records and tumor
specimens for research purposes.
EGFR mutation testing
Genomic DNA was isolated and purified from fresh
tumor specimens using TIANamp Genomic DNA kit (Taingen Biotech,
Beijing, People's Republic of China) according to the
manufacturer's instructions.
EGFR mutation analysis of DNA was performed using
ADx-ARMS® technology, a technology based on amplified
refractory mutation system (ARMS) (19). Quantitative polymerase chain
reaction (qPCR) was conducted on the MX3005P qPCR system
(Stratagene California; Agilent Technologies, Inc., Santa Clara,
CA, USA) using the AmoyDx human EGFR Gene Mutation Detection kit
(Amoy Diagnostics Co., Ltd., Xiamen, China) according to the
manufacturer's protocols.
A total of 26 mutations in exon 18 (G719A, G719S,
G719C), exon 20 (T790M, S768I), and exon 21 (L858R, L861Q), and
exon 19 (deletions, n=19) were detected. The primer sequences of
EGFR mutation testing were obtained from The Primer
Express® Software v3.0.1 (Thermo Fisher Scientific, MA,
Waltham, USA) was applied to design specific primers for these
common mutations of the EGFR gene; the primer catalogue numbers of
the 26 mutations of EGFR gene provided by the AmoyDx human EGFR
Gene Mutation Detection kit are presented in Table I.
 | Table I.Primer sequences for quantitative
polymerase chain reaction. |
Table I.
Primer sequences for quantitative
polymerase chain reaction.
| Primer | Catalogue
number |
|---|
| 18-F1 | SEQ ID NO:1 |
| 18-F2 | SEQ ID NO:2 |
| 18-F3 | SEQ ID NO:3 |
| 18-R | SEQ ID NO:4 |
| 18-P | SEQ ID NO:5 |
| 19-F1 | SEQ ID NO:6 |
| 19-F2 | SEQ ID NO:7 |
| 19-F3 | SEQ ID NO:8 |
| 19-F4 | SEQ ID NO:9 |
| 19-F5 | SEQ ID NO:10 |
| 19-P | SEQ ID NO:11 |
| 19-R | SEQ ID NO:12 |
| 20-F1 | SEQ ID NO:13 |
| 20-F2 | SEQ ID NO:14 |
| 20-F3 | SEQ ID NO:15 |
| 20-F4 | SEQ ID NO:16 |
| 20-F5 | SEQ ID NO:17 |
| 20-P | SEQ ID NO:18 |
| 20-R | SEQ ID NO:19 |
| 21-F1 | SEQ ID NO:20 |
| 21-F2 | SEQ ID NO:21 |
| 21-P | SEQ ID NO:22 |
| 21-R | SEQ ID NO:23 |
| CTRL-F | SEQ ID NO:24 |
| CTRL-R | SEQ ID NO:25 |
| CTRL-P | SEQ ID NO:26 |
The qPCR amplification program was performed as
follows: Initial denaturation at 95°C for 5 min, 15 cycles of
amplification (at 95°C for 25 sec, 64°C for 20 sec, and 72°C for 20
sec) and a final denaturation followed by 31 cycles of
amplification (at 93°C for 25 sec, 60°C for 35 sec, and 72°C for 20
sec), and the FAM and HEX signals were collected at 60°C.
According to the manufacturer's protocols of the
AmoyDx human EGFR Gene Mutation Detection kit described that
samples were defined as EGFR mutation-negative when the Cq
value≥34; when the sample mutation Cq value<31, the samples were
defined as EGFR mutation-positive. When the calculated ΔCq value
[ΔCq=Cq (sample)-Ct (control)], when ΔCq value <ΔCt (Cut-off)
was 31≤ the sample mutation Cq value ≤33, the samples were defined
as EGFR mutation-positive, and when the ΔCq value ≥ΔCq (Cut-off),
the samples were defined as EGFR mutation-negative.
Discordance rate of EGFR mutations
analysis
Patients with lung SCC (n=14) with EGFR activating
mutations were used for discordance rate of EGFR mutations
analysis. The tumor samples were fixed with 10% formaldehyde for 24
h at room temperature and embedded with paraffin. Sections were
sliced to 4-µm thickness, deparaffinized with a series of xylene
and rehydrated with a graded alcohol series. The sections were
stained with hematoxylin for 40 sec, then counterstained with eosin
for 1 min at room temperature. Finally, the sections were
dehydrated with a graded alcohol series and embedded in paraffin.
Inclusion in the study was based on the presence of morphologically
different tumor areas within the same tumor and a sufficient cancer
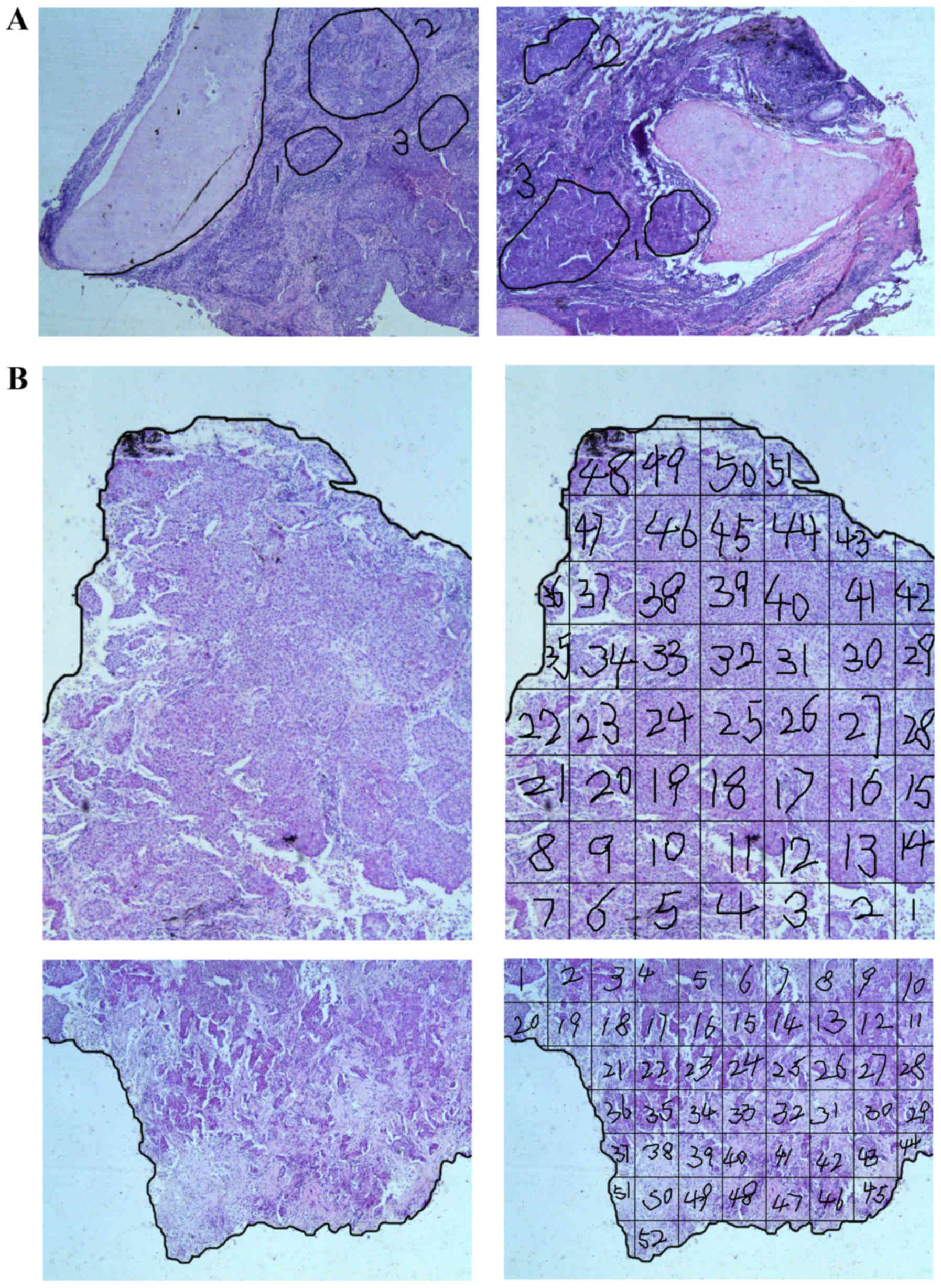
cell content (>30%) in each defined area. Subsequently, three
parts of each individual tumor were selected, and EGFR mutation
detection was performed. For every tumor, three areas were
identified by three pathologists to represent the most distinct and
variable histological patterns (Fig.
2).
Similarly, each of five such SCCs was divided into
>100 segments. The present study used two sections from each
tumor. Thus, each section yielded 50 or more pieces, which were 3×3
mm on average (Fig. 2). The EGFR
mutations of each piece were detected independently. To ensure that
the results were collected in an unbiased manner, mutations were
analyzed by a different technician from the one who had scratched
the tissues (20).
Statistical analyses
The correlations of EGFR mutations with
clinicopathological parameters were statistically analyzed using
the Mann-Whitney U test, and Kruskal-Wallis H (mainly used to
detect pathological differentiation). Kaplan-Meier survival
analysis was used to estimate the effect of the type of EGFR
mutation on the survival of patients with SCC. Statistical analyses
were performed using SPSS software version 18.0 (SPSS, Inc.,
Chicago, IL, USA). P-values were adjusted for multiple testing, and
P>0.05 was considered to indicate a statistically significant
difference.
Results
EGFR mutation
A total of 94 out of 1,359 patients with lung SCC
had EGFR mutations (6.92%), and 1,265 patients did not. All EGFR
mutations identified are present in Table II: Exon 19 (n=35, 37.2%); L858R
(n=37, 39.4%); T790M (n=5, 5.3%); G719X (n=4, 4.3%); L861Q (n=2,
2.1%); and other mutations (n=11, 11.7%).
 | Table II.Tumors examined in the present
study. |
Table II.
Tumors examined in the present
study.
| Variable | No. of tumors (n,
%) | Exon 19 (%) | L858R (%) | T790M (%) | G719X (%) | L861Q (%) | Others (%) |
|---|
| EGFR-mutated
SCC | 94 (1,359,
6.92) | 35 (37.2) | 37 (39.4) | 5 (5.3) | 4 (4.3) | 2 (2.1) | 11 (11.7) |
| Trans-sectional
analysis of number of areas in a tumor |
|
|
|
|
|
|
|
| 3 | 14 (94, 14.9) | 4 (28.6) | 7 (50) | 2 (14.3) | 0 | 0 | 1 (7.1) |
|
100 | 5 (14, 35.7) | 2 (40) | 3 (60) | 0 | 0 | 0 | 0 |
Patient characteristics
The clinicopathological characteristics of the
patients are summarized in Table
III. In 94 SCCs with EGFR mutations, there were 18 female and
76 male patients, with a median age of 59 years (range, 36–84
years). Histopathological diagnoses included well-differentiated
(10, 10.64%), moderate differentiation (60, 63.83%), and poor
differentiation (24, 25.53%). Postoperative staging evaluation
demonstrated stage I disease in 24 patients, stage II disease in 22
patients, stage III disease in 36 patients and stage IV disease in
12 patients. Metastatic sites included five brain metastases
(5.3%), one liver metastasis (1.1%), eight bone metastases (8.5%),
and one kidney metastasis (1.1%).
 | Table III.Patient characteristics. |
Table III.
Patient characteristics.
|
| Patients
(n=94) |
|---|
| Age, years |
|
|
| Median | 59 |
| Range | 36–84 |
|
|
| No. | % |
|
| Sex |
|
|
|
Male | 76 | 80.9 |
|
Female | 18 | 19.1 |
| Smoking
history |
|
|
|
Smoker | 69 | 73.4 |
|
Non-smoker | 25 | 26.6 |
|
Differentiation |
|
|
|
Well | 10 | 10.64 |
|
Moderate | 60 | 63.83 |
|
Poor | 24 | 25.53 |
| pTNM stage |
|
|
|
I–II | 46 | 48.9 |
|
III–IV | 48 | 51.1 |
| Primary tumor
location |
|
|
|
Central | 69 | 73.4 |
|
Peripheral | 25 | 26.6 |
| Primary tumor size,
cm |
|
|
|
<4.2 | 45 | 47.9 |
|
≥4.2 | 49 | 52.1 |
| Lymph node
metastasis |
|
|
|
Yes | 44 | 46.8 |
| No | 50 | 53.2 |
| No. of metastatic
sites analyzed |
|
|
|
Brain | 5 | 5.3 |
|
Liver | 1 | 1.1 |
|
Bone | 8 | 8.5 |
|
Kidney | 1 | 1.1 |
Clinical characteristics in smoker and
non-smoker patients with lung SCC with EGFR mutation
When comparing smoker and non-smoker patients in
terms of baseline clinical characteristics, significant differences
were identified in sex (smoker vs. non-smoker: 100% male vs. 28%,
respectively P>0.001), differentiation (smoker vs. non-smoker:
71.01% moderate vs. 44%; 18.84% poor vs. 44%, P=0.036) and pTNM
stage (smoker vs. non-smoker: 55.1% I–II vs. 32%, P=0.049). There
was no significant difference in age and lymph node metastasis
(Table IV).
 | Table IV.Clinical characteristics in smoker
and non-smoker patients with lung squamous cell carcinoma with EGFR
mutation. |
Table IV.
Clinical characteristics in smoker
and non-smoker patients with lung squamous cell carcinoma with EGFR
mutation.
| Variables | No. of cases, n=94
(%) | Smoker, n=69
(%) | Non-smoker, n=25
(%) | P-value |
|---|
| Age, years |
|
|
|
0.913 |
|
<59 | 46 (48.9) | 34 (49.3) | 12 (48) |
|
|
≥59 | 48 (51.1) | 35 (50.7) | 13 (52) |
|
| Sex |
|
|
| <0.001 |
|
Male | 76 (80.9) | 69 (100) | 7
(28) |
|
|
Female | 18 (19.1) | 0 (0) | 18 (72) |
|
|
Differentiation |
|
|
|
0.036 |
|
Well | 10 (10.64) | 7 (10.15) | 3
(12) |
|
|
Moderate | 60 (63.83) | 49 (71.01) | 11 (44) |
|
|
Poor | 24 (25.53) | 13 (18.84) | 11 (44) |
|
| pTNM stage |
|
|
|
0.049 |
|
I–II | 46 (48.9) | 38 (55.1) | 8
(32) |
|
|
III–IV | 48 (51.1) | 31 (44.9) | 17 (68) |
|
| Lymph node
metastasis |
|
|
|
0.546 |
|
Yes | 44 (46.8) | 31 (44.9) | 13 (52) |
|
| No | 50 (53.2) | 38 (55.1) | 12 (48) |
|
Clinical characteristics in patients
with early- and advanced-stage lung SCC with EGFR mutation
When comparing patients with early (I–II) and
advanced (III–IV) stage in baseline clinical characteristics,
significant differences were identified in smoking history
(patients with early vs. advanced stage: 82.6% smoker vs. 64.6%,
P=0.049), differentiation (patients with early vs. advanced stage:
71.74% moderate vs. 56.25%; 8.7% poor vs. 41.67%, P>0.001) and
lymph node metastasis (patients with early vs. advanced stage:
21.74% metastasis vs. 70.83%, P>0.001). There were no
significant differences in age and sex (Table V).
 | Table V.Clinical characteristics in early
stage and advanced stage lung squamous cell carcinoma patients with
EGFR mutation. |
Table V.
Clinical characteristics in early
stage and advanced stage lung squamous cell carcinoma patients with
EGFR mutation.
| Variables | No. of cases, n=94
(%) | TNM I–II, n=46
(%) | TNM III–IV, n=48
(%) | P-value |
|---|
| Age, years |
|
|
| 0.149 |
|
<59 | 46 (48.9) | 19 (41.3) | 27 (56.3) |
|
|
≥59 | 48 (51.1) | 27 (58.7) | 21 (43.7) |
|
| Sex |
|
|
| 0.143 |
|
Male | 76 (80.9) | 40 (87) | 36 (75) |
|
|
Female | 18 (19.1) | 6 (13) | 12 (25) |
|
| Smoking
history |
|
|
| 0.049 |
|
Smoker | 69 (73.4) | 38 (82.6) | 31 (64.6) |
|
|
Non-smoker | 25 (26.6) | 8 (17.4) | 17 (35.4) |
|
|
Differentiation |
|
|
| <0.001 |
|
Well | 10 (10.64) | 9 (19.56) | 1 (2.08) |
|
|
Moderate | 60 (63.83) | 33 (71.74) | 27 (56.25) |
|
|
Poor | 24 (25.53) | 4 (8.70) | 20 (41.67) |
|
| Lymph node
metastasis |
|
|
| <0.001 |
|
Yes | 44 (46.8) | 10 (21.74) | 34 (70.83) |
|
| No | 50 (53.2) | 36 (78.26) | 14 (29.17) |
|
Clinical characteristics in young,
middle-aged, and elderly patients with lung SCC with EGFR
mutation
When comparing young, middle-aged and elderly
patients in terms of baseline clinical characteristics, no
significant differences were identified in the clinical
characteristics of the patients (Table VI).
 | Table VI.Clinical characteristics in young and
elderly patients with lung squamous cell carcinoma with EGFR
mutation. |
Table VI.
Clinical characteristics in young and
elderly patients with lung squamous cell carcinoma with EGFR
mutation.
| Variables | No. of cases, n-94
(%) | ≤40 year of age,
n=2 (%) | 41–60 years of age,
n=56 (%) | 61–80 years of age,
n=33 (%) | >80 years of
age, n=3 (%) | P-value |
|---|
| Sex |
|
|
|
|
| 0.929 |
|
Male | 76 (80.9) | 0 (0) | 48 (85.7) | 25 (75.8) | 3 (100) |
|
|
Female | 18 (19.1) | 2 (100) | 8 (14.3) | 8 (24.2) | 0 (0) |
|
| Smoking
history |
|
|
|
|
| 0.613 |
|
Smoker | 69 (73.4) | 0 (0) | 45 (80.4) | 21 (63.6) | 3 (100) |
|
|
Non-smoker | 25 (26.6) | 2 (100) | 11 (19.6) | 12 (36.4) | 0 (0) |
|
|
Differentiation |
|
|
|
|
| 0.797 |
|
Well | 10 (10.64) | 0 (0) | 7 (12.5) | 3 (9.1) | 0 (0) |
|
|
Moderate | 60 (63.83) | 1 (50) | 37 (66.1) | 19 (57.6) | 3 (100) |
|
|
Poor | 24 (25.53) | 1 (50) | 12 (21.4) | 11 (33.3) | 0 (0) |
|
| pTNM stage |
|
|
|
|
| 0.303 |
|
I–II | 46 (48.9) | 0 (0) | 27 (48.2) | 16 (48.5) | 3 (100) |
|
|
III–IV | 48 (51.1) | 2 (100) | 29 (51.8) | 17 (51.5) | 0 (0) |
|
| Lymph node
metastasis |
|
|
|
|
| 0.108 |
|
Yes | 44 (46.8) | 2 (100) | 28 (50) | 14 (42.4) | 0 (0) |
|
| No | 50 (53.2) | 0 (0) | 28 (50) | 19 (57.6) | 3 (100) |
|
Overall discordance rate of EGFR
mutations in lung SCC
To determine the overall discordance rate of EGFR
mutations, EGFR mutations in 14 SCCs were detected and analyzed.
Three parts of each individual tumor were selected and examined for
the EGFR mutations subset, and identical mutations were
demonstrated in the three morphologically different tumor areas
(Table VII).
 | Table VII.Tumor cell content and epidermal
growth factor receptor mutation status detected in the three
histologically distinct tumor areas from each patient. |
Table VII.
Tumor cell content and epidermal
growth factor receptor mutation status detected in the three
histologically distinct tumor areas from each patient.
| Case no. | Area | Tumor cell content,
% | Predominant growth
patterna | Mutation |
|---|
| 1 | Tumor area 1 | 60 | Keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 2 | 60 | Keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 3 | 70 | Keratinizing | Exon 19 del
E746-A750 |
| 2 | Tumor area 1 | 80 |
Non-keratinizing | Exon 21 L858R |
|
| Tumor area 2 | 85 |
Non-keratinizing | Exon 21 L858R |
|
| Tumor area 3 | 90 | Keratinizing | Exon 21 L858R |
| 3 | Tumor area 1 | 55 | Basaloid | Exon 21 L858R |
|
| Tumor area 2 | 55 | Warty | Exon 21 L858R |
|
| Tumor area 3 | 50 | Warty | Exon 21 L858R |
| 4 | Tumor area 1 | 60 | Keratinizing | Exon 21 L858R |
|
| Tumor area 2 | 65 | Keratinizing | Exon 21 L858R |
|
| Tumor area 3 | 65 | Keratinizing | Exon 21 L858R |
| 5 | Tumor area 1 | 70 | Basaloid | Exon 21 L858R |
|
| Tumor area 2 | 75 | Basaloid | Exon 21 L858R |
|
| Tumor area 3 | 80 | Basaloid | Exon 21 L858R |
| 6 | Tumor area 1 | 40 | Basaloid | Exon 21 L858R |
|
| Tumor area 2 | 50 | Warty | Exon 21 L858R |
|
| Tumor area 3 | 60 | Basaloid | Exon 21 L858R |
| 7 | Tumor area 1 | 80 | Keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 2 | 70 | Keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 3 | 70 |
Non-keratinizing | Exon 19 del
E746-A750 |
| 8 | Tumor area 1 | 55 | Basaloid | Exon 21 L858R |
|
| Tumor area 2 | 45 | Warty | Exon 21 L858R |
|
| Tumor area 3 | 50 | Basaloid | Exon 21 L858R |
| 9 | Tumor area 1 | 45 | Keratinizing | Exon 20 Ins |
|
| Tumor area 2 | 35 | Keratinizing | Exon 20 Ins |
|
| Tumor area 3 | 40 | Keratinizing | Exon 20 Ins |
| 10 | Tumor area 1 | 70 | Basaloid | Exon 21 L858R |
|
| Tumor area 2 | 70 | Basaloid | Exon 21 L858R |
|
| Tumor area 3 | 70 | Basaloid | Exon 21 L858R |
| 11 | Tumor area 1 | 55 | Keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 2 | 60 | Keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 3 | 60 | Basaloid | Exon 19 del
E746-A750 |
| 12 | Tumor area 1 | 40 |
Non-keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 2 | 30 | Keratinizing | Exon 19 del
E746-A750 |
|
| Tumor area 3 | 50 | Keratinizing | Exon 19 del
E746-A750 |
| 13 | Tumor area 1 | 85 | Warty | Exon 20 T790M |
|
| Tumor area 2 | 85 | Basaloid | Exon 20 T790M |
|
| Tumor area 3 | 70 | Basaloid | Exon 20 T790M |
| 14 | Tumor area 1 | 60 |
Non-keratinizing | Exon 20 T790M |
|
| Tumor area 2 | 60 | Keratinizing | Exon 20 T790M |
|
| Tumor area 3 | 60 | Keratinizing | Exon 20 T790M |
As three parts may be insufficient to detect the
discordance rate of the EGFR mutations, five tumors were dissected
into >100 pieces, and each piece was examined for EGFR mutations
(Fig. 2). The results additionally
revealed identical mutations throughout each individual tumor.
Clinical characteristic-associated
prognosis of patients with lung SCC with EGFR mutations
Among clinicopathological factors, including age,
sex, smoking history, et al differentiation, pTNM stage and
lymph node metastasis were significantly associated with patient
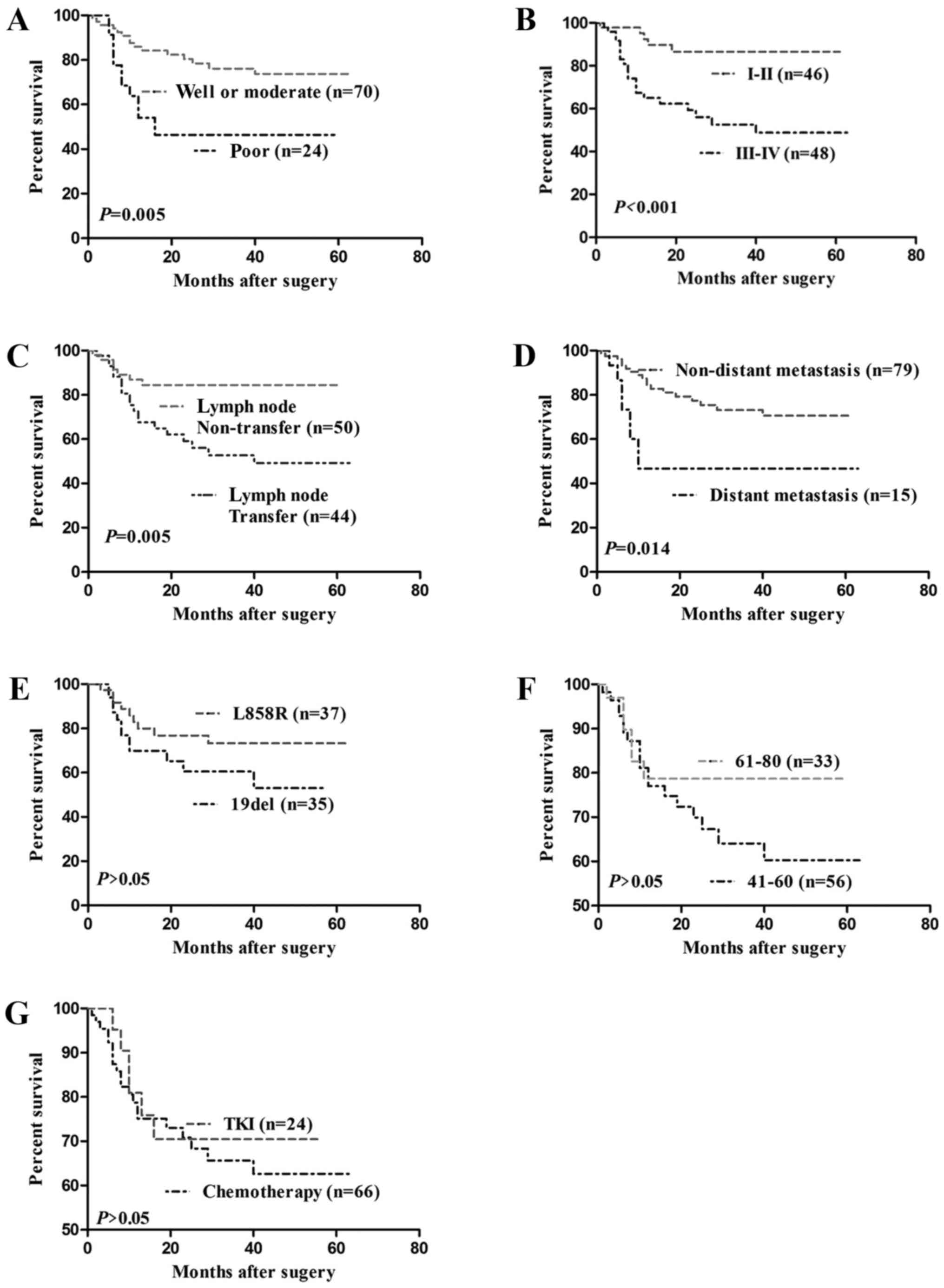
survival rates. Patients with well or moderately differentiated
tumors [n=70; 95% confidence interval (CI), 45.036–56.253 months]
exhibited longer durations of survival compared with those with
poorly differentiated tumors (n=24; 95% CI, 20.905–43.613 months;
P=0.005) (Fig. 3A). Patients with
pTNM I–II tumors (n=46; 95% CI, 49.091–60.002 months) exhibited a
longer duration of survival compared with those with pTNM III–IV
tumors (n=48; 95% CI, 29.621–45.614 months; P<0.001; Fig. 3B). Patients with no lymph node
metastasis (n=50; 95% CI, 46.783–58.485 months) exhibited a longer
duration of survival compared with those with lymph node metastasis
(n=44; 95% CI, 30.236–46.535 months; P=0.005; Fig. 3C).
The prognosis of patients with lung SCC with EGFR
mutations associated with distant metastases, EGFR mutations, and
postoperative treatment (chemotherapy and EGFR TKI) were
subsequently investigated. Patients with non-distant metastasis
(n=79; 95% CI, 42.350–53.076 months) exhibited a longer duration of
survival compared with those with distant metastasis (n=15; 95% CI,
19.069–47.515 months; P=0.014; Fig.
3D). A significant difference was not observed between patients
with L858R (n=37; 95% CI, 41.678–57.284 months) and patients with
Del 19 (n=35; 95% CI, 28.587–45.703 months; P>0.05; Fig. 3E). Additionally, a significant
difference between patients with aged 41–60 years (n=56; 95% CI,
37.213–51.322 months) and patients with aged 61–80 years was not
observed (n=33; 95% CI, 40.064–56.205 months; P>0.05; Fig. 3F). Furthermore, a significant
difference was observed between patients treated with TKI (n=24;
95% CI, 33.099–51.624 months) and patients treated with
chemotherapy (n=66; 95% CI, 38.160–51.387 months; P>0.05;
Fig. 3G).
Discussion
ADC, SCC, and large-cell undifferentiated carcinoma
are the principal subsets of non-small cell lung cancer (NSCLC),
and approximately 20–30% of cases of NSCLC are SCC (22). Historically, the subtype of NSCLC
has not been a major factor in determining patient therapy
management, and there is not been well established regarding the
fundamental difference in the molecular pathogenesis of ADC and SCC
(23). It is only in recent years
that driver oncogenes, including EGFR-activating mutations, and
subsequent corresponding therapies have been identified (7,24–26).
The majority of patients with NSCLC with EGFR mutations respond
well to EGFR TKIs (including gefitinib and erlotinib). EGFR
mutations are frequently observed in female, non-smoking, ADC and
Asian patients, but rare in SCC (9–12).
Research has identified that in pure SCC, there is the presence of
fibroblast growth factor receptor 1, phosphatase and tensin homolog
and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic
subunit á/AKT serine/threonine kinase 1 mutations, and an absence
of EGFR and KRAS proto-oncogene GTPase mutations (27). Compared with lung ADC, evidence
about the efficacy of EGFR TKIs and treatment progress in patients
with lung SCC is limited and controversial (4,28–30).
The present study performed ARMS analysis to
investigate the EGFR mutations subset in clinical lung SCC samples.
Statistical analysis revealed that 6.9% (94/1,359) of the tumor
samples were EGFR-activating mutations. The EGFR mutated SCC
samples were identified as follows: 37.2% (35/94) in exon 19; 39.4%
(37/94) in L858R; 5.3% (5/94) in T790M; 4.3% (4/94) in G719X; 2.1%
(2/94) in L861Q; and 11.7% (11/94) in other mutations (Table II). Due to the limited number in
the EGFR mutations subset, only the proportions of EGFR mutations
in exon 19 (Del 19) and exon 21 (L858R) were larger (~76.6% of the
total), although no significant difference in prognosis was
observed between the EGFR Del 19 and L858R groups in SCC.
In the present study, there were significant
differences between the smoking group, pTNM stage group and
baseline clinical characteristics. Recently, along with extended
life span, patients >80 years of age are increasing in number,
and differences in prognosis are significant in the age range 28–30
years (31–33). However, due to the limited sample
size, no significant difference was observed between very young and
very elderly patients.
Previous studies on the role of EGFR TKIs in SCC
have identified that EGFR TKIs may be an option for the treatment
of SCC, and the EGFR mutations subset may help to select a subgroup
of patients with best response to TKIs (34–36).
In the present study, among clinical characteristics, only the
differentiation, pTNM stage, lymph node metastasis and distant
metastases were significantly associated with patients' survival
(P>0.05; log-rank test). The SCC patients identified as having
EGFR activating mutations following surgery were treated as
follows: 70.2% (66/94) with chemotherapy; 25.5% (24/94) with EGFR
TKIs; 3.2% (3/94) with radiotherapy; and 1.1% (1/94) with
chemotherapy and EGFR TKIs. However, the difference in prognosis
was not marked between the chemotherapy and TKI therapy groups.
There may be specific reasons to explain these
results, and it was hypothesized that EGFR TKIs may prolong patient
survival in a way comparable to the function of chemotherapy.
However, the present study used a limited sample size, thus a study
with an expanded sample size is required. In addition, EGFR TKIs
are used for patients with EGFR mutation. Whether used in ADC or
SCC, EGFR TKIs are recommended as long as the EGFR site is mutated.
In the present study, the prognostic difference was not marked
between the chemotherapy and the TKIs therapy groups, which
indicated that EGFR TKIs were able to prolong patient survival in
way comparable to the function of chemotherapy; therefore, it was
hypothesized that EGFR TKIs may be an option for the treatment of
SCC with EGFR mutations.
In certain individual tumors, EGFR mutations were
not evenly distributed, and this may be one of the causes of
drug-resistance to EGFR TKIs. However, in previous studies, the
opposite results have been demonstrated in lung ADC (18,26).
In the present study, identical EGFR mutations were identified
throughout individual tumors by examining 14 tumors divided into
three parts and five tumors divided into 100 parts. However, the
limited sample size is a shortcoming of the present study, thus it
is intended to expand the sample size in the future.
The results of the present study suggested that EGFR
Del 19/L858R may be the main EGFR mutations subset in SCC. The
effect of EGFR TKIs on SCC patients' prognoses is the same as the
effect of chemotherapy, showing fewer complications and a higher
quality of life, so EGFR TKIs could be a worthwhile option for the
treatment of SCC. In addition, the heterogeneous distribution of
EGFR mutations in SCC is extremely rare.
Acknowledgements
The authors wish to thank Dr Xiao-li Li for
assistance with software assays, who from the laboratory of
Department of Cardiology, Tangdu Hospital, The Fourth Military
Medical University (Xi'an, China).
Funding
No funding was received.
Availability of data and materials
All data generated and analyzed during this study
are included in this published article.
Authors' contributions
YS, XY and MW conducted the EGFR mutation test; JZ
contributed to the follow-up; XW and JX interpreted the
clinicopathological information statistics; YZ conducted the
discordance rate of EGFR mutations analysis; and ZZ and XL
performed statistical analysis. The manuscript was drafted by YS
and edited by XY. All authors read and approved the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Regional
Ethics Committee for Clinical Research of the Fourth Military
Medical University. All patients provided written informed consent
for use of their medical records and tumor specimens for research
purposes.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
EGFR
|
epidermal growth factor receptor
|
|
TKIs
|
tyrosine kinase inhibitors
|
|
SCC
|
squamous cell carcinoma
|
|
ADC
|
lung adenocarcinoma
|
|
PFS
|
progression free survival
|
|
OS
|
overall survival
|
|
NSCLC
|
non-small cell lung cancer
|
References
|
1
|
Cohen S: Purification of the receptor for
epidermal growth factor from A-431 cells: Its function as a tyrosyl
kinase. Methods Enzymol. 99:379–387. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mendelsohn J and Baselga J: Status of
epidermal growth factor receptor antagonists in the biology and
treatment of cancer. J Clin Oncol. 21:2787–2799. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herbst RS, Maddox AM, Rothenberg ML, Small
EJ, Rubin EH, Baselga J, Rojo F, Hong WK, Swaisland H, Averbuch SD,
et al: Selective oral epidermal growth factor receptor tyrosine
kinase inhibitor ZD1839 is generally well-tolerated and has
activity in non-small-cell lung cancer and other solid tumors:
Results of a phase I trial. J Clin Oncol. 20:3815–3825. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thatcher N, Chang A, Parikh P, Pereira
Rodrigues J, Ciuleanu T, von Pawel J, Thongprasert S, Tan EH,
Pemberton K, Archer V and Carroll K: Gefitinib plus best supportive
care in previously treated patients with refractory advanced
non-small-cell lung cancer: Results from a randomised,
placebo-controlled, multicentre study (Iressa Survival Evaluation
in Lung Cancer). Lancet. 366:1527–1537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Han SW, Kim TY, Hwang PG, Jeong S, Kim J,
Choi IS, Oh DY, Kim JH, Kim DW, Chung DH, et al: Predictive and
prognostic impact of epidermal growth factor receptor mutation in
non-small-cell lung cancer patients treated with gefitinib. J Clin
Oncol. 23:2493–2501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang SF, Liu HP, Li LH, Ku YC, Fu YN,
Tsai HY, Chen YT, Lin YF, Chang WC, Kuo HP, et al: High frequency
of epidermal growth factor receptor mutations with complex patterns
in non-small cell lung cancers related to gefitinib responsiveness
in Taiwan. Clin Cancer Res. 10:8195–8203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pao W and Miller VA: Epidermal growth
factor receptor mutations, small-molecule kinase inhibitors, and
non-small-cell lung cancer: Current knowledge and future
directions. J Clin Oncol. 23:2556–2568. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kris MG, Johnson BE, Berry LD, Kwiatkowski
DJ, Iafrate AJ, Wistuba II, Varella-Garcia M, Franklin WA, Aronson
SL, Su PF, et al: Using multiplexed assays of oncogenic drivers in
lung cancers to select targeted drugs. JAMA. 311:1998–2006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jackman DM, Miller VA, Cioffredi LA, Yeap
BY, Jänne PA, Riely GJ, Ruiz MG, Giaccone G, Sequist LV and Johnson
BE: Impact of epidermal growth factor receptor and KRAS mutations
on clinical outcomes in previously untreated non-small cell lung
cancer patients: Results of an online tumor registry of clinical
trials. Clin Cancer Res. 15:5267–5273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rosell R, Moran T, Queralt C, Porta R,
Cardenal F, Camps C, Majem M, Lopez-Vivanco G, Isla D, Provencio M,
et al: Screening for epidermal growth factor receptor mutations in
lung cancer. N Engl J Med. 361:958–967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jukna A, Montanari G, Mengoli MC, Cavazza
A, Covi M, Barbieri F, Bertolini F and Rossi G: Squamous cell
carcinoma ‘Transformation’ concurrent with secondary T790M mutaton
in resistant EGFR-mutated adenocarcinomas. J Thorac Oncol.
11:e49–e51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kuiper JL, Ronden MI, Becker A, Heideman
DA, van Hengel P, Ylstra B, Thunnissen E and Smit EF:
Transformation to a squamous cell carcinoma phenotype of an
EGFR-mutated NSCLC patient after treatment with an EGFR-tyrosine
kinse inhibitor. J Clin Pathol. 68:320–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Levin PA, Mayer M, Hoskin S, Sailors J,
Oliver DH and Gerber DE: Histologic transformation from
adenocarcinoma to squamous cell carcinoma as a mechanism of
resistance to EGFR inhibition. J Thorac Oncol. 10:e86–e88. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Billah S, Stewart J, Staerkel G, Chen S,
Gong Y and Guo M: EGFR and KRAS mutations in lung carcinoma:
Molecular testing by using cytology specimens. Cancer
Cytopathology. 119:111–117. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pao W, Kris MG, Iafrate AJ, Ladanyi M,
Jänne PA, Wistuba II, Miake-Lye R, Herbst RS, Carbone DP, Johnson
BE and Lynch TJ: Integration of molecular profiling into the lung
cancer clinic. Clin Cancer Res. 15:5317–5322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goldstraw P, Crowley J, Chansky K, Giroux
DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V and Sobin L:
International Association for the Study of Lung Cancer
International Staging Committee; Participating Institutions: The
IASLC lung cancer staging project: Proposals for the revision of
the TNM stage groupings in the forthcoming (seventh) edition of the
TNM classification of malignant tumours. J Thorac Oncol. 2:706–714.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu J, Zhao R and Zhang J and Zhang J:
ARMS for EGFR mutation analysis of cytologic and corresponding lung
adenocarcinoma histologic specimens. J Cancer Res Clin Oncol.
141:221–227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yatabe Y, Matsuo K and Mitsudomi T:
Heterogeneous distribution of EGFR mutations is extremely rare in
lung adenocarcinoma. J Clin Oncol. 29:2972–2977. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ,
Van Schil PE, et al: International association for the study of
lung cancer/american thoracic society/european respiratory society
international multidisciplinary classification of lung
adenocarcinoma. J Thorac Oncol. 6:244–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu J, Wang W, Xu M, Li Y, Chen C and Wang
X: A global view of regulatory networks in lung cancer: An approach
to understand homogeneity and heterogeneity. Semin Cancer Biol.
42:31–38. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Petrelli F, Borgonovo K, Cabiddu M and
Barni S: Efficacy of EGFR tyrosine kinase inhibitors in patients
with EGFR-mutated non-small-cell lung cancer: A meta-analysis of 13
randomized trials. Clin Lung Cancer. 13:107–114. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rekhtman N, Paik PK, Arcila ME, Tafe LJ,
Oxnard GR, Moreira AL, Travis WD, Zakowski MF, Kris MG and Ladanyi
M: Clarifying the spectrum of driver oncogene mutations in
biomarker-verified squamous carcinoma of lung: Lack of EGFR/KRAS
and presence of PIK3CA/AKT1 mutations. Clin Cancer Res.
18:1167–1176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Ma S, Song X, Han B, Cheng Y,
Huang C, Yang S, Liu X, Liu Y, Lu S, et al: Gefitinib versus
placebo as maintenance therapy in patients with locally advanced or
metastatic non-small-cell lung cancer (INFORM; C-TONG 0804): A
multicentre, double-blind randomised phase 3 trial. Lancet Oncol.
13:466–475. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Clark GM, Zborowski DM, Santabarbara P,
Ding K, Whitehead M, Seymour L and Shepherd FA: National Cancer
Institute of Canada Clinical Trials Group: Smoking history and
epidermal growth factor receptor expression as predictors of
survival benefit from erlotinib for patients with non-small-cell
lung cancer in the National Cancer Institute of Canada Clinical
Trials Group study BR.21. Clin Lung Cancer. 7:389–394. 2016.
View Article : Google Scholar
|
|
30
|
Cappuzzo F, Ciuleanu T, Stelmakh L,
Cicenas S, Szczésna A, Juhász E, Esteban E, Molinier O, Brugger W,
Melezínek I, et al: Erlotinib as maintenance treatment in advanced
non-small-cell lung cancer: A multicentre, randomised,
placebo-controlled phase 3 study. Lancet Oncol. 11:521–529. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Owonikoko TK, Ragin CC, Belani CP, Oton
AB, Gooding WE, Taioli E and Ramalingam SS: Lung cancer in elderly
patients: An analysis of the surveillance, epidemiology, and end
results database. J Clin Oncol. 25:5570–5577. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Thomas A, Chen Y, Yu T, Jakopovic M and
Giaccone G: Trends and characteristics of young non-small cell lung
cancer patients in the United States. Front Oncol. 5:1132015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miyoshi S, Hamada H, Ito R, Hamaguchi N,
Kadowaki T and Higaki J: Successful treatment of non-small cell
lung cancer by gefitinib in an elderly patient with poor
performance status. Nihon Ronen Igakkai Zasshi. 45:338–342.
2008.(In Japanese). View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu J, Zhang Y, Jin B, Chu T, Dong X, Yang
H, Wu D, Lou Y, Zhang X, Wang H and Han B: Efficacy of EGFR
tyrosine kinase inhibitors for non-adenocarcinoma lung cancer
patients harboring EGFR-sensitizing mutations in China. J Cancer
Res Clin Oncol. 142:1325–1330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ameratunga M, Pavlakis N, Gebski V, Broad
A and Khasraw M: Epidermal growth factor receptor-tyrosine kinase
inhibitors in advanced squamous cell carcinoma of the lung: A
meta-analysis. Asia Pac J Clin Oncol. 10:273–278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cobo M, Gutiérrez V, Rodelo L, López O,
Ruiz M and Godoy A: Afatinib in patients with squamous cell
carcinoma of the lung: Current context and the option of oral
treatment. Med Clin (Barc). 146 Suppl 1:S25–S29. 2016.(In Spanish).
View Article : Google Scholar
|