Introduction
Saliva is an easily obtainable biological fluid,
which can reflect body metabolism (1). Certain components of saliva are
secreted by salivary glands, while other components originate from
plasma. Of human saliva proteins, 30% are of blood origin and the
majority of them exhibit antimicrobial activities, indicating that
composition of saliva is dependent on the composition of plasma
(2). Analysis of human saliva can
be used as a diagnostic tool to detect markers of diseases and
physiological states (3).
Certain reports have stated that several parameters
in human saliva resemble those in plasma. Öztürk et al
(4) examined lipid peroxidation
intensity during human gestation and postpartum, and identified an
association between the concentration of examined parameters in
plasma and saliva. Salivary glands express oestrogen receptors and
therefore, pregnancy may affect the composition of saliva (5).
Preterm delivery is defined as parturition before
the 37th week of gestation and is the most common cause of
perinatal morbidity and mortality. A general hormonal profile of
patients in the periparturient period has already been described,
however detailed mechanisms underlying the initiation of delivery
remain unclear (6). Elucidation
and annotation of protein profile in plasma and in saliva may be
useful for understanding the physiological processes underlying
delivery and potential pathologies (7). There are several reports describing
plasma and placental protein profile during human gestation
(8–10); however, data regarding salivary
proteins are scarce.
Menon et al (11), reviewed several co-regulated
processes associated with delivery, including the study of foetal
membranes, maternal signals, maturation of foetal organs and
hormonal profile. The study emphasized the role of proteins in the
aforementioned processes associated with delivery (11).
From a clinical point of view, it is necessary to
develop a rapid and accurate method for predicting the time of
delivery, and to identify women at risk for spontaneous preterm
birth or other alterations during pregnancy. The aim of the present
study was to evaluate protein profile in saliva from patients with
uncomplicated pregnancy and delivery at term, and to compare the
profile with that from premature delivery.
Materials and methods
Patients
The present study included 22 pregnant women
receiving treatment and delivering between February 2015 and
November 2016 at the Department of Obstetrics and Pathology of
Pregnancy at the Independent Clinical Hospital No. 1 in Lublin,
Medical University of Lublin (Lublin, Poland). The study was
approved by The Ethics Committee of the Medical University of
Lublin (KE-0254/77/2014).
The study group [partus praematurus imminens (PPI)]
included 12 patients with preterm uterine contractions and
diagnosis of threatened preterm delivery. Iatrogenic preterm birth
occurred in 1 patient as a result of foetal distress and 3 patients
delivered at term despite symptoms of threatened preterm delivery.
These 4 patients were excluded from further analysis. Saliva was
collected at the time of admission to the Independent Clinical
Hospital No 1 in Lublin, Poland.
The control group (C) included 10 patients with
uncomplicated pregnancies. During routine control visits between
27–32 weeks of gestation in the outpatients clinic, saliva was
collected form each patient. One patient from the control group
declined to continue participation in the study and another
delivered in a different hospital; thus, these patients were
excluded from the study. Therefore, data from 16 women were further
analysed, including 8 C group patients and 8 PPI patients.
Inclusion criteria was a follows: Maternal age, >18; delivery
prior to 35 weeks of gestation; intact membranes; and regular
uterine contractions with cervical effacement ≥50%. Exclusion
criteria were as follows: Multiple pregnancy; no confirmation of
gestational age by ultrasound examination performed in the first
trimester; iatrogenic preterm birth; and smoking during pregnancy.
Written informed consent was obtained from all participants at the
time of enrolment. Participation in the study was voluntary. The
characteristics of study participants are presented in Table I. Mean gestational age at the time
of sample collection in both groups was 31.5±2.8 weeks. Saliva was
collected in the morning prior to eating.
 | Table I.Characteristics of the patients and
controls. |
Table I.
Characteristics of the patients and
controls.
| Characteristic | Experimental group
(n=8) | Control group
(n=8) |
|---|
| Mean maternal age
(years) | 27.8±3.6 | 28.4±5.8 |
| Mean gestational age
at delivery (weeks) | 31.5±2.8 | 39.2±1.3 |
| Number
primiparous | 5 (62%) | 3 (37%) |
| Mean birth weight
(g) | 1952±980 | 3608±505 |
| Maternal BMI
(kg/m2) | 28.4±7.2 | 29.6±6.8 |
Preparation of saliva
Saliva samples were collected in Salivette tubes
(Sarstedt, Inc., Newton, NC, USA). Samples were centrifuged for 15
min at 5,000 × g in 4°C. Protease inhibitor cocktail (10 µl; cat.
no. I3911; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added
to 1,000 µl of supernatant. The samples were stored at −20°C prior
to further analysis.
Then thawed saliva (700 µl) was concentrated to ~50
µl using Vivaspin columns for ultrafiltration (polyethersulfone
membrane, molecular weight cutoff 3 kDa; Sartorius AG, Göttingen,
Germany), washed twice with 100 µl 7.0 pH 20 mM PBS and suspended
in 330 µl rehydration buffer containing 8 M urea, 2% CHAPS, 50 mM
dithiothreitol (DTT), 0.2% Bio-Lyte 3/10 ampholyte and 0.001%
bromophenol blue.
Electrophoresis
Isoelectric focusing was performed using strips with
linear gradient of pH 3–10 and length of 17 cm. Each saliva sample
was loaded onto one strip. Preparation of SDS-PAGE (16×20 cm; 1.0
mm; T=11%, C=2.6%) was performed as previously described by Laemmli
(12). The second dimension was
performed using a Protean II XL cell (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) according to the manufacturer's protocol. Gels
were stained with silver nitrate according to MS compatible
protocol (13).
Identification of spots
Selected spots of interest (27 spots) were removed
from the gels manually (in accordance to significance and intensity
of staining), cut into small pieces and transferred to 0.5 ml
Eppendorf tubes. The gel pieces were subsequently washed three
times with 100 µl 100 mM NH4HCO3 buffer (pH
8.5; Sigma-Aldrich; Merck KGaA) for 5 min. Gel pieces were
dehydrated by adding 100 µl acetonitrile (ACN) and dried for 15 min
at room temperature in vacuum concentrator CentriVap with 220 × g
(Labconco Corporation, Kansas City, MO, USA). Samples were
resuspended in 100 µl of 10 mM DTT in 50 mM
NH4HCO3 buffer in order to perform reduction
at 56°C for 60 min. Following cooling to room temperature, the
solution was replaced by 100 µl of 50 mM iodoacetamide in 50 mM
NH4HCO3 buffer and samples were incubated in
the dark for 45 min at room temperature. Trypsin digestion of
proteins was carried out on ice by gradual addition of 10 µl 12.5
ng/ml enzyme solution (Trypsin Gold; mass spectrometry grade;
Promega Corporation, Madison, WI, USA) in 50 mM
NH4HCO3 buffer until proteins were completely
rehydrated. Finally, 30 µl 50 mM NH4HCO3
buffer was added to each sample and digestion reaction was allowed
to proceed at 37°C overnight. Following digestion, the supernatant
was collected and peptides were extracted three times with 50 µl
70% ACN and 1.5% trifluoroacetic acid (TFA) by sonication for 15
min at room temperature in an ultrasonic water bath (Ultron U-507;
Ultron, Dywity, Poland). The supernatant was collected and dried in
the CentriVap (Labconco Corporation) for 45 min at 40°C and 220 ×
g.
The obtained pellet of peptides was resuspended in
10 µl 0.1% TFA and purified with µC18 ZipTip columns (Eppendorf,
Hamburg, Germany) in accordance with the manufacturer's protocol.
After purification, 1 µl peptide solution (1.5% TFA, 30% ACN) was
applied to α-cyano-4-hydroxycinnamic acid-Prespotted AnchorChip
frame (Bruker Corporation, Billerica, MA, USA) and left to dry at
room temperature. Mass spectra were obtained using Ultraflex III
matrix-assisted laser desorption/ionization
time-of-flight/time-of-flight spectrometer (MALDI; Bruker
Corporation). Acquisition was performed in positive ion reflector
mode with a 25 kV acceleration voltage. External calibrations were
prepared using the peptide calibration standard (Bruker
Corporation). FlexAnalysis 3.0 software (Bruker Corporation) was
used for selection of monoisotopic peptide masses.
Identification of peptides and proteins from mass
spectrometry data was performed using Mascot algorithm (Mascot 2.2
software; Matrix Science, Ltd., London, UK) and Swiss-Prot database
(UniProt release 2016_07; uniprot.org/statistics/Swiss-Prot) restricted to the
human taxonomy. Search parameters were set in the following mode:
Enzyme-trypsin, up to 1 missed cleavages, fixed
modification-carboamoinodmethylation cysteine, variable
modification-oxidation of methionine, error of 50 ppm. Analyses and
comparisons with the database were made by the BioTools 3.2
software (Bruker Corporation).
Statistical analysis
Delta2D software (version 4.6; Decodon GmbH,
Greifswald, Germany) was used to differentiate the presence of
particular spots and intensity of staining within examined saliva
samples using one-way analysis of variance, as previously described
(14). As a result, a list of most
common and differential spots between groups C and PPI (P<0.05)
was created and spots for further analysis were selected. A
principal components analysis scatterplot (PCA 3D) of all spots was
constructed using Delta2D software (15).
Spot selection for identification was based on the
presence of spots in one or both of the examined groups as well as
their intensity of staining that reflects the amount of protein
necessary for further identification analysis (highly visible spots
with amount of protein more that 5–10 ng). The number of analysed
saliva samples followed proteomic requirements (16,17).
The specificity and sensitivity of used laboratory methods
(16,17) allowed for reliable statistical
analysis. Data is presented as the mean ± standard deviation.
P<0.05 was considered to indicate a statistically significant
difference.
The protein identification of spots was based on the
comparison with UniProt databases and confirmed by Mascot scores
(http://www.matrixscience.com/help/scoring_help.html).
Scores >67 were considered to indicate a statistical
significance.
Results
Out of 1,393 detected spots, 59 were significantly
different between the C and PPI groups. Statistical analysis
demonstrated that 32 spots exhibited increased intensity of
staining in PPI group samples compared with the C group, while 27
had the opposite trend (Table
II). Out of 59 spots that differed significantly between
examined groups 9 were identified and are shown in Table III. Three spots exhibited
increased intensity of staining in the PPI group compared with the
C group: Dedicator of cytokinesis protein 1 (DOCK1),
metallothionein-2 (MT2A) and guanylyl cyclase-activating protein 1
(GUCA1A). Six spots exhibited an opposite pattern in PPI and C
groups: Epithelial-stromal interaction protein 1 (EPSTI1), serum
albumin, tyrosine--tRNA ligase, cytoplasmic (YARS), protein chibby
homolog 3, leukemia inhibitory factor receptor (LIFR) and
adenosylhomocysteinase 3. DOCK1 is a haemostasis-associated
protein, and GUCA1A and EPSTI1 are proteins associated with
programmed cell death. YARS is associated with gene expression and
LIFR has a role in the immune system. MT2A involved in
antioxidative defence.
 | Table II.Statistical analysis of significantly
different (P<0.05) spots between examined groups of patients, C
and PPI. |
Table II.
Statistical analysis of significantly
different (P<0.05) spots between examined groups of patients, C
and PPI.
| Spot label | PPI mean | Control mean | C/PPI ratio | P-value |
|---|
| 1 | 0.053 | 0.001 | 0.027 | 0.024 |
| 2 | 0.045 | 0.002 | 0.035 | 0.021 |
| 3 | 0.011 | 0.002 | 0.181 | 0.015 |
| 4 | 0.024 | 0.005 | 0.225 | 0.024 |
| 5 | 0.015 | 0.002 | 0.118 | 0.034 |
| 6 | 0.145 | 0.219 | 1.509 | 0.048 |
| 7 | 0.307 | 0.597 | 1.948 | 0.031 |
| 8 | 0.107 | 0.283 | 2.639 | 0.045 |
| 9 | 0.025 | 0.005 | 0.189 | 0.004 |
| 10 | 0.015 | 0.003 | 0.197 | 0.011 |
| 11 | 0.064 | 0.229 | 3.574 | 0.011 |
| 12 | 0.207 | 0.364 | 1.759 | 0.048 |
| 13 | 1.233 | 2.411 | 1.956 | 0.023 |
| 14 | 0.068 | 0.235 | 3.434 | 0.016 |
| 15 | 0.009 | 0.002 | 0.184 | 0.044 |
| 16 | 0.01 | 0.002 | 0.174 | 0.03 |
| 17 | 0.032 | 0.003 | 0.105 | 0.015 |
| 18 | 0.1 | 0.037 | 0.368 | 0.024 |
| 19 | 0.05 | 0.139 | 2.793 | 0.028 |
| 20 | 0.028 | 0.008 | 0.273 | 0.033 |
| 21 | 0.014 | 0.062 | 4.396 | 0.019 |
| 22 | 0.037 | 0.011 | 0.307 | 0.014 |
| 23 | 0.017 | 0.053 | 3.071 | 0.028 |
| 24 | 0.015 | 0.002 | 0.14 | 0.021 |
| 25 | 0.008 | 0.059 | 7.127 | 0.012 |
| 26 | 0.096 | 0.025 | 0.262 | 0.016 |
| 27 | 0.818 | 0.311 | 0.38 | 0.003 |
| 28 | 0.335 | 0.042 | 0.125 | 0.036 |
| 29 | 1.116 | 0.542 | 0.485 | 0.026 |
| 30 | 0.023 | 0.002 | 0.099 | 0.044 |
| 31 | 0.118 | 0.318 | 2.681 | 0.017 |
| 32 | 0.009 | 0.002 | 0.238 | 0.041 |
| 33 | 0.011 | 0.001 | 0.119 | 0.042 |
| 34 | 0.027 | 0.003 | 0.13 | 0.0005 |
| 35 | 0.021 | 0.005 | 0.215 | 0.044 |
| 36 | 0.084 | 0.254 | 3.03 | 0.006 |
| 37 | 0.347 | 1.062 | 3.06 | 0.002 |
| 38 | 0.018 | 0.074 | 4.14 | 0.024 |
| 39 | 0.135 | 0.035 | 0.26 | 0.047 |
| 40 | 0.013 | 0.003 | 0.218 | 0.043 |
| 41 | 0.009 | 0.025 | 2.606 | 0.03 |
| 42 | 0.31 | 0.975 | 3.144 | 0.009 |
| 43 | 0.017 | 0.003 | 0.18 | 0.022 |
| 44 | 0.021 | 0.005 | 0.254 | 0.039 |
| 45 | 0.02 | 0.086 | 4.383 | 0.006 |
| 46 | 0.058 | 0.138 | 2.386 | 0.007 |
| 47 | 0.07 | 0.862 | 12.254 | 0.01 |
| 48 | 0.475 | 0.069 | 0.144 | 0.016 |
| 49 | 0.103 | 0.357 | 3.473 | 0.0005 |
| 50 | 0.045 | 0.006 | 0.133 | 0.049 |
| 51 | 0.046 | 0.011 | 0.244 | 0.026 |
| 52 | 0.776 | 1.185 | 1.526 | 0.045 |
| 53 | 0.143 | 0.363 | 2.539 | 0.005 |
| 54 | 0.049 | 0.019 | 0.384 | 0.041 |
| 55 | 0.056 | 0.175 | 3.13 | 0.006 |
| 56 | 0.04 | 0.127 | 3.144 | 0.013 |
| 57 | 0.207 | 0.692 | 3.338 | 0.026 |
| 58 | 0.029 | 0.134 | 4.617 | 0.025 |
| 59 | 0.105 | 0.024 | 0.228 | 0.001 |
 | Table III.The list of identified spots in
saliva of the examined groups of patients. |
Table III.
The list of identified spots in
saliva of the examined groups of patients.
| Spot label | Protein | UniProt entry | Mascot Score | Protein sequence
coverage (%) | Molecular weight
(kDa) | pI |
|---|
| 6 | Epithelial-stromal
interaction protein 1 | Q96J88
(ESIP1_HUMAN) | 58 | 16 | 36.80 | 9.90 |
| 12 | Serum albumin | P02768
(ALBU_HUMAN) | 61 | 21 | 69.37 | 5.92 |
| 18 | Dedicator of
cytokinesis protein 1 | Q14185
(DOCK1_HUMAN) | 87 | 8 | 21.53 | 7.29 |
| 19 | Tyrosine-tRNA
ligase, cytoplasmic | P54577
(SYYC_HUMAN) | 80 | 18 | 59.14 | 6.61 |
| 21 | Protein chibby
homolog 3 | A6NI87
(CBY3_HUMAN) | 59 | 33 | 27.34 | 10.65 |
| 22 |
Metallothionein-2 | P02795
(MT2_HUMAN) | 65 | 54 | 6.04 | 8.23 |
| 23 | Leukemia inhibitory
factor receptor | P42702
(LIFR_HUMAN) | 133 | 16 | 123.74 | 5.50 |
| 25 |
Adenosylhomocysteinase 3 | Q96HN2
(SAHH3_HUMAN) | 160 | 19 | 66.72 | 7.13 |
| 29 | Guanylyl
cyclase-activating protein 1 | P43080
(GUC1A_HUMAN) | 62 | 57 | 22.92 | 4.34 |
In addition, 18 spots that did not differ in
intensity of staining between groups were identified. The
identified proteins included, carboxypeptidase E (Cpe),
fucose-1-phosphate guanylyltransferase (FKGP), E3 ubiquitin-protein
ligase TRIM56 (TRIM56), myosin light chain kinase 2,
skeletal/cardiac muscle (Mylk2),
CMP-N-acetylneuraminate-β-galactosamide-α-2,3-sialyltransferase 4
(ST8SIA4), methyltransferase-like protein 13 (METTL13) and
pancreatic lipase-related protein 3 (PNLIPRP3). The above proteins
have roles in cell metabolism, but are not associated with
pregnancy and were not considered as clinically significant.
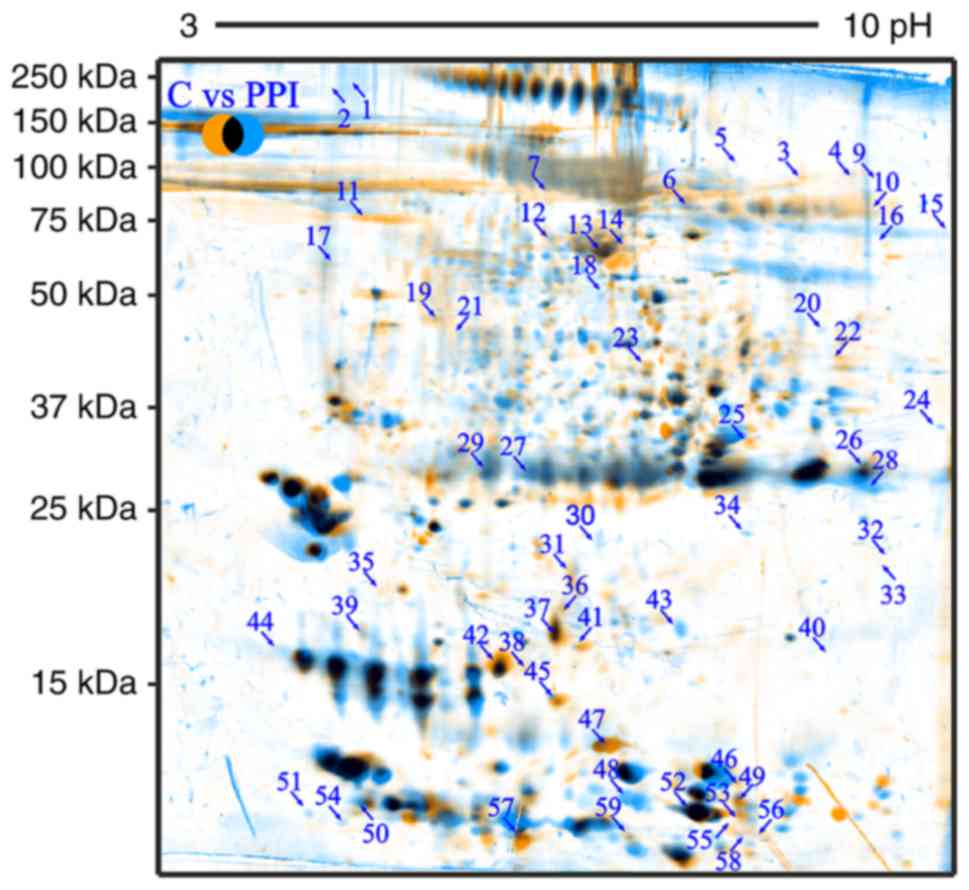
Fig. 1 presents
fused image of spots from both examined groups, which were
overlapped by use of Delta2D software. Orange spots are from C
group, blue from PPI group while common spots for both groups are
marked with black. The figure demonstrates the profile and location
of the spots distributed in accordance to the isoelectric point and
molecular weight of proteins, which were in examined the biological
fluids.
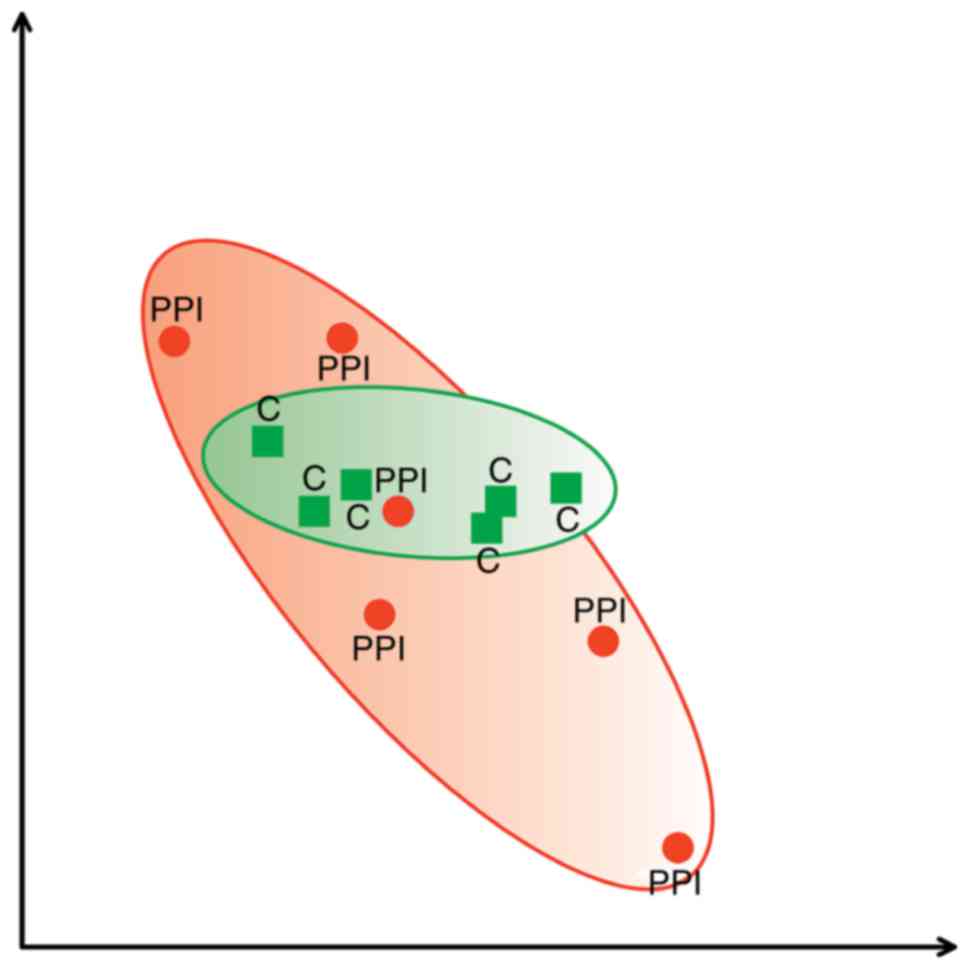
Fig. 2 presents PCA
scatterplot from C group marked with green area and PPI group
marked with red area. The location of areas represents the
association between the obtained spots and indicates that the
majority of proteins are similar in examined samples as they
overlay.
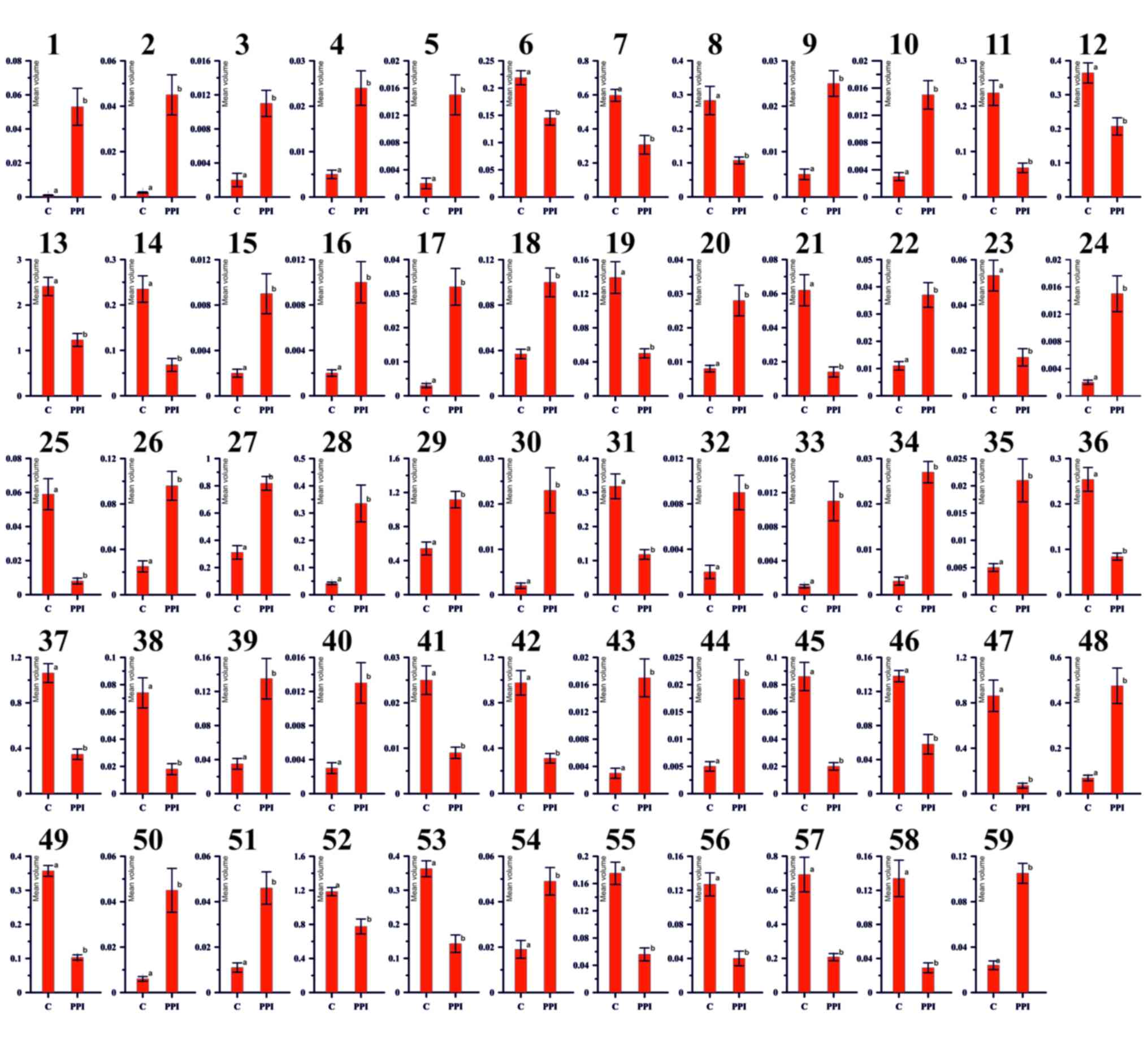
Fig. 3 represents
histograms of all spots that differ significantly in accordance to
intensity of staining what indirectly reflects the amount of
protein. All 59 pairs are significantly different between each
other at (P<0.05). Y-axis reflects the mean spot volume
(intensity of staining × spot area). X axis-experimental
groups.
Discussion
The present study compared the proteomic pattern of
saliva from patients that delivered at term, with those that
delivered prematurely. Statistical analysis enabled identification
of the spots that differed significantly in intensity of staining
between the examined groups of patients. As the intensity of
staining reflects the amount of protein in spot, this analysis
indicated quantitative differences in saliva protein profile
between examined spots. The majority of detected spots expressed
similar intensity. Proteins corresponding to certain spots were
identified.
A preliminary report featuring a plasma protein
profile in humans during pregnancy was published in 1978 by Joseph
et al (8). Term 2D protein
profile of human placenta was described by Mushahary et al
(9). Pregnancy-associated
proteins, which have a role in maintenance of human placenta
include proteins detected in maternal serum, foetal proteins and
soluble, solubilized or membrane-associated placental proteins
(18).
Yuan et al (10) described protein profile of human
placental blood plasma obtained from patients that underwent
elective caesarean section (not in labour), spontaneous vaginal
delivery, induction of labour vaginal delivery, spontaneous
delivery, emergency caesarean section and induction of delivery.
Apolipoproteins A-IV, E and C-III were considered to be associated
with the onset of delivery while hepatocyte nuclear factor 1
homeobox A was common for human parturition regardless of type of
delivery (10).
Preterm delivery may cause perinatal mortality and
morbidity, and is defined as birth prior to 37 weeks of gestation
(19). Although several factors
are known to be involved in spontaneous preterm termination of
pregnancy, biochemical markers of preterm delivery remain to be
elucidated. Previous studies indicated that intrauterine
inflammation may be one of the initiating factors of preterm
delivery (20,21). There are several other possible
factors associated with this pathological condition, including
uteroplacental ischemia, decidual haemorrhage, failure in the
maternal immunological tolerance of the foetus, allergies, abnormal
uterine size, cervical incompetence, maternal and foetal stress and
hormonal irregularities (22).
Numerous studies associated with preterm birth have
demonstrated that alterations in the expression of particular
molecules are associated with certain disturbances but none of them
were considered diagnostic tools and markers (19,21–24).
These molecules include membrane and soluble proteins present or
absent in the placental tissue of patients with preterm birth
(23). Proteins that are only
detectable in preterm placenta compared with placenta from
full-term births include, vimentin, β-actin and γ-actin as
representatives of structural proteins, and soluble proteins,
including lactoylglutathione lyase, transgelin and 78 kDa
glucose-regulated protein. Preterm labour is also characterized by
lack of certain placental proteins, which are present in normal
term placenta. These include membrane proteins annexin A4 and type
I keratin, and proteins present in the soluble fraction, including
endoplasmin, and β- and γ-actin (24).
Elevated levels of MT2A in saliva of PPI patients
compared with C patients detected in the present study may indicate
oxidative stress (25). Oxidative
stress alters protein molecules via peroxidative damage and
complicate preterm births (26).
These results are supported by the present study where, LIFR, which
has a role in ovulation, implantation and fertility, was
downregulated in the PPI group compared with the C group,
suggesting potential alterations in protein metabolism.
Identification of proteins that exhibit similar
patterns of expression in the examined saliva may aid in
elucidating the sequence of events that leads to premature
delivery. Molecules identified in the present study belong to
different functional groups, including enzymes, and may indicate
the presence of reactions that take place regardless of
pregnancy-associated alterations and appear to be necessary both in
physiological and pathological conditions. Among identified
proteins were enzymes, including, Cpe, FKGP, TRIM56, Mylk2,
ST8SIA4, METTL13 and PNLIPRP3.
As described in the review by Menon et al
(11), the onset of parturition
requires a complex and simultaneous transduction of several signals
from the foetus, foetal membranes and the mother via local and
general systems. Certain proteins may have roles in modulation of
reactions during delivery in physiological and pathological
conditions. Previous studies indicated that saliva may be a source
of parameters of diagnostic use in diseases of the oral cavity,
diabetes or breast cancer; however, the use of saliva samples for
diagnostics in obstetrics requires further investigation (27–29).
The present investigated saliva in the context of threatened
preterm delivery.
In conclusion, although saliva is an easy obtainable
biological fluid, its use in diagnostic procedures is currently
limited, due to the lack of information regarding its content. The
results of the present study suggested that MT2A in saliva, which
has a role in oxidative stress, may be a potential marker
indicating risk of premature delivery. Further studies with an
increased number of patients are necessary for verification and
validation of the results. The present study suggests a potential
for targeted proteomic analysis of saliva to identify biomarkers
that may predict premature delivery and other alterations
associated with the periparturient period.
Acknowledgements
Not applicable.
Funding
The present study was supported by Statutory
Activity of University of Life Sciences in Lublin (WKB-DS-4) and
Statutory Activity of Medical University of Lublin
(DS/122/2015-2017).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MŁ and MK conceived the presented idea. TG and AM
developed the theory and performed the computations. MŁ and MK
verified the analytical methods. JW and MF expressed and purified
all proteins. TG and AM analysed the data. JW designed the figures.
MŁ, TG and AM supervised the findings of this work. All authors
discussed the results and contributed to the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
participants at the time of enrolment. The study was approved by
the Ethics Committee of the Medical University of Lublin
(KE-0254/77/2014).
Consent for publication
Informed patient consent was obtained from all
participants at the time of enrolment.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lima DP, Diniz DG, Moimaz SA, Sumida DH
and Okamoto AC: Saliva: Reflection of the body. Int J Infect Dis.
14:e184–e188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schulz BL, Cooper-White J and Punyadeera
CK: Saliva proteome research: Current status and future outlook.
Crit Rev Biotechnol. 33:246–259. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xiao H and Wong DT: Proteomics and its
applications for biomarker discovery in human saliva.
Bioinformation. 5:294–296. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Öztürk LK, Akyüz S, Yarat A, Koç S, Gül N
and Doğan BN: Salivary lipid peroxidation and total sialic acid
levels during healthy gestation and postpartum: A longitudinal
study. Clin Biochem. 43:430–434. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valimaa H, Savolainen S, Soukka T,
Silvoniemi P, Mäkelä S, Kujari H, Gustafsson JA and Laine M:
Estrogen receptor-beta is the predominant estrogen receptor subtype
in human oral epithelium and salivary glands. J Endocrinol.
180:55–62. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Catalano RD, Lannagan TR, Gorowiec M,
Denison FC, Norman JE and Jabbour HN: Prokineticins: Novel
mediators of inflammatory and contractile pathways at parturition?
Mol Hum Reprod. 16:311–319. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shankar R, Gude N, Cullinane F, Brennecke
S, Purcell AW and Moses EK: An emerging role for comprehensive
proteome analysis in human pregnancy research. Reproduction.
129:685–696. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Joseph JC, Baker C, Sprang ML and Bermes
EW: Changes in plasma proteins during pregnancy. Ann Clin Lab Sci.
8:130–141. 1978.PubMed/NCBI
|
|
9
|
Mushahary D, Gautam P, Sundaram CS and
Sirdeshmukh R: Expanded protein expression profile of human
placenta using two-dimensional gel electrophoresis. Placenta.
34:193–196. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yuan W, Heesom K, Phillips R, Chen L,
Trinder J and Bernal López A: Low abundance plasma proteins in
labour. Reproduction. 144:505–518. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Menon R, Bonney EA, Condon J, Mesiano S
and Taylor RN: Novel concepts on pregnancy clocks and alarms:
Redundancy and synergy in human parturition. Hum Reprod Update.
22:535–560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Laemmli UK: Cleavage of structural
proteins during the assembly of the head of bacteriophage T4.
Nature. 227:680–685. 1970. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shevchenko A, Wilm M, Vorm O and Mann M:
Mass spectrometric sequencing of proteins silver-stained
polyacrylamide gels. Anal Chem. 68:850–858. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jerrold HZ: Biostatistical Analysis. 4th
edition. Prentice Hall; New Jersey: 1999
|
|
15
|
Jolliffe IT: Principal Component Analysis.
2nd edition. Springer; 2002
|
|
16
|
Smit S, Hoefsloot HC and Smilde AK:
Statistical data processing in clinical proteomics. J Chromatogr B
Analyt Technol Biomed Life Sci. 866:77–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chich JF, David O, Villers F, Schaeffer B,
Lutomski D and Huet S: Statistics for proteomics: Experimental
design and 2-DE differential analysis. J Chromatogr B Analyt
Technol Biomed Life Sci. 849:261–272. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mine K, Katayama A, Matsumura T, Nishino
T, Kuwabara Y, Ishikawa G, Murata T, Sawa R, Otsubo Y, Shin S and
Takeshita T: Proteome analysis of human placentae: Pre-eclampsia
versus normal pregnancy. Placenta. 28:676–687. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Parry S, Zhang H, Biggio J, Bukowski R,
Varner M, Xu Y, Andrews WW, Saade GR, Esplin MS, Leite R, et al:
Maternal serum serpin B7 is associated with early spontaneous
preterm birth. Am J Obstet Gynecol. 211:678.e1–e12. 2014.
View Article : Google Scholar
|
|
20
|
Nelson DM, Sadovsky Y, Robinson JM, Croy
BA, Rice G and Kniss DA: Advanced techniques in placental
biology-workshop report. Placenta. 27 Suppl A:S87–S90. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim MA, Lee YS and Seo K: Assessment of
predictive markers for placental inflammatory response in preterm
births. PLoS One. 9:e1078802014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pařízek A, Koucký M and Dušková M:
Progesterone, inflammation and preterm labor. J Steroid Biochem Mol
Biol. 139:159–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Romero R, Espinoza J, Gotsch F, Kusanovic
JP, Friel LA, Erez O, Mazaki-Tovi S, Than NG, Hassan S and Tromp G:
The use of high-dimensional biology (genomics, transcriptomics,
proteomics, and metabolomics) to understand the preterm parturition
syndrome. BJOG. 113:118–135. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Butt RH, Lee MW, Pirshahid SA, Backlund
PS, Wood S and Coorssen JR: An initial proteomic analysis of human
preterm labor: Placental membranes. J Proteome Res. 5:3161–3172.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vašák M and Meloni G: Chemistry and
biology of mammalian metallothioneins. J Biol Inorg Chem.
16:1067–1078. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Menon R: Oxidative stress damage as a
detrimental factor in preterm birth pathology. Front Immunol.
5:5672014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang A, Sun H, Wang P and Wang X:
Salivary proteomics in biomedical research. Clin Chim Acta.
415:261–265. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Al Kawas S, Rahim ZH and Ferguson DB:
Potential uses of human salivary protein and peptide analysis in
the diagnosis of disease. Arch Oral Biol. 57:1–9. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Al-Tarawneh SK, Border MB, Dibble CF and
Bencharit S: Defining salivary biomarkers using mass
spectrometry-based proteomics: A systematic review. OMICS.
15:353–361. 2011. View Article : Google Scholar : PubMed/NCBI
|