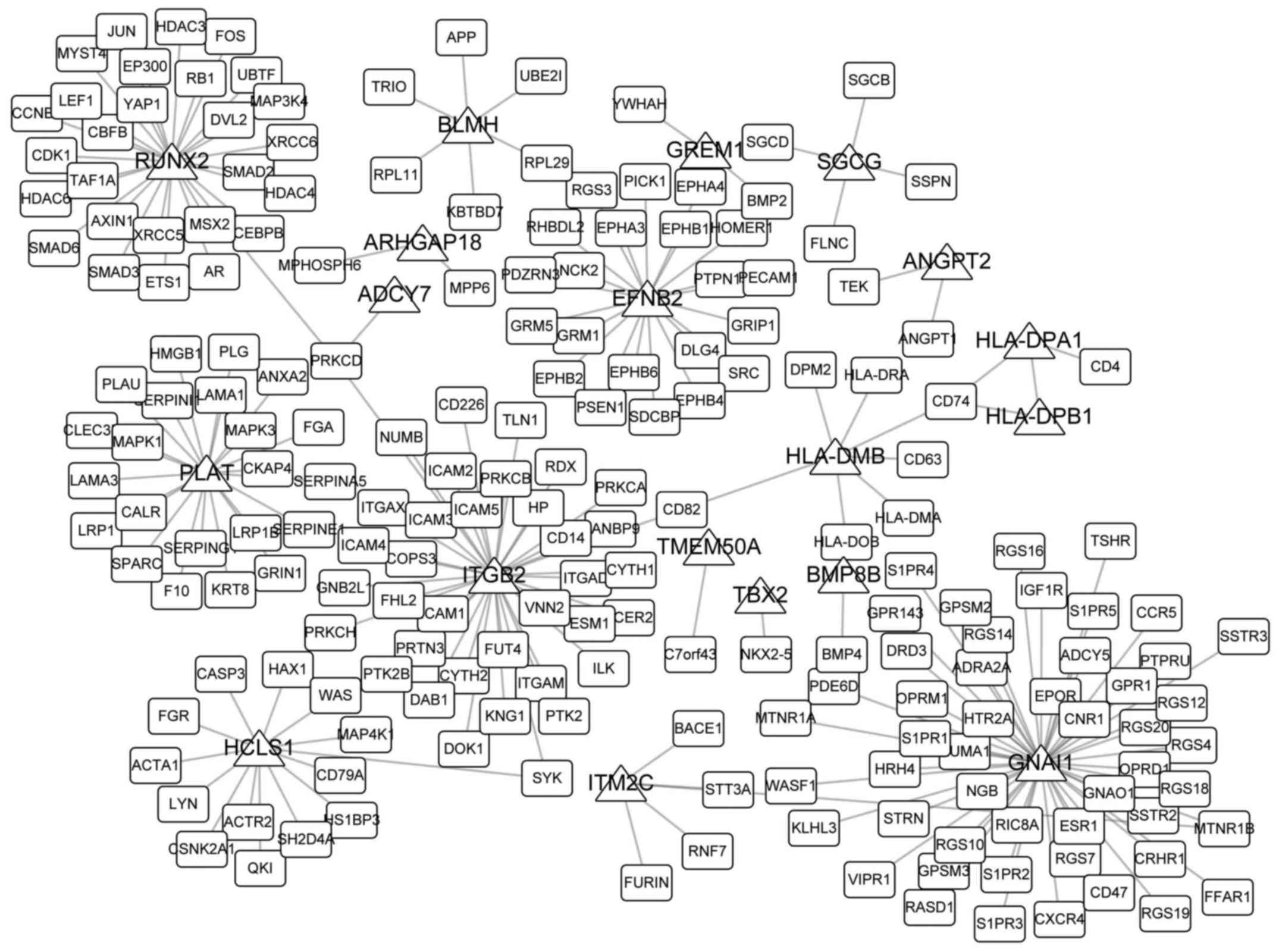
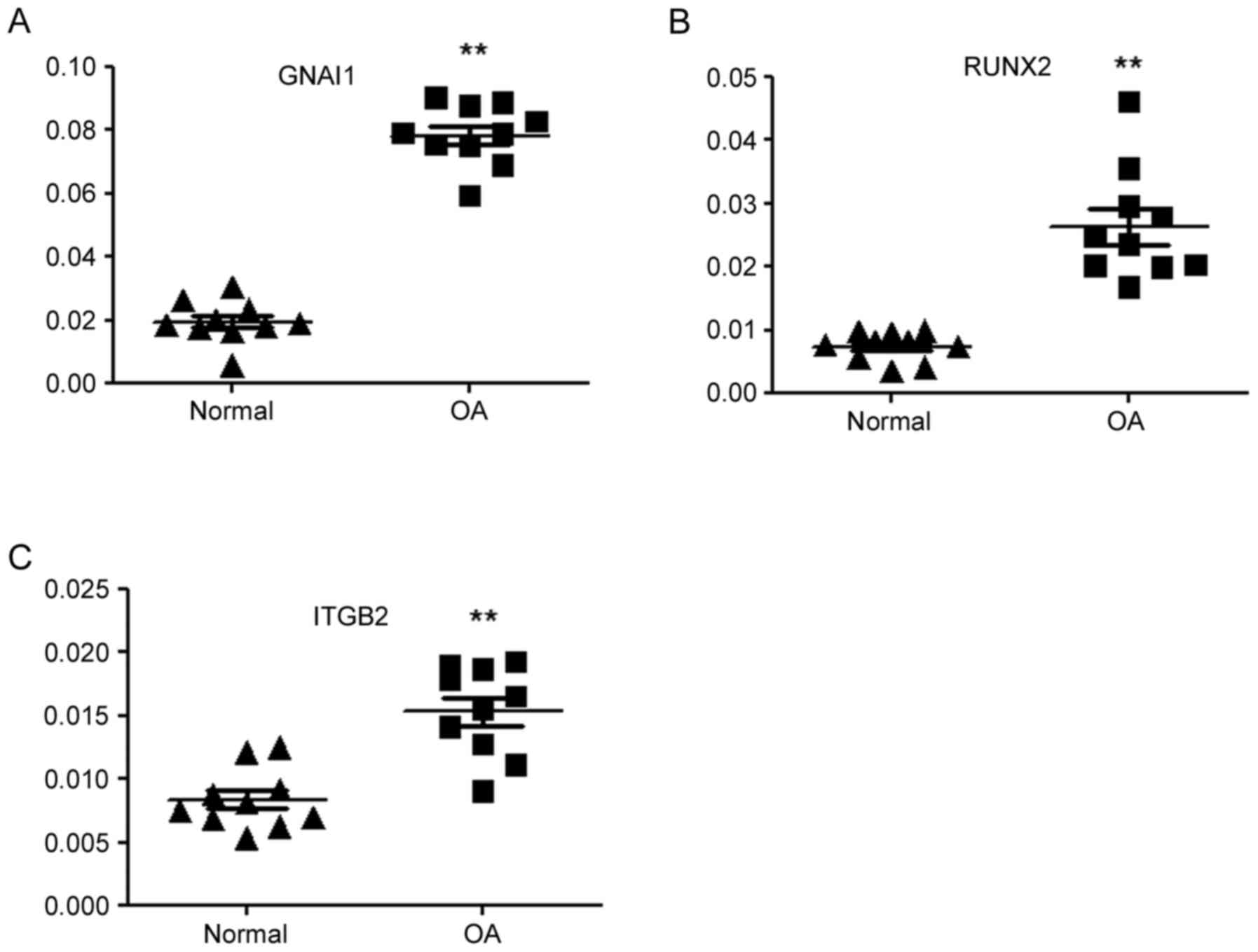
|
1
|
Pan Q, O'Connor MI, Coutts RD, Hyzy SL,
Olivares-Navarrete R, Schwartz Z and Boyan BD: Characterization of
osteoarthritic human knees indicates potential sex differences.
Biol Sex Differ. 7:272016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Young IC, Chuang ST, Hsu CH, Sun YJ, Liu
HC, Chen YS and Lin FH: Protective effects of aucubin on
osteoarthritic chondrocyte model induced by hydrogen peroxide and
mechanical stimulus. BMC Complement Altern Med. 17:912017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rhee J, Park SH, Kim SK, Kim JH, Ha CW,
Chun CH and Chun JS: Inhibition of BATF/JUN transcriptional
activity protects against osteoarthritic cartilage destruction. Ann
Rheum Dis. 76:427–434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang M, Egan B and Wang J: Epigenetic
mechanisms underlying the aberrant catabolic and anabolic
activities of osteoarthritic chondrocytes. Int J Biochem Cell Biol.
67:101–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schwager J, Hoeller U, Wolfram S and
Richard N: Rose hip and its constituent galactolipids confer
cartilage protection by modulating cytokine, and chemokine
expression. BMC Complement Altern Med. 11:1052011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nair A, Gan J, Bush-Joseph C, Verma N,
Tetreault MW, Saha K, Margulis A, Fogg L and Scanzello CR: Synovial
chemokine expression and relationship with knee symptoms in
patients with meniscal tears. Osteoarthritis Cartilage.
23:1158–1164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hashimoto S, Rai MF, Gill CS, Zhang Z,
Sandell LJ and Clohisy JC: Molecular characterization of articular
cartilage from young adults with femoroacetabular impingement. J
Bone Joint Surg Am. 95:1457–1464. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park R and Ji JD: Unique gene expression
profile in osteoarthritis synovium compared with cartilage:
Analysis of publicly accessible microarray datasets. Rheumatol Int.
36:819–827. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rasheed Z, Al-Shobaili HA, Rasheed N, Al
Salloom AA, Al-Shaya O, Mahmood A, Alajez NM, Alghamdi AS and
Mehana el-SE: Integrated study of globally expressed micrornas in
il-1beta-stimulated human osteoarthritis chondrocytes and
osteoarthritis relevant genes: A microarray and bioinformatics
analysis. Nucleosides Nucleotides Nucleic Acids. 35:335–355. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun J, Yan B, Yin W and Zhang X:
Identification of genes associated with osteoarthritis by
microarray analysis. Mol Med Rep. 12:5211–5216. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Loeser RF, Olex AL, McNulty MA, Carlson
CS, Callahan MF, Ferguson CM, Chou J, Leng X and Fetrow JS:
Microarray analysis reveals age-related differences in gene
expression during the development of osteoarthritis in mice.
Arthritis Rheum. 64:705–717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Housman G, Havill LM, Quillen EE, Comuzzie
AG and Stone AC: Assessment of DNA methylation patterns in the bone
and cartilage of a nonhuman primate model of osteoarthritis.
Cartilage. 1:19476035187591732018.
|
|
13
|
Peffers M, Balaskas P and Smagul A:
Osteoarthritis year in review 2017: Genetics and epigenetics.
Osteoarthritis Cartilage. 26:304–311. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Fukui N, Yahata M, Katsuragawa Y,
Tashiro T, Ikegawa S and Lee MT: Genome-wide DNA methylation
profile implicates potential cartilage regeneration at the late
stage of knee osteoarthritis. Osteoarthritis Cartilage. 24:835–843.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rushton MD, Young DA, Loughlin J and
Reynard LN: Differential DNA methylation and expression of
inflammatory and zinc transporter genes defines subgroups of
osteoarthritic hip patients. Ann Rheum Dis. 74:1778–1782. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reynard LN, Bui C, Syddall CM and Loughlin
J: CpG methylation regulates allelic expression of GDF5 by
modulating binding of SP1 and SP3 repressor proteins to the
osteoarthritis susceptibility SNP rs143383. Hum Genet.
133:1059–1073. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Klinger P, Beyer C, Ekici AB, Carl HD,
Schett G, Swoboda B, Hennig FF and Gelse K: The transient
chondrocyte phenotype in human osteophytic cartilage: A role of
pigment epithelium-derived factor? Cartilage. 4:249–255. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aref-Eshghi E, Zhang Y, Liu M, Harper PE,
Martin G, Furey A, Green R, Sun G, Rahman P and Zhai G: Genome-wide
DNA methylation study of hip and knee cartilage reveals embryonic
organ and skeletal system morphogenesis as major pathways involved
in osteoarthritis. BMC Musculoskelet Disord. 16:2872015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of affymetrix genechip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang D, Yan L, Hu Q, Sucheston LE, Higgins
MJ, Ambrosone CB, Johnson CS, Smiraglia DJ and Liu S: IMA: An R
package for high-throughput analysis of Illumina's 450K Infinium
methylation data. Bioinformatics. 28:729–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Prasad Keshava TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37:D767–D772. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang F, Zhou S, Wang C, Huang Y, Li H,
Wang Y, Zhu Z, Tang J and Yan M: Epigenetic modifications of
interleukin-6 in synovial fibroblasts from osteoarthritis patients.
Sci Rep. 7:435922017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou X, Chen L, Grad S, Alini M, Pan H,
Yang D, Zhen W, Li Z, Huang S and Peng S: The roles and
perspectives of microRNAs as biomarkers for intervertebral disc
degeneration. J Tissue Eng Regen Med. 11:3481–3487. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Suzuki MM and Bird A: DNA methylation
landscapes: Provocative insights from epigenomics. Nat Rev Genet.
9:465–476. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang J and Wang N: Genome-wide expression
and methylation profiles reveal candidate genes and biological
processes underlying synovial inflammatory tissue of patients with
osteoarthritis. Int J Rheum Dis. 18:783–790. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Johnson AA, Akman K, Calimport SR, Wuttke
D, Stolzing A and de Magalhaes JP: The role of DNA methylation in
aging, rejuvenation, and age-related disease. Rejuvenation Res.
15:483–494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ezura Y, Sekiya I, Koga H, Muneta T and
Noda M: Methylation status of CpG islands in the promoter regions
of signature genes during chondrogenesis of human synovium-derived
mesenchymal stem cells. Arthritis Rheum. 60:1416–1426. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rogers EL, Reynard LN and Loughlin J: The
role of inflammation-related genes in osteoarthritis.
Osteoarthritis Cartilage. 23:1933–1938. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takahashi A, de Andres MC, Hashimoto K,
Itoi E and Oreffo RO: Epigenetic regulation of interleukin-8, an
inflammatory chemokine, in osteoarthritis. Osteoarthritis
Cartilage. 23:1946–1954. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Imagawa K, de Andrés MC, Hashimoto K, Pitt
D, Itoi E, Goldring MB, Roach HI and Oreffo RO: The epigenetic
effect of glucosamine and a nuclear factor-kappa B (NF-κB)
inhibitor on primary human chondrocytes-implications for
osteoarthritis. Biochem Biophys Res Commun. 405:362–367. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fernández-Tajes J, Soto-Hermida A,
Vázquez-Mosquera ME, Cortés-Pereira E, Mosquera A, Fernández-Moreno
M, Oreiro N, Fernández-López C, Fernández JL, Rego-Pérez I and
Blanco FJ: Genome-wide DNA methylation analysis of articular
chondrocytes reveals a cluster of osteoarthritic patients. Ann
Rheum Dis. 73:668–677. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiao JL, Meng JH, Gan YH, Zhou CY and Ma
XC: Association of GDF5, SMAD3 and RUNX2 polymorphisms with
temporomandibular joint osteoarthritis in female Han Chinese. J
Oral Rehabil. 42:529–536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ji Q, Xu X, Xu Y, Fan Z, Kang L, Li L,
Liang Y, Guo J, Hong T, Li Z, et al: miR-105/Runx2 axis mediates
FGF2-induced ADAMTS expression in osteoarthritis cartilage. J Mol
Med (Berl). 94:681–694. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Leijten JC, Bos SD, Landman EB, Georgi N,
Jahr H, Meulenbelt I, Post JN, van Blitterswijk CA and Karperien M:
GREM1, FRZB and DKK1 mRNA levels correlate with osteoarthritis and
are regulated by osteoarthritis-associated factors. Arthritis Res
Ther. 15:R1262013. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han EJ, Yoo SA, Kim GM, Hwang D, Cho CS,
You S and Kim WU: GREM1 is a key regulator of synoviocyte
hyperplasia and invasiveness. J Rheumatol. 43:474–485. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Valverde-Franco G, Lussier B, Hum D, Wu J,
Hamadjida A, Dancause N, Fahmi H, Kapoor M, Pelletier JP and
Martel-Pelletier J: Cartilage-specific deletion of ephrin-B2 in
mice results in early developmental defects and an
osteoarthritis-like phenotype during aging in vivo. Arthritis Res
Ther. 18:652016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kaya AI, Lokits AD, Gilbert JA, Iverson
TM, Meiler J and Hamm HE: A conserved hydrophobic core in Gαi1
regulates G protein activation and release from activated receptor.
J Biol Chem. 291:19674–19686. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Diehl SA, McElvany B, Noubade R, Seeberger
N, Harding B, Spach K and Teuscher C: G proteins galphai1/3 are
critical targets for bordetella pertussis toxin-induced vasoactive
amine sensitization. Infect Immun. 82:773–782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rivetti S, Lauriola M, Voltattorni M,
Bianchini M, Martini D, Ceccarelli C, Palmieri A, Mattei G, Franchi
M, Ugolini G, et al: Gene expression profile of human colon cancer
cells treated with cross-reacting material 197, a diphtheria toxin
non-toxic mutant. Int J Immunopathol Pharmacol. 24:639–649. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mabey T, Honsawek S, Saetan N, Poovorawan
Y, Tanavalee A and Yuktanandana P: Angiogenic cytokine expression
profiles in plasma and synovial fluid of primary knee
osteoarthritis. Int Orthop. 38:1885–1892. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao W, Sweeney C, Walsh C, Rooney P,
McCormick J, Veale DJ and Fearon U: Notch signalling pathways
mediate synovial angiogenesis in response to vascular endothelial
growth factor and angiopoietin 2. Ann Rheum Dis. 72:1080–1088.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yi J, Jin Q, Zhang B, Wu X and Ge D:
Gremlin-1 concentrations are correlated with the severity of knee
osteoarthritis. Med Sci Monit. 22:4062–4065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fan Y, Chen J, Yang Y, Lin J and Wu Z:
Genome-wide expression and methylation profiling reveal candidate
genes in osteoarthritis. Clin Exp Rheumatol. 35:983–990.
2017.PubMed/NCBI
|
|
47
|
Hopwood B, Tsykin A, Findlay DM and
Fazzalari NL: Microarray gene expression profiling of
osteoarthritic bone suggests altered bone remodelling, WNT and
transforming growth factor-β/bone morphogenic protein signalling.
Arthritis Res Ther. 9:R1002007. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liao L, Zhang S, Gu J, Takarada T, Yoneda
Y, Huang J, Zhao L, Oh CD, Li J, Wang B, et al: Deletion of Runx2
in articular chondrocytes decelerates the progression of
DMM-induced osteoarthritis in adult mice. Sci Rep. 7:23712017.
View Article : Google Scholar : PubMed/NCBI
|