Introduction
Prostate cancer is more prevalent in Western
countries (1). However, in recent
years increasing evidence suggests that it has started to become
prevalent in the aging male Chinese population too (2). Androgen receptor (AR) is regarded as
the main etiologic factor in prostate cancer initiation, and
androgen deprivation therapy (ADT) serves as the most effective
modality at the early stage of the disease. However, in time the
majority of patients will eventually develop resistance, denoted as
castration-resistant prostate cancer (CRPC), concurring with
distant metastasis and lethal prognosis. Thus, understanding the
in-depth mechanisms associated with AR signaling and its tumor
promoting effects is useful for the control of this disease.
A recent study suggested that post-transcriptional
regulations by various microRNA and mRNA-binding proteins appeared
to be the important regulators in normal cell development and that
their aberrant expression levels may participate in carcinogenesis
in various ways, including affecting mRNA stability, alternative
splicing of mRNA and translation efficiency (3).
Quaking (QKI) is a RNA-binding protein encoded by
the quaking gene, which belongs to the signal transduction and
activation of RNA family of proteins with important signal
transduction and RNA activation functions (4). There are three isoforms, QKI-5, QKI-6
and QKI-7. The QKI-5 isoform possesses a noncanonical nuclear
localization signal not present in the other QKI isoforms. This may
assist in it being regulated by the AR gene, which is a nuclear
receptor gene. Therefore, the QKI-5 isoform was the main subject of
the present study. At the cellular level, it significantly
modulates cell proliferation, apoptosis and differentiation.
Notably, previous studies have identified its tumor suppressor
roles (5) and the mechanical
dysregulation of cell cycle with the involvement of
apoptosis-associated signaling pathways (6,7). In
colon epithelium, it positively promotes the expression of 26S
proteasome non-ATPase regulatory subunit 9 (p27) by suppressing the
transcription of β-catenin, which in turn suppresses cell
proliferation and promotes cell differentiation (8). During carcinogenesis in colon and
gastric tissues, hypermethylation of QKI promoter regions are
responsible for silenced QKI expression (8). In addition, aberrant alternative
splicing, or even circular RNA changes, has been reported to be the
unique mechanisms contributing to QKI-associated carcinogenesis
(9,10).
In prostate cancer, our previous study demonstrated
that QKI repression was also observed via hypermethylation in high
staging prostate cancer samples and cell lines (11). There may be an association between
QKI expression and positive androgen-associated signals in
early-stage prostate cancer. In terms of AR-positive prostate
cancer, the C4-2 cell line, denoted as CRPC cell line, developed an
androgen agonist independent cell growth by having more
constitutive active splicing isoforms of AR, including AR-V7 and
ARv567es (12). In
addition, heat-shock protein (HSP)90 is known to be one of the most
important HSPs responsible for various tumor invasion and
metastasis via regulating cell growth and apoptosis processes
(13). It is also a coactivator
and molecular chaperone of AR, capable of stabilizing and enhancing
AR functions (14). The
combination of a heat shock protein (HSP)90 inhibitor with the
anti-androgenic drug Casodex exhibits a synergistic
anti-proliferative capacity on CRPC cells (15).
The present study explored the association between
AR and QKI expression in prostate cancer, particularly the role of
QKI in AR-induced carcinogenesis and anti-AR treatment effects.
Materials and methods
Tissue sample collection
The present study recruited 16 male unrelated
individuals diagnosed with prostate cancer (age, 52–78 years; mean
age, 68.25±5.39 years). Frozen tumor samples were collected from
radical prostatectomy specimens from Xijing Hospital, the
University of the Fourth Military Medical University (Xi'an, China)
between May 2014 and December 2016. Tissue was frozen in liquid
nitrogen within 30 min of resection. Non-neoplastic mucosa from
colon was dissected free of muscle and histologically confirmed to
be tumor-free by frozen sections. For cancer tissue, tumor purity
of over 70% was confirmed by frozen sections for each case before
submission for DNA and protein extraction. Information on patient
age and sex in addition to Duke's tumor stage was collected. This
study was approved by the Ethics Committee of the Fourth Military
Medical University and the Institutional Review Board of Xijing
Hospital. Informed written consent was obtained from patients prior
to the study.
Cell lines and reagents
The LNCaP human prostate cancer cell line was
sourced from the Chinese Academy of Sciences Shanghai Cell
Repository (Shanghai, China), and the C4-2 human prostate cancer
cell line was sourced from Dr Donghui Han (Department of Urologic
Surgery, Xijing Hospital, Fourth Military Medical University,
Xi'an, China), who obtained from the China Center for Type Culture
Collection (Wuhan, China). Fetal bovine serum (FBS) and charcoal
stripped serum were purchased from Gibco (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Gene-specific primers were synthesized by
Sangon Biotech Co., Ltd. (Shanghai, China). The cells were cultured
in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 2 mmol/l glutamine, 0.06 g/l penicillin, 0.1 g/l
streptomycin and 10% FBS (Invitrogen; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere of 5% CO2.
Small interfering (si)RNA synthesis
and transfection
All AR- and QKI-specific siRNA sequences were
synthesized according to the literature by Shanghai GenePharma Co.,
Ltd. (Shanghai, China). AR siRNA interference sequences (16): AR RNAi-1 forward,
5′-AAGAAGGCCAGUUGUAUGGAC-3′ and reverse,
5′-GUCCAUACAACUGGCCUUCUU-3′; AR RNAi-2 forward,
5′-AAGACGCUUCUACCAGCUCAC−3′ and reverse,
5′-GUGAGCUGGUAGAAGCGUCUU−3′. QKI siRNA interference sequences
(8): QKI RNAi-1 forward,
5′-GGCACCUACAGAGAUGCCAACAUUA-3′ and reverse,
5′-UAAUGUUGGCAUCUCUGUAGGUGCC-3′; QKI RNAi-2 forward,
5′-CCUUGAGUAUCCUAUUGAACCUAGU-3′ and reverse,
5′-ACUAGGUUCAAUAGGAUACUCAAGG-3′.
The quality was confirmed by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR), as
detailed in a subsequent section. The transfection procedure was
carried out in accordance with the protocol of the Liposome 2000
Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
for adherent cells. At 4–6 h post-transfection, the cells were
resuspended in fresh medium containing 10% FBS. For the Casodex
sensitivity test, 5×103 cells were seeded per well into
a 96-well plate (3599; Corning Incorporated, Corning, NY, USA), in
100 µl fresh medium containing 10% FBS per well, and after 24 h of
culture, a final concentration of 10−5 mol/l Casodex
(98%; Sigma-Aldrich; Merck KGaA; Darmstadt, Germany) was added to
the appropriate treatment groups as a 10−4 mol/l stock
solution in phosphate buffered solution (PBS). After 48 h, a final
concentration of 10% Cell Counting kit-8 (CCK-8) reagent (Dojindo
Molecular Technologies, Inc., Shanghai, China) was added to
determine proliferative capacity.
Androgen deprivation treatment
RPMI-1640 medium with the addition of 20% charcoal
stripped serum was used to cultivate the LNCaP and C4-2 cells. The
cells were first cultured for 24 h, following which the medium was
removed to prevent any remaining androgens from influencing the
experiment. Subsequently, the cells were transferred into 6-well
plates (3596; Corning Incorporated) with 2 ml medium per well, and
assigned to experimental and control groups. After 36 h, protein
and RNA samples were obtained, and the remaining cells were
transferred to fresh 1640 medium with 20% charcoal stripped serum
and a physiological concentration of 10−9 mol/l
dihydrotestosterone (DHT) (98%; Sigma-Aldrich; Merck KGaA) for
another 36 h of continued cultivation, following which the final
protein and RNA samples were measured.
Western blotting and determination of
protein contents
Cells were lysed on ice for 30 min using NP-40 cell
lysis solution (Abiocenter, Shanghai, China) and the resultant
lysate harvested by centrifugation at 13,400 × g and 4°C for 5 min.
The Bicinchoninic Acid assay (Thermo Fisher Scientific, Inc.) was
used to determine protein concentrations with 10% bovine serum
albumin (CWBio, Beijing, China) as a standard, following which the
measured protein concentrations were adjusted to 2.5 µg/µl with SDS
(Guidechem, Shanghai, China) loading buffer. Aliquots comprising 50
µg total protein were separated using 12% SDS-PAGE, the proteins
were then transferred onto a nitrocellulose membrane (0.2 µm;
Invitrogen; Thermo Fisher Scientific, Inc.), which was blocked with
5% skim milk powder in tris-buffered saline (TBS) at room
temperature for 1 h. Detection was conducted using rabbit primary
antibodies directed against AR (cat. no. A9853; 1:500),
prostate-specific antigen (PSA) (cat. no. SAB4501531, 1:1,000), QKI
(cat. no. HPA019123; 1:300) purchased from Sigma-Aldrich (Merck),
p27 (cat. no. D121177; 1:500), p21 (cat. no. D153391, 1:250)
purchased from Sangon Biotech Co., Ltd. (Shanghai, China) and mouse
primary antibodies directed against caspase-12 (cat. no. ab10455;
1:500) purchased from Abcam (Shanghai, China). All membranes were
incubated with the primary antibodies overnight at 4°C. Following
washing with TBST (0.1% Tween-20), the membranes were incubated
with an IgG-IRDye® 800CW fluorescent secondary antibody
solution (cat. no. ab216773; 1:15,000 in TBS; Abcam) for 1 h at
37°C. The protein bands were visualized using an Odyssey Infrared
Imaging Laser scanning imaging system (LI-COR Biosciences, Lincoln,
NE, USA). Monoclonal mouse anti-β-actin antibodies (cat. no.
D190606; 1:500; Sangon Biotech Co., Ltd.) were incubated at 4°C
overnight and used as an internal reference.
RT-qPCR determination of mRNA
levels
A total of 1×106 cells were used for
RT-qPCR analysis. Total RNA was isolated using the TRIzol kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The cDNA for AR, QKI and PSA was
synthesized using SYBR PrimeScript RT Reagent kit (Takara
Biotechnology Co., Ltd., Dalian, China). The cDNA for all other
mRNA were synthesized using GoScript™ Reverse Transcriptase system
(Promega Corporation, Madison, WI, USA). The RT reaction was as
follows: 37°C for 15 min and 85°C for 5 sec. qPCR analyses were
performed using the SYBR Premix Ex Taq II (Takara Biotechnology
Co., Ltd.); the thermocycler used for AR, QKI and PSA mRNA analysis
was the LightCycler 480 system (Roche, Basel, Switzerland) and the
thermocycler used for the analysis of all other mRNAs was the
Applied Biosystems® 7500 Fast Real-time PCR system
(Thermo Fisher Scientific, Inc.). The thermal cycling conditions
were as follows: 95°C for 10 min, followed by 40 cycles at 95°C for
10 sec, 60°C for 35 sec and 72°C for 45 sec, followed by a final
elongation step at 72°C for 10 min. The PCR primers were as
follows: Human AR gene forward, 5′-GACTTCACCGCACCTGATGT-3′ and
reverse, 5′-GCAGTCTCCAAACGCATGTC-3′; human PSA gene forward,
5′-ATCTGTGGAGCTGGATTCTGG-3′ and reverse,
5′-AAGACCCAGTGTGCCCTAAG-3′; human QKI gene (8) forward, 5′-GGGGAAATGGAAACGAAGG-3′ and
reverse, 5′-TTGAGCCTTTGCCTCGGAC-3′; human β-actin gene (8) forward, 5′-AGCGGGAAATCGTGCGTGAC-3′ and
reverse, 5′-TGGAAGGTGGACAGCGAGGC-3′; human ARv567es splicing
variant (17) forward,
5′-CCTTGCTCTCTAGCCTCAATGAA-3′ and reverse,
5′-CTTGATTAGCAGGTCAAAAGTGAACT-3′; human AR-V7 splicing variant
forward, 5′-CCATCTTGTCGTCTTCGGAAATGTTATGAAGC-3 and reverse,
5′-TTTGAATGAGGCAAGTCAGCCTTTCT-3′; human UBE2C gene forward,
5′-TGGTCTGCCCTGTATGATGT-3 and reverse, 5′-AAAAGCTGTGGGGTTTTTCC-3′;
human HSP90 gene forward, 5′-GGGGGATCCCCAGCTATGAACTCCTTCTCC-3′ and
reverse, 5′-GGGGTCGACCTACATTTGCCGAAGAGCCCT-3′. Each sample was
replicated in three wells of a 96-well plate (3599; Corning
Incorporated). Relative mRNA expression was calculated using
2−ΔΔCq as previously described (18).
Flow cytometry for the determination
of cell cycle status and apoptosis
LNCaP and C4-2 cells (1×106) were
trypsinized, washed twice with PBS and fixed in 70% ice-cold
ethanol (Sangon Biotech Co., Shanghai, China) for 1 h at 4°C. The
samples were centrifuged at 300 × g for 5 min at 4°C, the ethanol
was removed and they were incubated with 100 mg/ml RNaseA
(Sigma-Aldrich; Merck KGaA) for 30 min at 37°C. For cell-cycle
distribution analysis, 400 ul PI solution (BestBio Co., Ltd.,
Shanghai, China) was added to the cells. The cells were vortexed
and incubated in the dark for 30–60 min at 2–8°C. For cell
apoptosis analysis, cellular DNA was stained with the Annexin
V-FITC/PI Apoptosis Detection kit (cat. no. BB-4101-2; BestBio Co.,
Ltd., Shanghai, China). Briefly, the cells were centrifuged at 300
× g for 5 min at room temperature and resuspended in Annexin V
Binding Buffer. A total of 5 µl FITC Annexin V was added to the
suspension, they were then gently vortexed and incubated for 15 min
at 2–8°C in the dark. The cells were then incubated with 10 µl of
PI solution, gently vortexed and incubated for 5 min at 2–8°C in
the dark. Cell-cycle distributions and cell apoptosis were
determined by flow cytometry using a BD FACSCalibur system (BD
Biosciences, Franklin Lakes, NJ, USA) and data was analyzed using
the ModFit software version 4.1 (Verity Software House, Inc.,
Topsham, ME, USA).
MTT cell proliferation assay
Following siRNA transfection, cells were transferred
into 96-well plates (3599; Corning Incorporated) in triplicate
wells. After 48 h, 10 µl CCK-8 was added to each well, and the
cells incubated at 37°C in an atmosphere comprising 5%
CO2 for 1–4 h, following which the reaction product,
which accumulated in proportion to cell viability, was dissolved in
RPMI-1640 medium and measured using a 96-well plates reader
(Sunrise; Tecan Group Ltd., Männedorf, Switzerland) at 450 nm.
Luciferase reporter assay
To detect the interaction between AR 3UTR (forward,
5′-CAGTACAACTTGGC-3′ and reverse, 5′-GCCAAGTTGTACTG-3′; Shanghai
GenePharma Co., Ltd.) and QKI, LNCaP cells were seeded in 24-well
plates and transfected with 400 ng pGL3-AR in combination with 400
ng pcDNA3.1-QKI and the internal control vector 40 ng pRL-TK,
respectively, using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. siRNAs
targeting AR were synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.) and were dissolved in diethypyrocarbonate-treated
H2O at a concentration of 20 µmol/l as a stock. Cells
were co-transfected with 50 ng pBIND (Promega Corporation, Madison,
WI, USA), a plasmid constitutively expressing Renilla
luciferase, to normalize for the transfection efficiency. After 48
h transfection, cells were lysed using passive lysis buffer and
analyzed for firefly and Renilla luciferase activities using
the Dual-Luciferase Reagent Assay kit (Promega Corporation)
according to the manufacturer's protocols.
Statistical analysis
Data are expressed as the mean ± standard deviation.
All experiments were performed at least in triplicate. SPSS
software version 18.0 (SPSS, Inc., Chicago, IL, USA) was used to
perform a direct-probability t-test, one-way analysis of variance
with a Dunnett post hoc test. P<0.05 was considered to indicate
a statistically significant difference.
Results
Associations of QKI expression with AR
level in prostate cancer clinical samples and different prostate
cancer cell lines
The present study used 16 clinical prostate cancer
samples and 2 prostate cancer cell lines; one an androgen-dependent
prostate cancer cell line (LNCaP) and the other a CRPC cell line
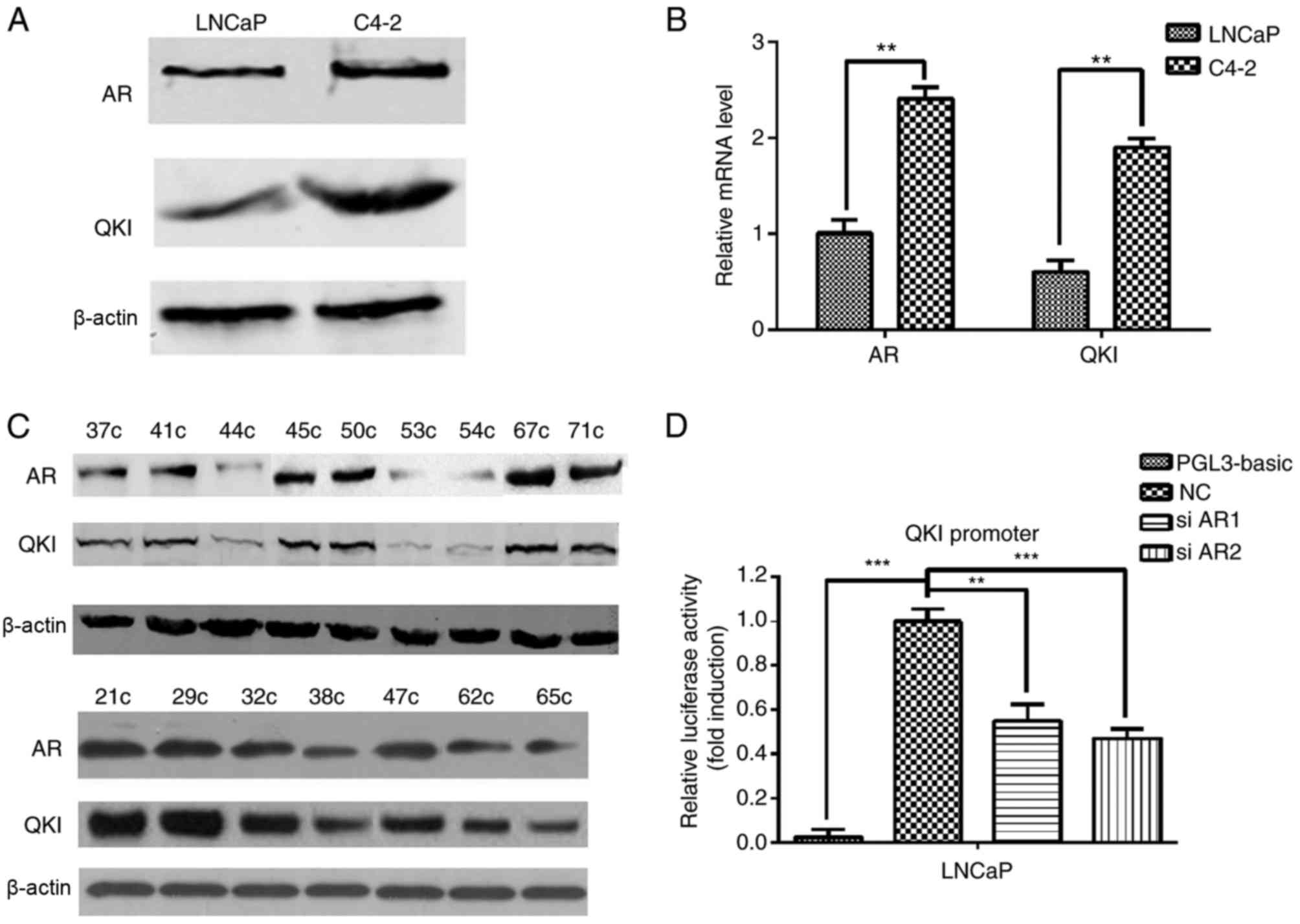
(C4-2). In order to determine the expression levels of AR and QKI
in the clinical samples and cell lines, protein and mRNA expression
levels were detected by using western blot analysis and RT-qPCR
assays, respectively. C4-2 cells had increased AR and QKI protein
(Fig. 1A) and mRNA (Fig. 1B) expression levels compared with
LNCaP cells, indicating their possible association in prostate
cancer development. Meanwhile, in clinical samples, positive AR
expression often co-existed with higher QKI expression levels too
(Fig. 1C). Dual-luciferase
reporter demonstrated that there are AR binding elements in the QKI
promoter region, and AR can regulate the expression of QKI
positively (Fig. 1D). These data
suggested that there are positive associations between AR and QKI
expression levels in prostate cancer development.
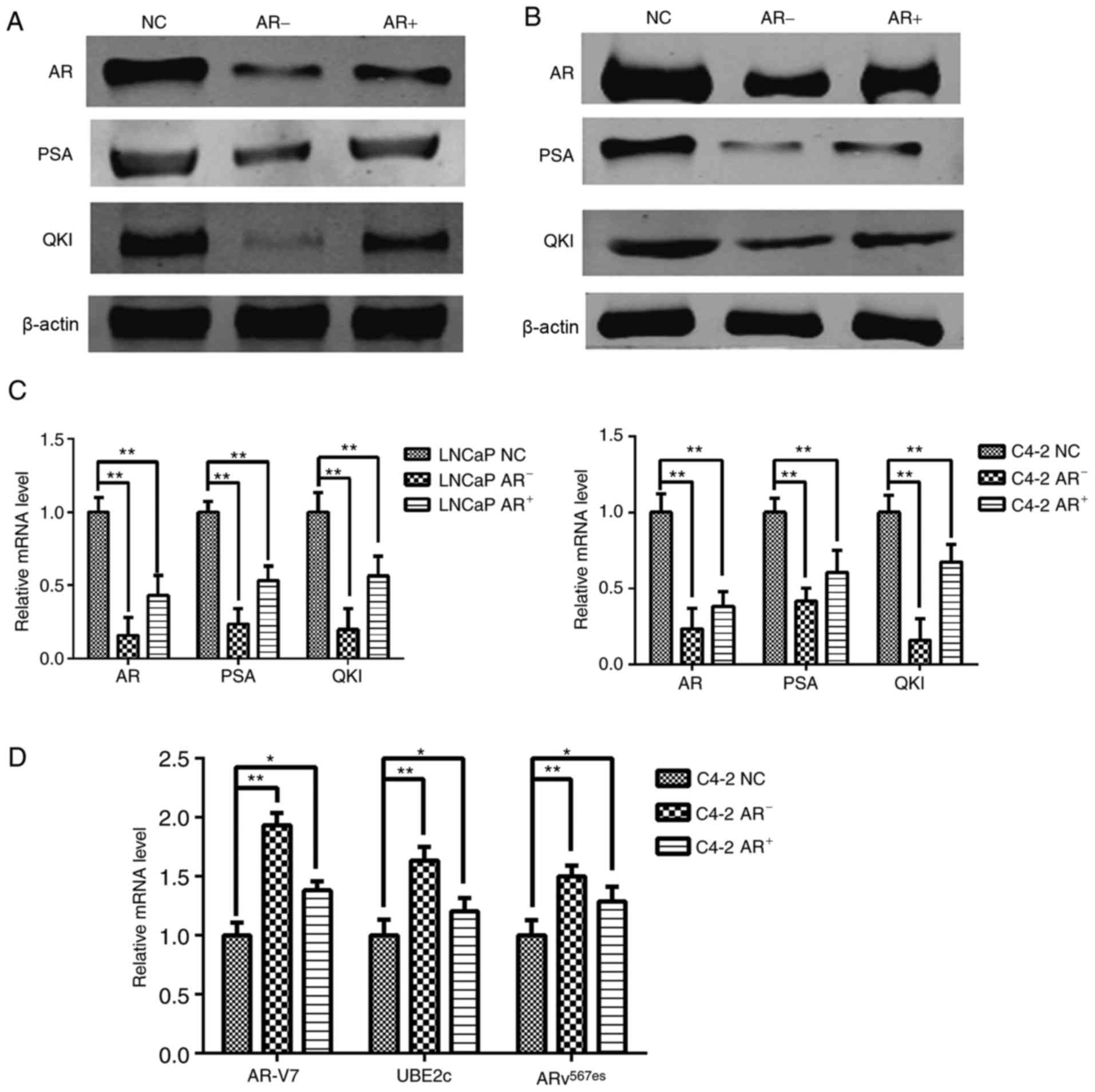
Dynamic changes of QKI with ADT
As ADT is an optional treatment for prostate cancer,
following androgen deprivation and re-addition, positive altered
expression levels of QKI and PSA were observed in parallel with
androgen levels, since PSA is the known positive target gene of AR
(Fig. 2A-C). These phenomena
suggested that QKI may function as novel target gene in AR
signaling pathways. Notably, even castration-resistant C4-2 cells
still retained the partial sensitivity to exogenous AR alterations,
although to a lesser extent, compared with LNCaP cells. In
addition, the aberrant splice variants AR, including AR-V7 and
ARv567es mutations, contribute to the progression of
prostate cancer ending with the castration-resistant state
(19). Therefore the RNA levels of
ARv567es and AR-V7 were determined, in addition to the
target gene UBE2c; the results demonstrated that RNA levels of all
three genes were significantly increased following androgen
removal, in contrast with reductions following androgen
replenishment (Fig. 2D). These
results demonstrated there are negative associations between
androgen levels and the aberrant AR splicing forms in C4-2
cells.
Influential effects of AR silencing on
the expression of QKI and prostate cancer proliferation and
apoptosis
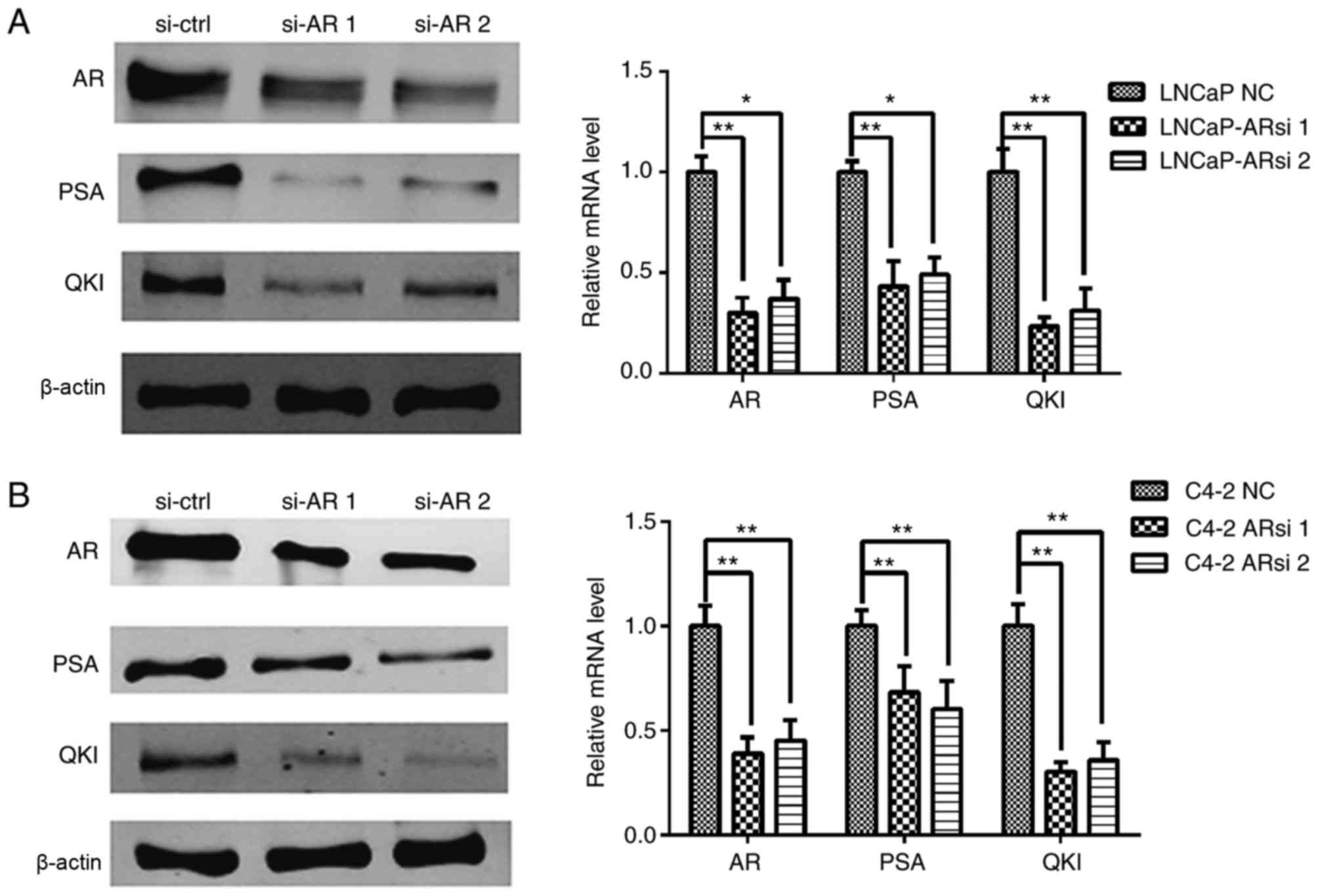
The above data demonstrated the positive
associations between AR and QKI. In order to validate this, an
siRNA sequence was used to specifically silence AR expression and
then observe its regulatory role on QKI (16). As a control, the expression of the
AR known target gene, PSA, was inhibited following AR gene
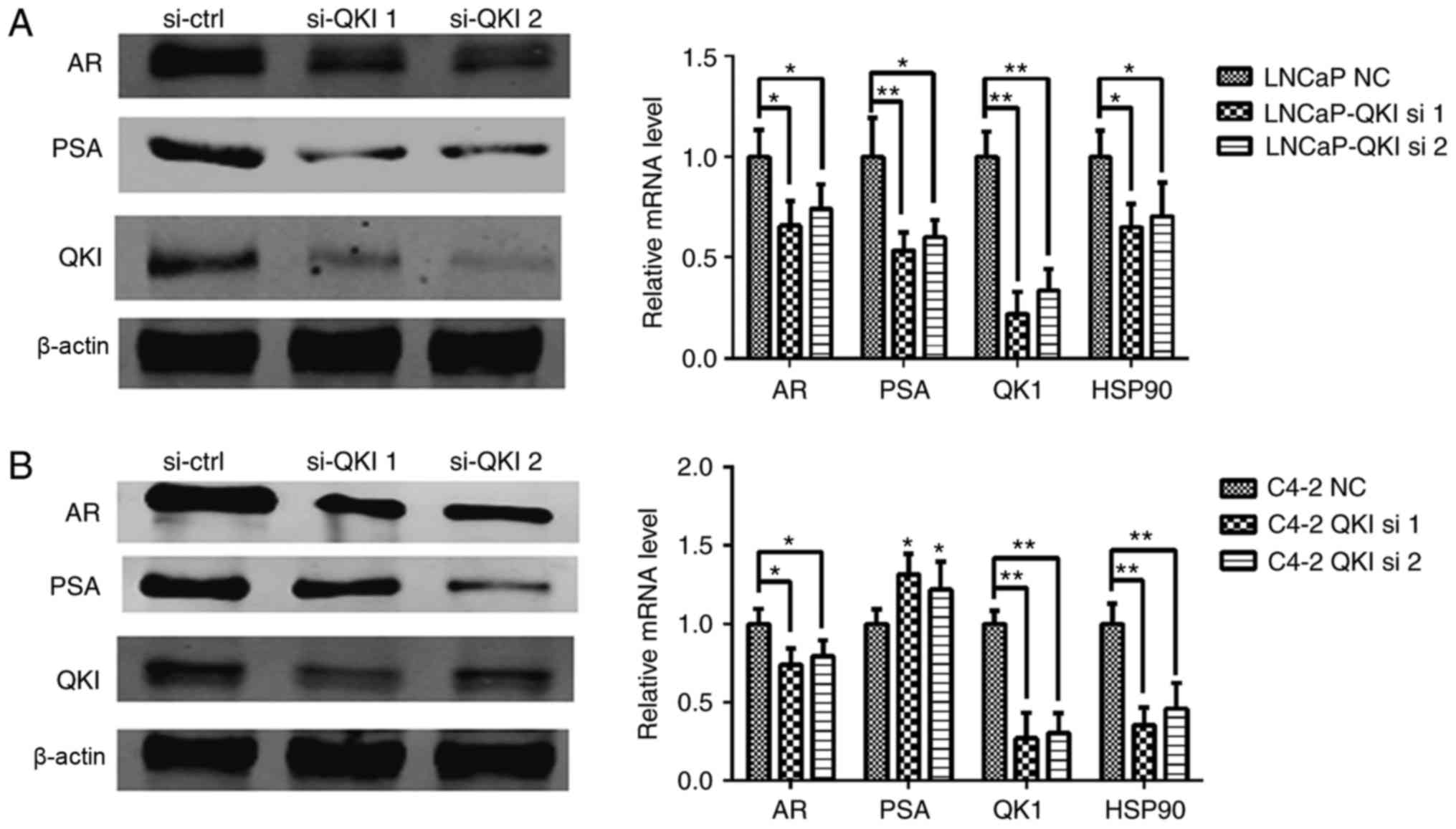
silencing. The expression levels of QKI were demonstrated to be
markedly inhibited in both cell lines (Fig. 3). On the whole, the positive
association observed between QKI and AR expression level, combined
with the concomitant changes of QKI in androgen deprivation and AR
silencing conditions, suggesting that androgen and the relevant AR
signals aid the enhancement of QKI expression at mRNA and protein
levels.
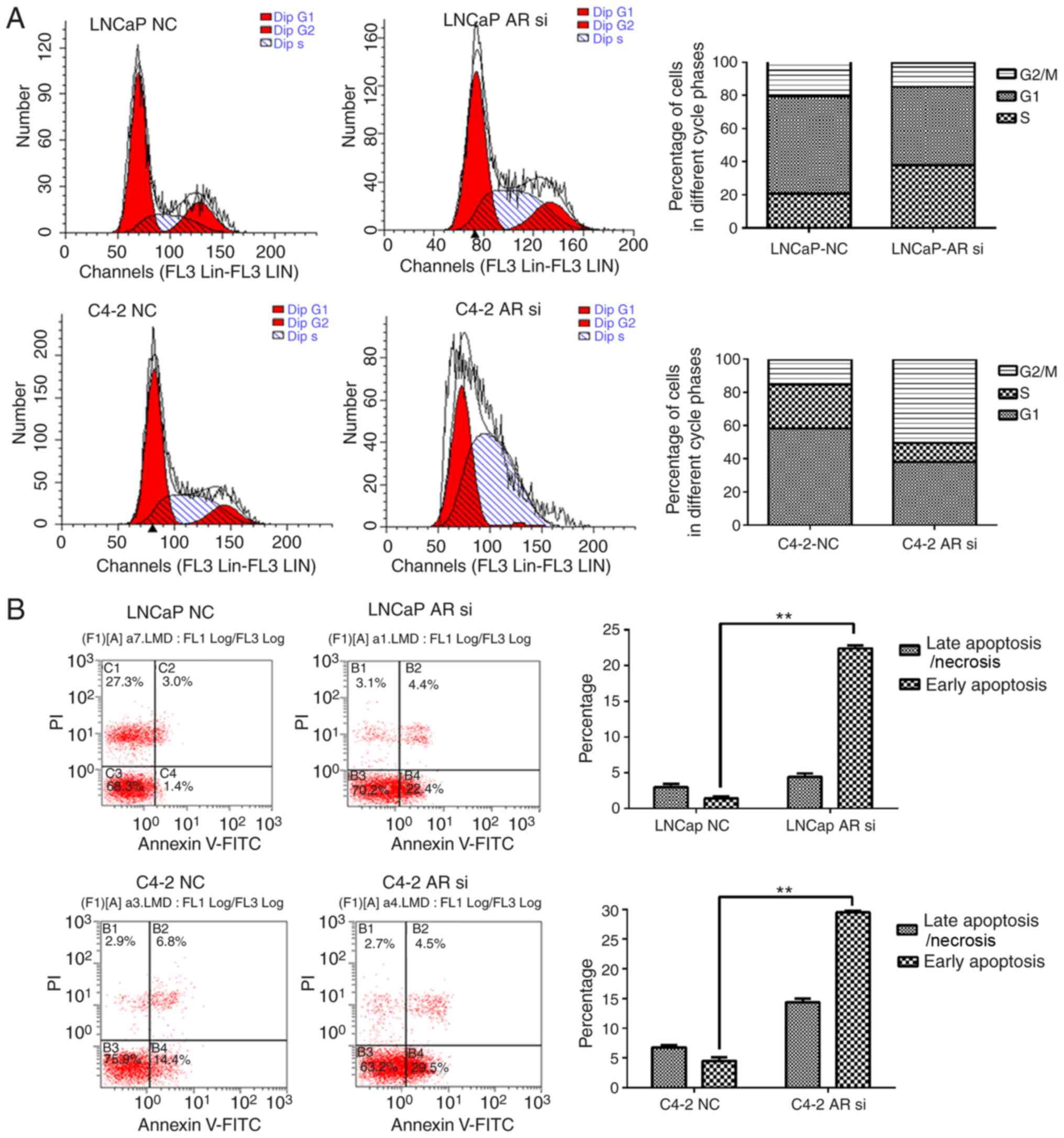
Prior to considering the role of QKI in AR related
tumorigenesis, the present study first investigated AR effects in
prostate cancer cells. Flow cytometry demonstrated that in LNCaP
cells, AR silencing arrested cell cycle at G1/S phase, while in
C4-2 cells, knocking down AR effectively retained the cell cycle at
G2/M phase. The cell-cycle G2/M phase gene UBE2C is over-expressed
in various solid tumors including CRPC, whereas UBE2C is a target
gene of AR spliced variants, which is a significant difference
between LNCaP and C4-2. In addition, the two cell lines
demonstrated increased rates of apoptosis (Fig. 4). These data further supported the
anti-apoptosis and proliferation-promoting effects of AR.
Effects of QKI gene silencing on
prostate cancer cells and AR antagonist drug Casodex
In agreement with the above data, QKI was confirmed
as a novel target gene of AR, although its functions in AR-positive
prostate cancer remain to be elucidated. In the present study, two
differing sets of RNA interference sequences against QKI were
employed, effectively knocking down the QKI gene (8), and expression levels of PSA and AR
were mildly reduced (Fig. 5). In
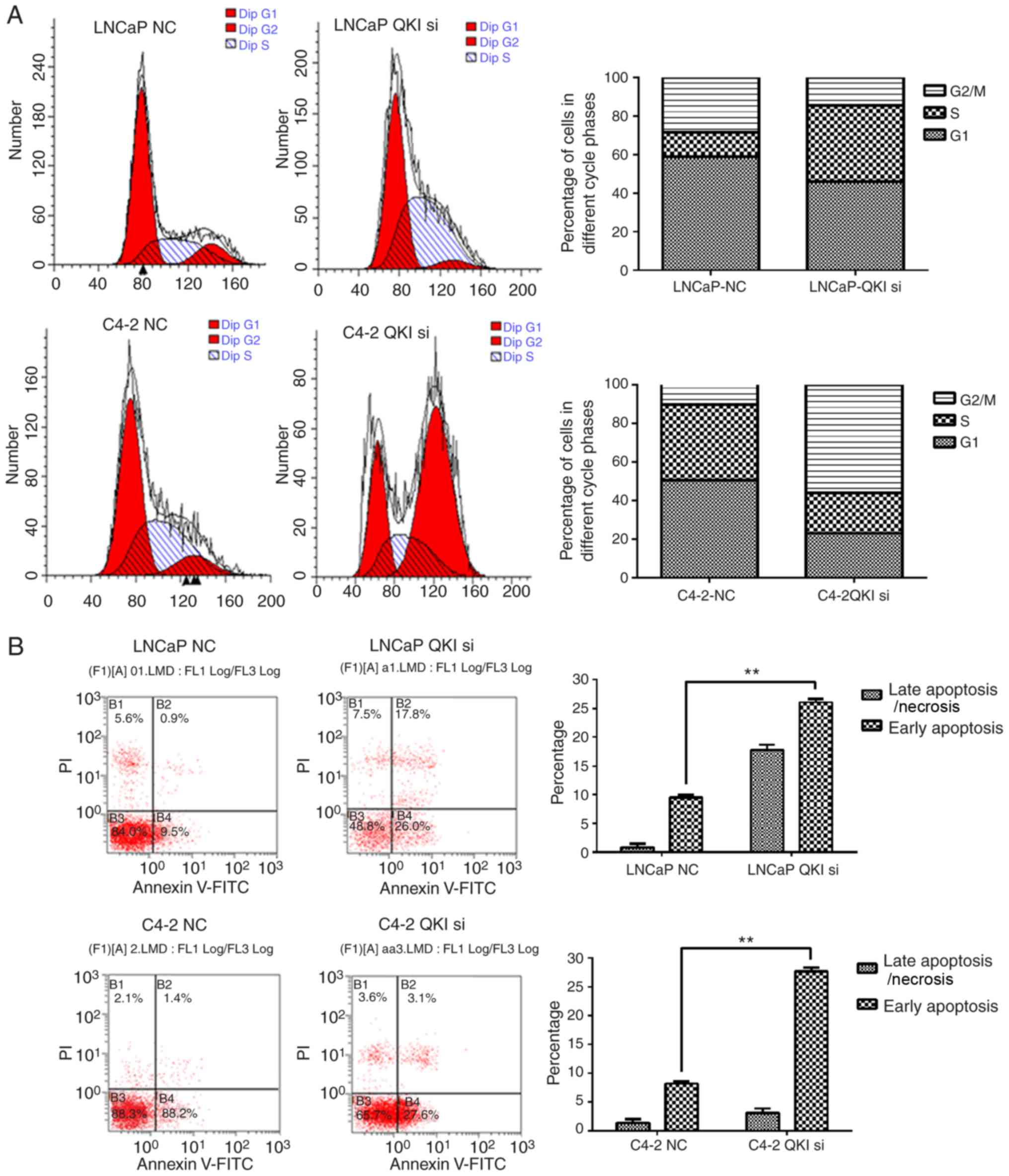
addition, identical cell cycle patterns were detected in LNCaP and
C4-2 cells following QKI reductions (Fig. 6), as was the case in AR silencing.
In particular, LNCaP cells mainly exhibited G1/S phase arrest,
while C4-2 cells exhibited a G2/M retention. Corresponding
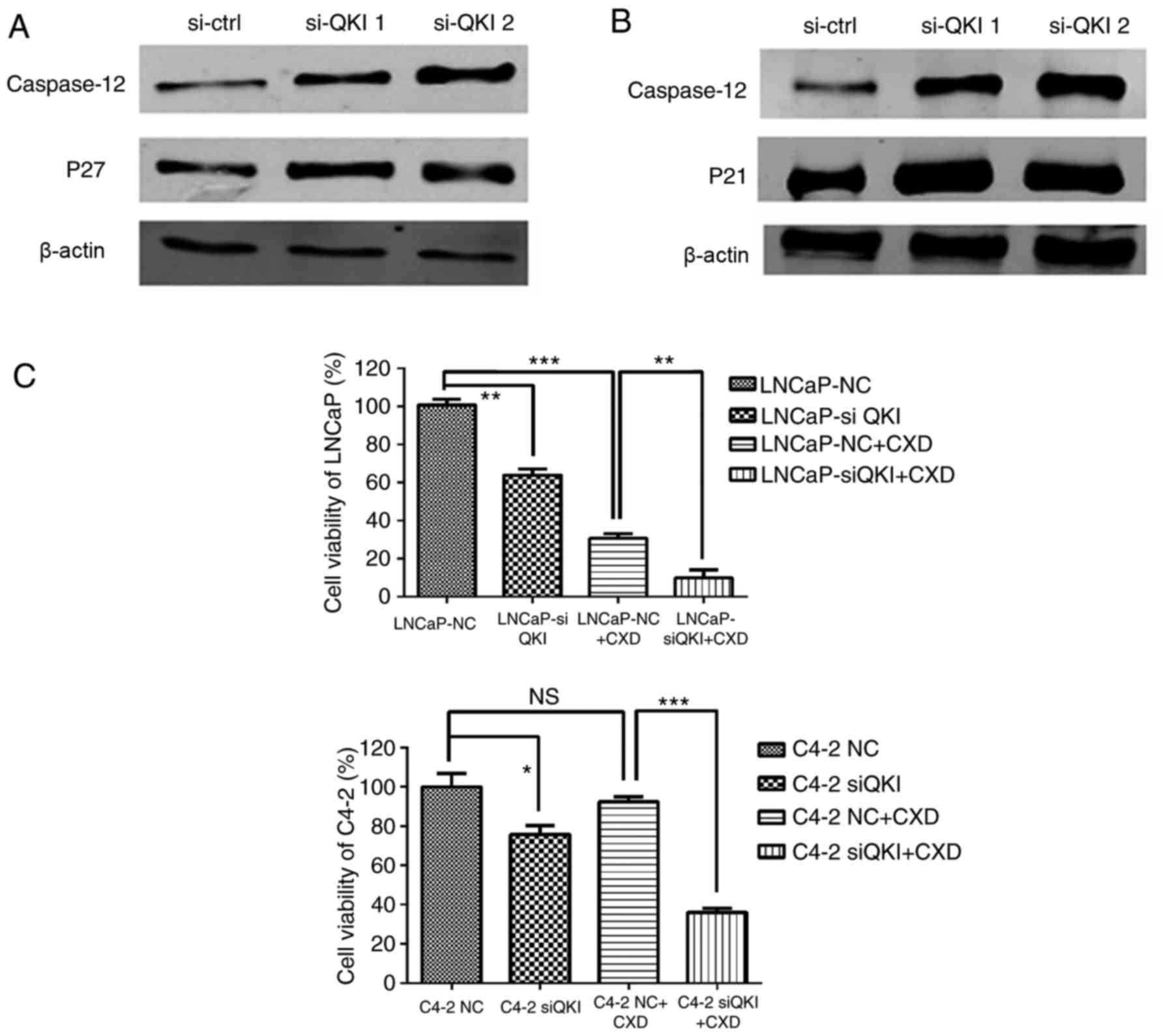
cell-cycle regulators, p27 and p21 demonstrated upregulations
additionally (Fig. 7A and B).
Furthermore, knock down QKI enhanced the susceptibility of prostate
cancer cells to death in fluorescence-activated cell sorting
assays, accompanied with higher level of caspase-12, an apoptosis
indicator (Fig. 7A and B). In
conclusion, these data indicated that QKI exerted a tumor promoting
effect in AR-associated prostate cancers.
Finally, a Casodex-sensitivity assay was conducted;
prostate cancer cells were treated with QKI siRNA 24 h in advance,
and the AR antagonist drug Casodex was added and cultivated for
another 48 h. MTT results demonstrated that C4-2 cells were more
resistant to Casodex treatment compared with LNCaP cells, whereas
QKI reduction synergistically sensitized androgen resistant cells
C4-2 to Casodex treatment (Fig.
7C). This may be associated with HSP90 repression; QKI
silencing dramatically reduced HSP90 expression.
Discussion
The incidence of prostate cancer is increasing among
Chinese males, and studies on its underlying mechanism are becoming
more important. QKI is an RNA-binding protein closely associated
with cell cycle and differentiation regulation (20). Aberrant hypermethylation at
promoter GC island regions of QKI is responsible for the reduced
expression in the higher staging prostate cancers, particularly in
those AR negative cell lines, such as PC-3 and DU-145. Previous
functional characterizations in prostate cancer have suggested
tumor suppressing effects for QKI.
The present study demonstrated a positive
association between QKI and AR in AR positive cell lines and
clinical samples. Either altered AR or androgen levels were able to
affect QKI expression levels positively in androgen sensitive LNCaP
and resistant C4-2 cells. Dual-luciferase reporter gene results
demonstrated that QKI promoter activity could be regulated by AR,
and future studies may elucidate this interaction further. This is
the first study, to the best of our knowledge, to report that QKI
is a positive target gene associated with AR signals in prostate
cancer.
The data from the current study suggested that QKI
mediated tumor-promoting effects in AR positive prostate cancer
deriving from the cell cycle and cell apoptosis assays and relevant
molecular markers. The present study indicated that QKI has a tumor
suppressor role in inhibiting cell proliferation and enhancing
apoptosis (8,20). The level of QKI expression has been
demonstrated to be high in ER-β positive breast cancer cells
(21), increasing with the degree
of malignancy. However, QKI may not serve a tumor suppressor
function in all cancer cells. The results of the present study
demonstrated that QKI may facilitate ADPC to CRPC in prostate
cancer progress. The reason for this phenomenon remains to be
elucidated. More importantly, HSP90, known to be responsible for AR
relevant signals and a number of cell cycle regulations, was
significantly diminished following QKI silencing. AR protein
stability is known to be also affected by the presence of HSP90.
Therefore, it is possible that QKI exerted tumor promoting effects
by improving the anti-stress efficiency in response to AR signals.
Further experiments are required to clarify this.
Casodex resistance is a severe problem in prostate
cancer treatment (22), and the
findings of the present study provided a novel option for
overcoming that resistance. QKI reduction markedly enhanced the
efficiency of Casodex in the resistant C4-2 cell line, as did a
combination of Casodex and HSP90 inhibitors (23); therefore, HSP90 repression
following QKI silencing through a mechanism yet to be elucidated,
may contribute to the increased sensitivity of the C4-2 cells to
Casodex.
The present study reported that QKI, defined as a
novel target gene downstream of AR signaling, functions as a
positive regulator in AR signaling to induce cell growth and
anti-apoptosis effects in the early stage of prostate cancer.
Knocking down QKI may enhance the anti-AR efficiency.
Acknowledgements
The authors thank Dr Donghui Han for providing the
C4-2 cells, and the Pharmacogenomics Department of Fourth Military
Medical University for providing the laboratory. The present study
was supported by the Scientific Innovative Project of Shaanxi
Province (grant no. 2012KTCL03-03), Collaborative Innovation
Projects of Shaanxi Province (grant no. 2015XT-53), the National
Science Foundation of China (grant no. 31571215), the Military
Medical Innovation Project (grant no. 16CXZ023) and Xijing Hospital
Subject Booster Plan Translational Medicine Research Projects
(grant no. XJZT13Z05).
Glossary
Abbreviations
Abbreviations:
|
AR
|
androgen receptor
|
|
QKI
|
quaking
|
|
PSA
|
prostate-specific antigen
|
|
CRPC
|
castration-resistant prostate
cancer
|
|
PBS
|
phosphate buffered solution
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
siRNA
|
small interfering RNA
|
|
ADT
|
androgen deprivation therapy
|
|
DHT
|
dihydrotestosterone
|
|
HSP90
|
heat shock protein 90
|
|
CXD
|
Casodex
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Babu Suresh S, Joladarashi D, Jeyabal P,
Thandavarayan RA and Krishnamurthy P: RNA-stabilizing proteins as
molecular targets in cardiovascular pathologies. Trends Cardiovasc
Med. 25:676–683. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Feng Y and Bankston A: The star family
member QKI and cell signaling. Adv Exp Med Biol. 693:25–36. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bohnsack BL, Lai L, Northrop JL, Justice
MJ and Hirschi KK: Visceral endoderm function is regulated by
quaking and required for vascular development. Genesis. 44:93–104.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao L, Ku L, Chen Y, Xia M, LoPresti P
and Feng Y: QKI binds MAP1B mRNA and enhances MAP1B expression
during oligodendrocyte development. Mol Biol Cell. 17:4179–4186.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mulholland PJ, Fiegler H, Mazzanti C,
Gorman P, Sasieni P, Adams J, Jones TA, Babbage JW, Vatcheva R,
Ichimura K, et al: Genomic profiling identifies discrete deletions
associated with translocations in glioblastoma multiforme. Cell
Cycle. 5:783–791. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang G, Fu H, Zhang J, Lu X, Yu F, Jin L,
Bai L, Huang B, Shen L, Feng Y, et al: RNA-binding protein quaking,
a critical regulator of colon epithelial differentiation and a
suppressor of colon cancer. Gastroenterology. 138(231–240): e1–e5.
2010.
|
|
9
|
Zong FY, Fu X, Wei WJ, Luo YG, Heiner M,
Cao LJ, Fang Z, Fang R, Lu D, Ji H and Hui J: The RNA-binding
protein QKI suppresses cancer-associated aberrant splicing. PLoS
Genet. 10:e10042892014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao Y, Zhang G, Wei M, Lu X, Fu H, Feng
F, Wang S, Lu W, Wu N, Lu Z and Yuan J: The tumor suppressing
effects of QKI-5 in prostate cancer: A novel diagnostic and
prognostic protein. Cancer Biol Ther. 15:108–118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu G, Sprenger C, Sun S, Epilepsia KS,
Haugk K, Zhang X, Coleman I, Nelson PS and Plymate S: AR variant
ARv567es induces carcinogenesis in a novel transgenic mouse model
of prostate cancer. Neoplasia. 15:1009–1017. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garrido C, Paul C, Seigneuric R and
Kampinga HH: The small heat shock proteins family: The long
forgotten chaperones. Int J Biochem Cell Biol. 44:1588–1592. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaul G and Thippeswamy H: Role of heat
shock proteins in diseases and their therapeutic potential. Indian
J Microbiol. 51:124–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao M, Geng XP and Xiang HP: HSP90 and
SIRT3 expression in hepatocellular carcinoma and their effect on
invasive capability of human hepatocellular carcinoma cells. Asian
Pac J Trop Med. 8:305–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liao X, Tang S, Thrasher JB, Griebling TL
and Li B: Small-interfering RNA-induced androgen receptor silencing
leads to apoptotic cell death in prostate cancer. Mol Cancer Ther.
4:505–515. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gillis JL, Selth LA, Centenera MM, Townley
SL, Sun S, Plymate SR, Tilley WD and Butler LM:
Constitutively-active androgen receptor variants function
independently of the HSP90 chaperone but do not confer resistance
to HSP90 inhibitors. Oncotarget. 4:691–704. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Penel N: Splicing variant of androgen
receptors (AR-V7): New paradigms. Bull Cancer. 103:711–713.
2016.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu X and Feng Y: QKI-5 suppresses cyclin
D1 expression and proliferation of oral squamous cell carcinoma
cells via MAPK signalling pathway. Int J Oral Maxillofac Surg.
44:562–567. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu F, Jin L, Yang G, Ji L, Wang F and Lu
Z: Post-transcriptional repression of FOXO1 by QKI results in low
levels of FOXO1 expression in breast cancer cells. Oncol Rep.
31:1459–1465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Colabufo NA, Pagliarulo V, Berardi F,
Contino M, Inglese C, Niso M, Ancona P, Albo G, Pagliarulo A and
Perrone R: Bicalutamide failure in prostate cancer treatment:
Involvement of multi drug resistance proteins. Eur J Pharmacol.
601:38–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Daniels G, Jha R, Shen Y, Logan SK and Lee
P: Androgen receptor coactivators that inhibit prostate cancer
growth. Am J Clin Exp Urol. 2:62–70. 2014.PubMed/NCBI
|