Introduction
Smoking is a strong risk factor for various oral
diseases and particularly for periodontitis (1,2),
which primarily presents as alveolar bone resorption. A higher
prevalence and more severe form of periodontitis has been
associated with smokers compared with non-smokers (3). Nicotine is the major toxic component
of tobacco and regulates bone metabolism by accelerating the loss
of periodontal tissue attachment and alveolar bone absorption,
exacerbating periapical disease, inducing peri-implantitis and
decreasing the rate of bone regeneration following tooth extraction
or surgery (4–7). The inhalation of tobacco leads to the
accumulation of toxic components in the circulatory system and
local oral tissue (8). Our
unpublished data indicated that nicotine is directly deposited on
tooth surfaces and in alveolar bone; therefore, we inferred that
the deposition of nicotine may serve an important role in
regulating bone metabolism.
Bone metabolism depends on the dynamic balance
between bone formation and absorption. Osteoblasts have an
important role in bone formation. En-Nosse et al (9) identified a number of types of
nicotinic acetylcholine receptors, to which nicotine can
specifically bind, on the surface of osteoblasts. Therefore, the
effects of nicotine on alveolar bone may be partially due to its
effects on osteoblasts. In an in vitro osteogenesis process,
osteoblasts adhere to the bone defect zone, synthesize and secrete
collagen I to form bone matrix, and subsequently secrete various
non-collagen matrix proteins, including alkaline phosphatase (ALP),
bone osteocalcin (OCN), bone sialoprotein (BSP) and osteopontin
(OPN) (10), which serve important
roles in the mineralization of bone matrix.
Previous studies have indicated the effects of
nicotine on the proliferation and differentiation of osteoblasts in
different types of models (11–15).
Few of the studies used primary osteoblasts. Additionally, the
results remain controversial, and the underlying mechanisms
unclear. A number of the studies have demonstrated that nicotine
has an inhibitory effect on bone metabolism, while others
demonstrated a biphasic effect. In the present study, primary
Sprague-Dawley (SD) rat osteoblasts were cultured and the effects
of different concentrations of nicotine on osteogenic activity and
the whole genome expression profile were investigated to clarify
the molecular mechanisms and to provide a theoretical basis for
diseases caused by smoking and associated with the imbalance of
bone metabolism.
Materials and methods
Cell culture
The current study was approved by the Ethics
Committee of West China Hospital of Stomatology of Sichuan
University (Chengdu, China). A total of 80 male newborn SD rats
(<72 h old, 6–8 g) were obtained from the West China Animal
Experiment Center of Sichuan University and bred in constant
specific-pathogen-free conditions (relative humidity, 50%;
temperature, 25°C; 12-h light/dark cycle) and had free access to
breast milk. The newborn rats were sacrificed by cervical
dislocation. Primary osteoblasts were obtained from the parietal
bone; the bone was chopped into 1 mm2 fragments, rinsed
with PBS (Hyclone; GE Healthcare Life Sciences; Logan, UT, USA),
digested with 5 ml trypsin (0.25%; Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 10 min at 37°C and incubated with 10 ml
collagenase I (1 g/l; Sigma-Aldrich; Merck KGaA) at 37°C for 90
min. The fragments were seeded into 25-cm2 flasks at a
density of 15 fragments per flask, mixed with Dulbecco's modified
Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences)
containing 10% fetal bovine serum (Hyclone; GE Healthcare Life
Sciences), 100 units/ml penicillin and 100 µg/ml streptomycin
(Sigma-Aldrich; Merck KGaA) and then cultured at 37°C with 5%
CO2. The medium was changed every other day. Osteoblasts
from passages 2–4 were used in subsequent experiments.
Cell identification
Cells were cultured at 37°C with 5% CO2
in osteogenic induction medium containing DMEM with 10% fetal
bovine serum, β-glycerophosphate (10 mmol/l) and L-ascorbate (50
µg/ml), all purchased from Sigma-Aldrich (Merck KGaA), in 6-well
plates at a density of 5×104 cells/ml. When the cells
reached 80% confluence, they were thrice rinsed with PBS and fixed
with 95% ethanol for 10 min at the room temperature. ALP was
stained with a BCIP/NBT kit (Beyotime Institute of Biotechnology,
Haimen, China) and then observed using an inverted microscope
(magnification, ×200). Following culture for 3 weeks, cells were
fixed with 4% polyoxymethylene for 30 min at the room temperature,
stained using alizarin red was performed at 37°C (Cyagen
Biosciences Guangzhou, Inc., Guangzhou, China) and observed by an
inverted microscope (magnification, ×40) to characterize the
osteoblasts.
Cell viability and apoptosis
Osteoblasts cultured in 96-well plates at 37°C with
5% CO2 at a density of 5×104 cells/ml were
exposed to nicotine (Sigma-Aldrich; Merck KGaA) at various
concentrations (0, 1×10−6, 1×10−5,
1×10−4 and 1×10−3 mol/l) for 1, 3, 5 and 7
days. Cell proliferation was evaluated using a Cell Counting kit-8
(CCK-8) kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
in accordance with the manufacturer's protocols. The optical
absorbance of samples was measured using a microplate reader at a
wavelength of 450 nm. Following the culturing of cells
(5×104 cells/ml) with nicotine-containing medium in 25
cm2 flasks at 37°C with 5% CO2 for 7 days,
apoptosis was evaluated using an Annexin V-Fluorescein
Isothiocyanate (FITC)/Propidium Iodide (PI) Apoptosis Detection kit
(Dojindo Molecular Technologies, Inc.), in accordance with the
manufacturer's protocols, and detection was performed on a flow
cytometer (Beckman Coulter, Inc., Brea, CA, USA). CytExpert
software (version 1.0.135.1; Beckman Coulter, Inc.) was used for
analysis. The results were interpreted as follows: Active cells
[Annexin V-FITC(−)/PI(−)], early-phase apoptotic cells [Annexin
V-FITC(+)/PI(−)], and necrotic and late-phase apoptotic cells
[Annexin V-FITC(+)/PI(+)].
ALP activity assay
Osteoblasts were cultured in 12-well plates at a
density of 5×104 cells/ml at 37°C with 5%
CO2. When cells reached 70–80% confluence,
nicotine-containing medium was added at 0, 1×10−6,
1×10−5, 1×10−4 and 1×10−3 mol/l
concentrations. ALP activity was evaluated after days 4, 7 and 10
using an ALP test kit (Nanjing Jiancheng Bioengineering Institute,
Nanjing, China) according to the manufacturer's protocol. The
optical absorbance was measured by a microplate reader at a
wavelength of 520 nm. The protein concentrations of samples were
determined by a BCA kit (Beyotime Institute of Biotechnology)
according to the manufacturer's protocol. ALP levels were
normalized to the total protein content at the end of the
experiment.
Alizarin red staining and quantitative
analysis
Osteoblasts were plated in 6-well plates at a
density of 5×104 cells/ml. Following incubation with
nicotine-containing medium (0, 1×10−6,
1×10−5, 1×10−4 and 1×10−3 mol/l)
at 37°C with 5% CO2 for 21 days, 4% paraformaldehyde
(Shanghai Chemical Reagent Co., Ltd., Shanghai, China) and 0.1%
alizarin red staining solution (Cyagen Biosciences Guangzhou, Inc.)
were consecutively added with room temperature incubations of 30
and 5 min, respectively. A digital camera (D610; Nikon Corporation,
Tokyo, Japan) was used to capture images. Subsequently, 1 ml PBS
and 1 ml OF 10% cetylpyridinium chloride (Sigma-Aldrich; Merck
KGaA) were successively added to each well and incubated at 37°C
for 15 min. The optical absorbance of samples was measured using a
microplate reader at a wavelength of 562 nm.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Osteoblasts were plated in 6-well plates at a
density of 5×104 cells/ml. Following incubation with
nicotine-containing medium (0, 1×10−6,
1×10−5, 1×10−4 and 1×10−3 mol/l)
at 37°C with 5% CO2 for 7 days, the total RNA was extracted using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). Following quality confirmation of the extracted RNAs, cDNAs
were synthesized using a Revert Aid™ First Strand cDNA Synthesis
kit (Fermentas; Thermo Fisher Scientific, Inc., Pittsburgh, PA,
USA) for 10 min at 20°C, for 60 min at 42°C and 10 min at 70°C. The
primers used for the amplification are given in Table I. qPCR was conducted with
SYBR® Premix Ex Taq™ II (Tli RNaseH Plus) kit
(Takara Biotechnology Co., Ltd., Dalian, China). The reaction
mixture (30 µl) contained 3 µl buffer (Mg2+ free, 10X),
3 µl MgCl2 (25 mM), 0.36 µl dNTP (25 mM), 1 µl each of
10 µM forward and reverse primers, 1 µl SYBR-Green I (10X), 0.3 µl
Taq enzyme (5 U/µl), 5 µl cDNA and 15.34 µl ddH2O.
Amplification was performed using a real-time PCR detection system
(Funglyn Biotech, Inc., Richmond Hill, Canada) with temperature
cycles set as follows: 94°C for 2 min, 45 cycles of 94°C for 20 sec
and 56°C [for ALP, collagen type I α1 (Col1-α1), OCN and OPN], 52°C
(for BSP) or 54°C (for β-actin) for 20 sec; and 72°C for 30 sec.
Following normalizing the mean quantification cycle (Cq) values,
the changes in the expression of ALP, Col1-α1, OCN, OPN and BSP
genes were evaluated relative to the expression of β-actin as an
internal control gene using the 2−∆∆Cq method, as
previously described (16).
 | Table I.Primer sequences for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences for reverse
transcription-quantitative polymerase chain reaction.
|
| Sequence |
|
|---|
|
|
|
|
|---|
| Gene | Forward | Reverse | Product size
(bp) |
|---|
| ALP |
CATCATGTTCCTGGGAGATG |
GGTGTTGTACGTCTTGGAGA | 148 |
| Col1-α1 |
GTGCTAAGGGTGAAGCTGGT |
CATCAGCACCAGGGTTTCCAG | 126 |
| OCN |
CCTCTCTGACCTGGCAGGT |
GGCTCCAAGTCCATTGTTGA | 124 |
| OPN |
CACTCAGATGCTGTAGCCACTT |
GTTGCTTGGAAGAGTTTCTTGCT | 126 |
| BSP |
CTGAAGAAAACGGGGTCTTTAAG |
GAACTATCGCCATCTCCATT | 136 |
| Notch1 |
TCAGCGGGATCCACTGTGAG |
ACACAGGCAGGTGAAGTTG | 175 |
| Fgf21 |
AGATCAGGGAGGATGGAACA |
TCAAAGTGAGGCGATCCATA | 126 |
| β-actin |
GAAGATCAAGATCATTGCTCCT |
TACTCCTGCTTGCTGATCCACA | 111 |
Western blot analysis
Cells were incubated with nicotine-containing medium
(0, 1×10−6, 1×10−5, 1×10−4 and
1×10−3 mol/l) for 7 days and then lysed on ice for 30
min using 150 µl radioimmunoprecipitation buffer (Cell Signaling
Technology, Inc., Danvers, MA, USA) to extract the protein. Protein
concentrations were measured using a BCA kit. Protein samples (50
µg/lane) were subjected to SDS-PAGE on 12% polyacrylamide gels.
Gels were run at 80 V for 20 min and 120 V for 50 min.
Gel-separated proteins were transferred onto polyvinylidene
difluoride membranes at 100 V for 90 min. The transfer membrane was
blocked with PBS-0.1% Tween-20 containing 5% bovine serum albumin
(Cell Signaling Technology, Inc.) at 4°C overnight. Following
incubation at 4°C overnight with rabbit or mouse polyclonal IgG
antibodies against ALP (cat. no. 11187-1-AP; 1:300), Col1-α1 (cat.
no. 14695-1-AP; 1:300), OPN (cat. no. 25715-1-AP; 1:300), β-actin
(cat. no. 66009-1-Ig; 1:5,000; all Wuhan Sanying Biotechnology,
Wuhan, China), OCN (cat. no. ab13420; 1:300) or BSP (cat. no.
ab52128; 1:300; both Abcam, Cambridge, MA, USA), the membranes were
incubated at 37°C for 1 h with biotin-conjugated goat anti-rabbit
IgG (cat. no. SA00004-2; 1:20,000; Wuhan Sanying Biotechnology) and
biotin-conjugated goat anti-mouse IgG (cat. no. SA00004-1;
1:20,000; Wuhan Sanying Biotechnology). Immunoreactive proteins
were visualized using a DAB color reagent kit (OriGene
Technologies, Inc., Beijing, China) in the dark. Following image
acquisition, the protein bands were quantitatively analyzed by
Quantity One analysis software (version 4.6.6; Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and the relative expression
levels of target proteins were calculated according to the ratio of
gray value of target proteins to the gray value of the internal
reference protein, β actin.
mRNA microarray analysis
To reveal the differentially expressed gene profiles
following nicotine treatment, cells (5×104 cells/ml)
were cultured with nicotine-containing medium (1×10−3
mol/l) and medium alone in 25 cm2 flasks at 37°C with an
atmosphere of 5% CO2 for 7 days. Following this,
osteoblast samples were subjected to whole genome microarray
analysis using an Agilent Whole Rat Genome Oligonucleotide
Microarray (Agilent Technologies, Inc., Santa Clara, CA, USA). The
entire analysis procedure was performed by KangChen Bio-tech, Inc.
(Shanghai, China). Using a 2.0-fold cutoff and P<0.05 for mRNA
expression upregulation or downregulation, the key genes associated
with nicotine treatment were identified. The results of microarray
analysis were validated by RT-qPCR to detect the expression of
Notch1 and Fgf21, which demonstrated significant down- and
upregulation, respectively, by microarray analysis. The primers
used for amplification are given in Table I. Amplification was performed with
annealing and extension at 56°C for Notch1 and Fgf21 and 54°C for
β-actin. Gene expression was calculated as described above for
RT-qPCR.
Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway and Gene Ontology (GO) enrichment analysis. To further
investigate the functions of the differentially expressed genes, GO
terms were analyzed using the GO database (17) (http://www.geneontology.org/) and P-values for all
genes were calculated in all GO categories, including Cellular
Component, Molecular Function and Biological Process terms. The
threshold of significance was defined as P<0.05. The data on
signaling pathways were obtained from the KEGG pathway database
(18) (http://www.kegg.jp/kegg/kegg2.html), and the threshold
of significance was defined as P<0.05.
Statistical analysis
All experiments were performed at least in
triplicate (n=3 independent experiments). Data are presented as the
mean ± standard deviation. The data were analyzed by SPSS software
version 16.0 (SPSS, Inc., Chicago, IL, USA). The differences
between groups were analyzed by one-way analysis of variance and
Student-Newman-Keuls tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
Cell identification
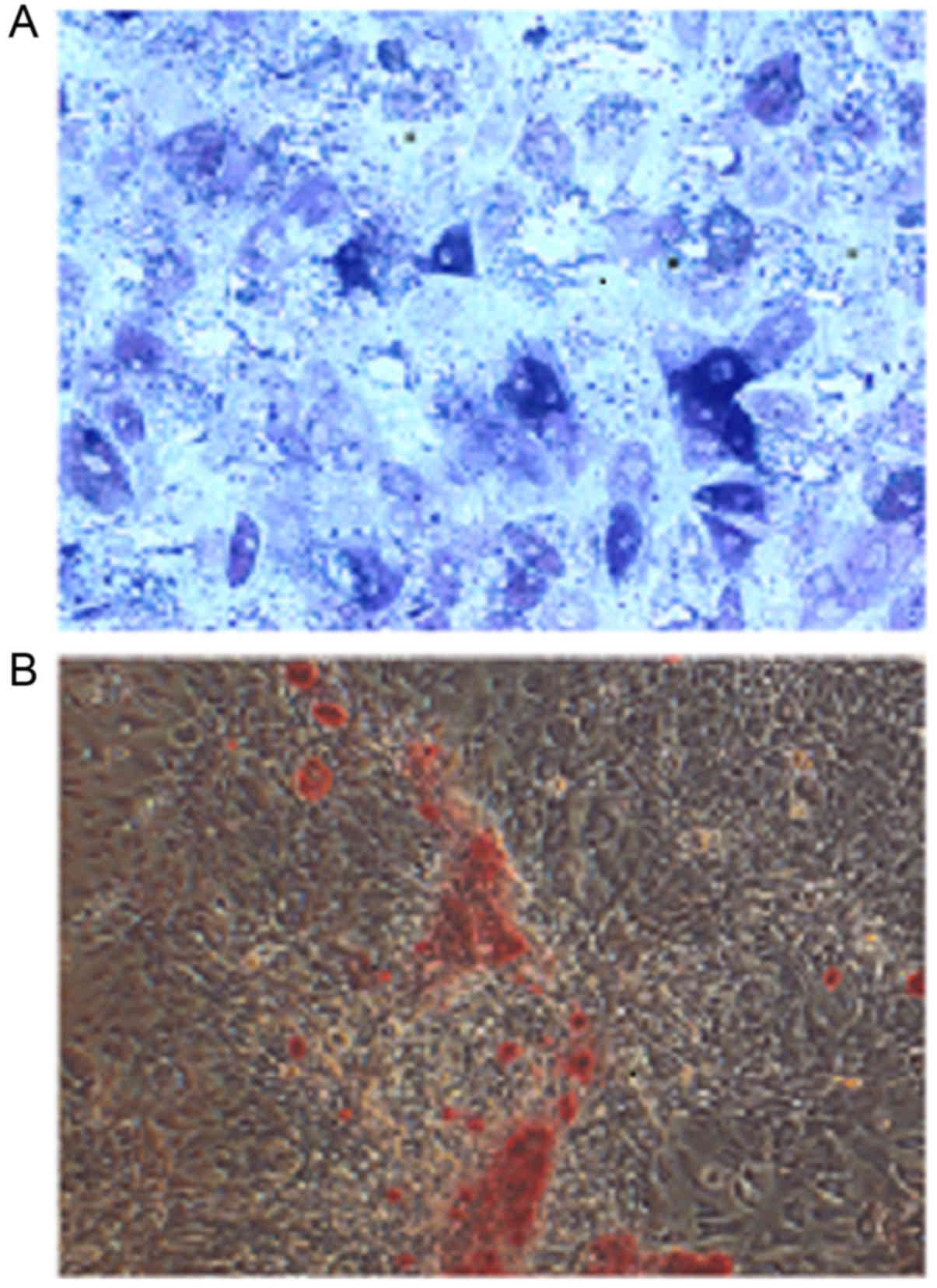
Cultured osteoblasts were stained using the BCIP/NBT
ALP staining kit and observed on an inverted microscope. A large
number of positively stained cells with blue-violet cell membranes
and cytoplasmic granules were observed (Fig. 1A). Alizarin red-stained calcified
nodules appeared red (Fig. 1B).
This result confirmed that the cultured cells were in fact
osteoblasts.
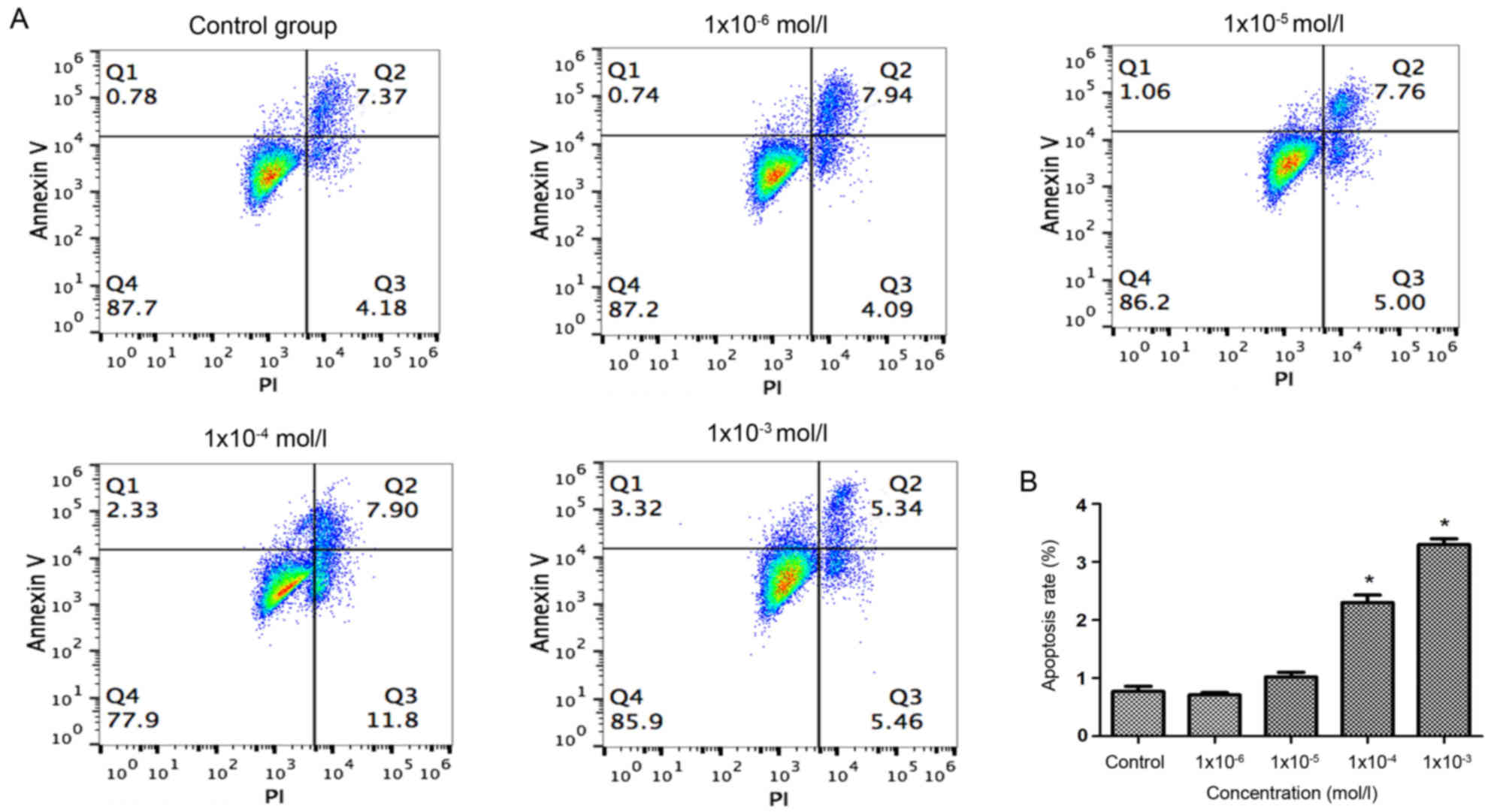
Effects of nicotine on early
apoptosis
The results of Annexin V-FITC/PI double staining was
performed to investigate the early apoptosis of osteoblasts.
Quadrant 1 was considered to indicate early apoptotic cells and
thus was used for the quantification of apoptosis rates. The
results indicated that the early apoptosis of osteoblasts was only
significantly promoted by treatment with nicotine concentrations of
1×10−4 and 1×10−3 mol/l, compared with the
control group (Fig. 2). These
results suggested that nicotine promoted the early apoptosis of
osteoblasts in a dose-dependent manner.
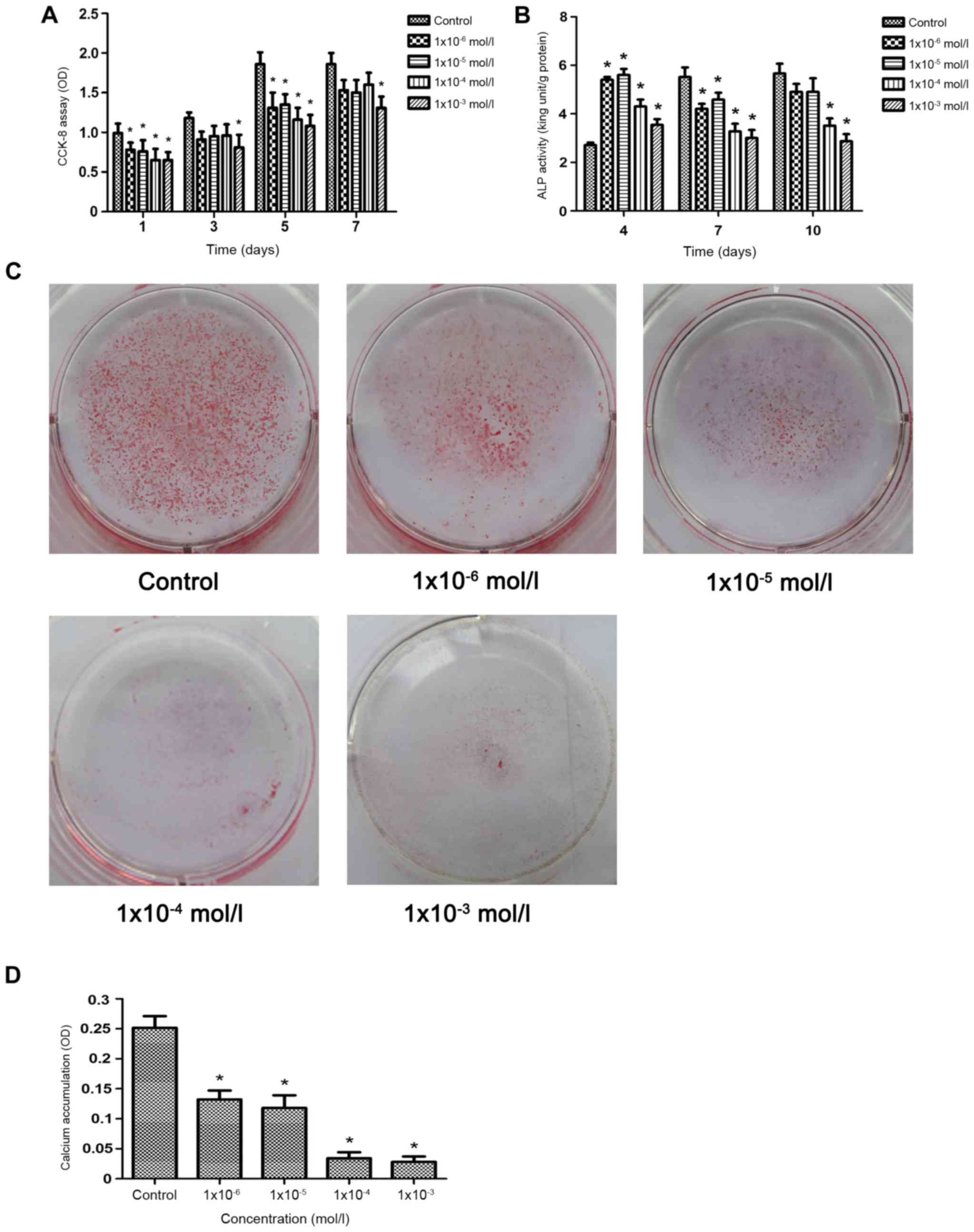
Effects of nicotine on osteoblast
proliferation and ALP activity
As demonstrated in Fig.
3A, compared with the control group, the proliferation of
osteoblasts was significantly decreased in all nicotine groups
(P<0.05) after 1 and 5 days, whereas proliferation was decreased
only in the 1×10−3−mol/l group after 3 and 7 days
(P<0.05). The inhibition of osteoblast proliferation was
enhanced by increased nicotine concentrations. Compared with the
control group, the ALP activity of all groups was significantly
higher compared with the control group on the 4th day (P<0.05;
Fig. 3B). On the 7th day, the ALP
activity of the cells in all experimental groups was significantly
lower compared with the control group (P<0.05; Fig. 3B). However, on the 10th day, only
the ALP activity of the 1×10−4 and 1× 10−3
mol/l groups was significantly lower compared with the control
group (P<0.05; Fig. 3B). This
suggested demonstrated that nicotine inhibited the proliferation of
osteoblasts and ALP activity in a dose-dependent manner.
Effects of nicotine on the mineralized
nodule formation of osteoblasts
The results of alizarin red staining revealed
decreased numbers of mineralized nodules following treatment with
nicotine in a dose-dependent manner (Fig. 3C). Quantification of alizarin red
staining demonstrated that the number of mineralized nodules
stained in the experimental groups decreased significantly compared
with the control group as the nicotine concentration increased
(Fig. 3D). These results
demonstrated that nicotine suppressed the bone formation of
osteoblasts in a dose-dependent manner.
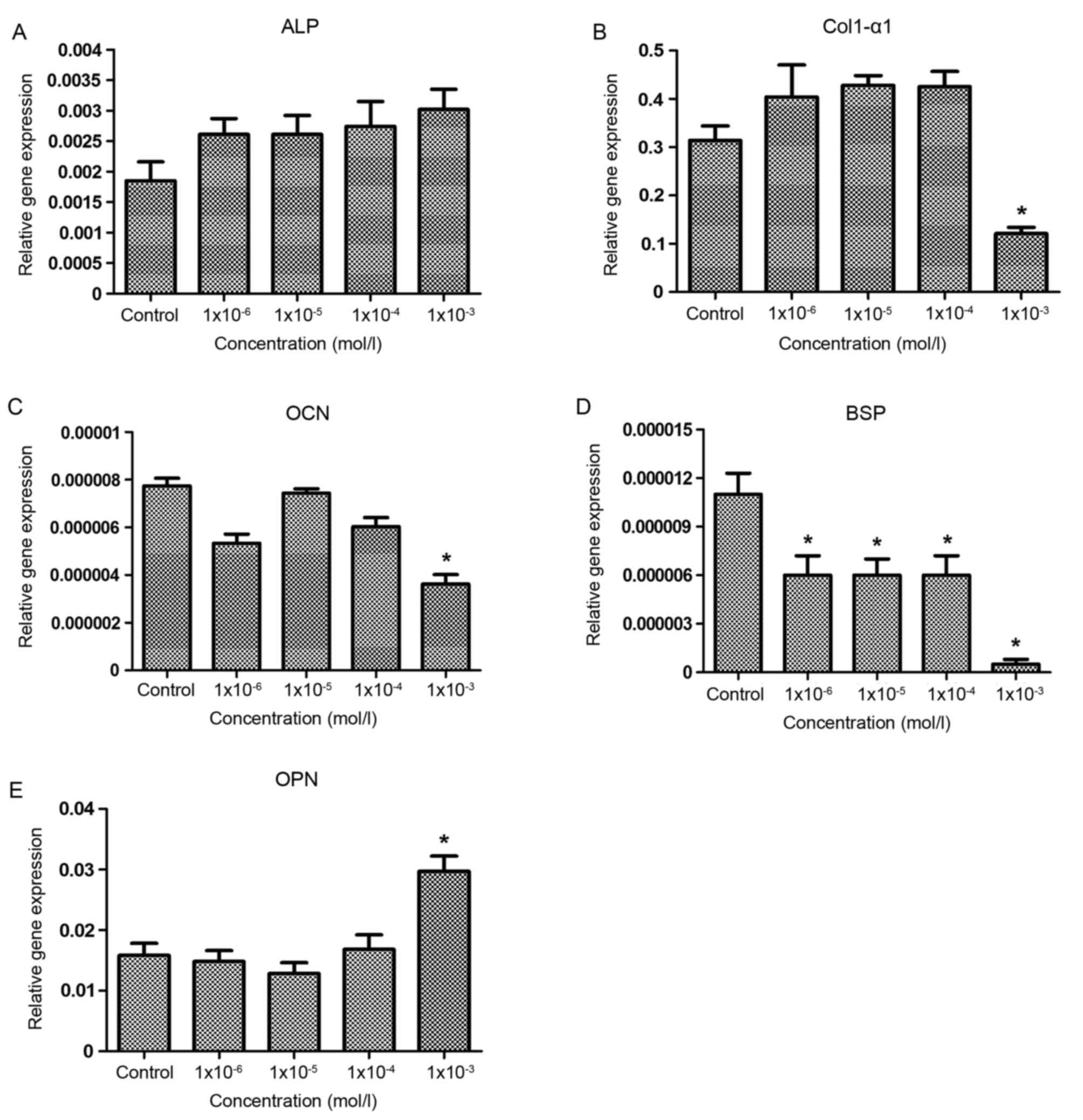
Effects of nicotine on osteoblast
metabolism-associated mRNA expression
The effect of nicotine on the mRNA expression of
genes associated with osteoblast metabolism was investigated by
RT-qPCR (Fig. 4). As demonstrated
in Fig. 4A, nicotine exhibited no
significant effect on the gene expression of ALP in the osteoblasts
of SD rats. In addition, there were no significant differences in
the expression of Col1-α1 and OCN genes among the control group and
the 1×10−6, 1×10−5 and 1×10−4
mol/l groups; however, in the 1×10−3 mol/l group, the
expression level of Col1-α1 and OCN were significantly lower
compared with the control group (P<0.05; Fig. 4B and C). The expression of the BSP
gene in each experimental group was significantly lower compared
with the control group (P<0.05; Fig. 4D). Furthermore, although no
significant differences in OPN expression were observed between the
control and the 1×10−6, 1×10−5 and
1×10−4 mol/l groups, OPN expression was significantly
higher compared with the control group in the 1×10−3
mol/l group (Fig. 4E). The results
suggested that the inhibitory effect of nicotine on bone formation
was regulated by the upregulation of OPN mRNA as well as the
downregulation of Col1-α1, BSP and OCN mRNA in osteoblasts.
Effects of nicotine on osteoblast
metabolism-associated protein expression
As demonstrated in Fig.
5, the ALP and BSP protein levels were significantly decreased
in the 1×10−5, 1×10−4 and 1×10−3
mol/l groups compared with the control group (P<0.05), the
expression levels of Col1-α1 and OCN protein in the
1×10−4 and 1×10−3 mol/l groups were
significantly lower compared with the control group (P<0.05) and
the OPN protein level was significantly higher compared with the
control group at 1×10−4 and 1×10−3 mol/l
nicotine (P<0.05). The results indicated that the inhibitory
effect of nicotine on bone formation was regulated by the
upregulation of OPN protein levels as well as the downregulation of
ALP, BSP, Col1-α1 and OCN protein levels in osteoblasts.
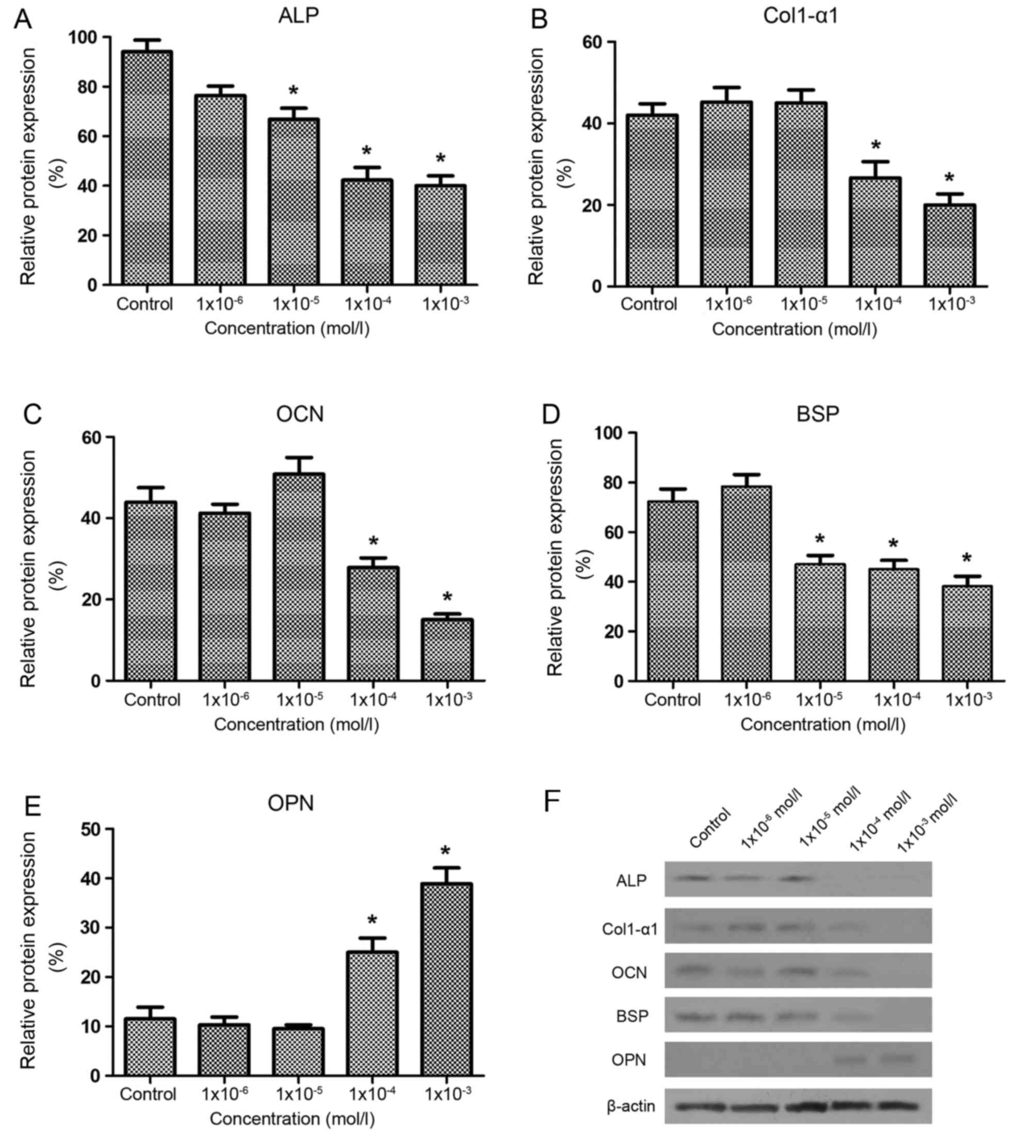 | Figure 5.Effects of nicotine on the expression
of Sprague-Dawley rat osteoblast metabolism-associated proteins at
day 7 of culture. (A) Nicotine significantly decreased ALP protein
expression in the 1×10−5, 1×10−4 and
1×10−3 mol/l nicotine groups compared with the control
group. The protein levels of (B) Col1-α1 and (C) OCN were
significantly decreased by nicotine in the 1×10−4 and
1×10−3 mol/l groups compared with the control group. (D)
BSP proteins levels were significantly decreased by all
concentrations of nicotine, excluding 1×10−6 mol/l,
compared with the control group. (E) OPN protein was significantly
increased by 1×10−3 and 1×10−4 mol/l nicotine
compared with the control group. (F) Representative western blot
bands for the protein expression of ALP, Col1-α1, OCN, BSP and OPN
in Sprague-Dawley rat osteoblasts. Each bar represents the mean ±
standard deviation (n=3) *P<0.05 vs. control group. ALP,
alkaline phosphatase; Col1-α1, collagen type I α1; OCN,
osteocalcin; BSP, bone sialoprotein; OPN, osteopontin. |
Effects of nicotine on the whole
genome expression profile of SD rat osteoblasts
Microarray analysis revealed 277 genes with at least
2-fold altered expression and P<0.05 following nicotine
treatment, including 121 upregulated genes and 156 downregulated
genes. The top 10 upregulated and downregulated genes are presented
in Tables II and III, respectively. As demonstrated in
Table IV, the expression of genes
associated with bone metabolism, including matrix metallopeptidase
3, SMAD family member 3 and latent transforming growth factor
b-binding protein 2, were significantly altered. In order to
further verify the microarray results, the upregulated and
downregulated genes with greatest fold changes were chosen and
further analyzed using RT-qPCR. The results demonstrated that the
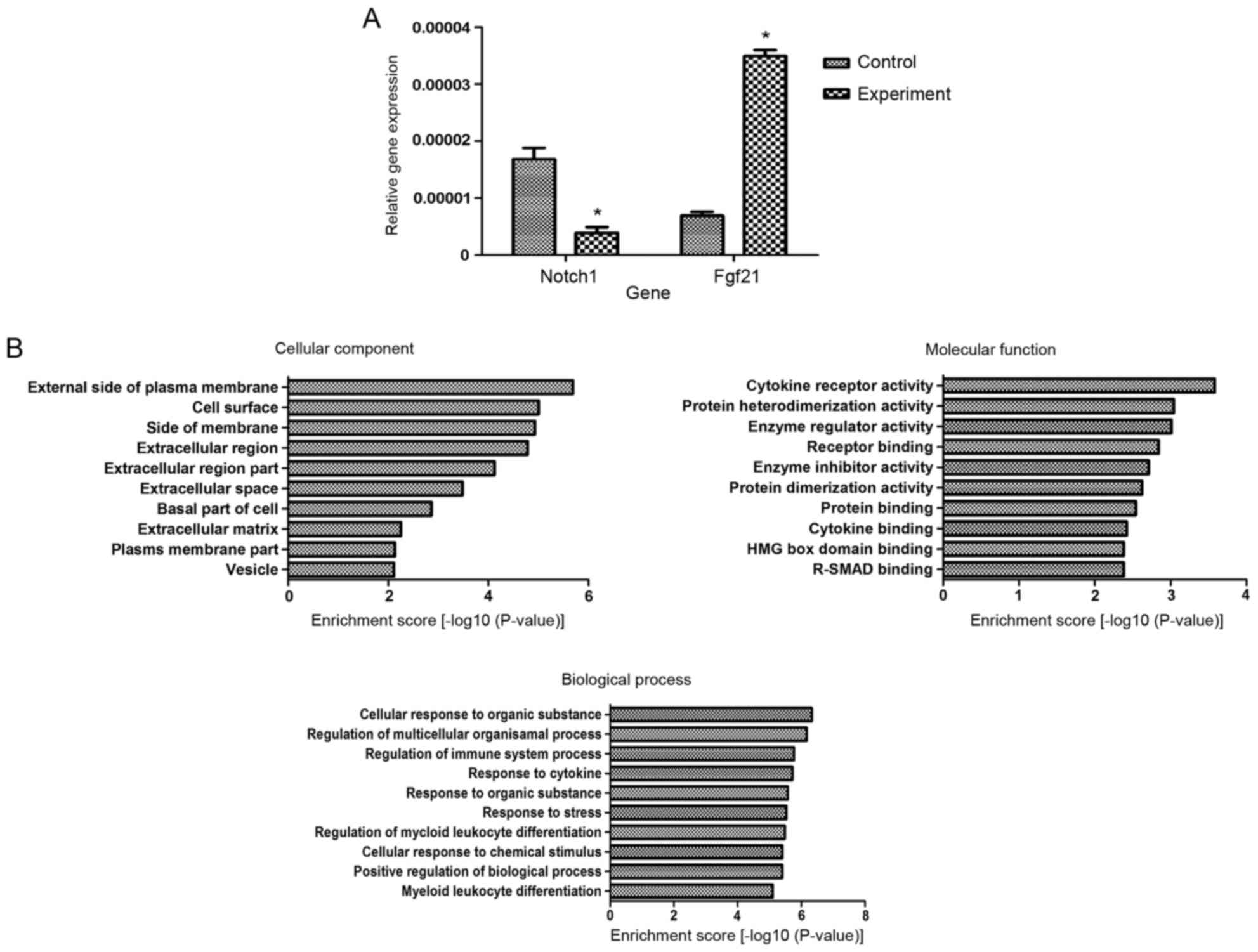
expression of Notch1 was downregulated 5.67-fold and that Fgf21
expression was upregulated 5.94-fold in the osteoblasts of SD rats
treated with 1×10−3 mol/l nicotine compared with the
control group, which was consistent with the microarray results
(Fig. 6A). Gene Ontology analysis
indicated that 1×10−3 mol/l nicotine altered several
functions of the osteoblasts of SD rats in the three main aspects
of Cellular Component, Molecular Function and Biological Process.
The differentially expressed genes associated with various
functions are presented in Fig.
6B. Pathway analysis indicated that differentially expressed
genes were primarily associated with signal transduction pathways.
The top 10 up and downregulated pathways are presented in Tables V and VI. Among the pathways identified, the
Hedgehog and Notch pathways have been previously revealed to
directly regulate osteoblast differentiation (19–21).
The upregulated and downregulated genes associated with the
Hedgehog and Notch pathways are presented in Table VII. These results revealed the
potential underlying mechanisms associated with the effect induced
by nicotine on the osteogenic activity of osteoblasts.
 | Table II.The top 10 upregulated genes that
were differentially expressed in nicotine-treated osteoblasts
compared with control cells. |
Table II.
The top 10 upregulated genes that
were differentially expressed in nicotine-treated osteoblasts
compared with control cells.
| Gene name | Fold change | P-value |
|---|
| Fgf21 | 6.212 | 0.0442 |
| RGD1562667 | 6.067 | 0.0289 |
| RGD1559482 | 6.057 | 0.0127 |
| Fam25a | 5.948 | 0.0021 |
| Cd38 | 5.214 | 0.0035 |
| Pla2g10 | 4.315 | 0.0074 |
| Akr1c3 | 4.285 | 0.0043 |
| Serpinb10 | 4.193 | 0.0005 |
| Ccr1 | 4.125 | 0.0083 |
| Ebi3 | 4.013 | 0.0011 |
 | Table III.The top 10 downregulated genes that
were differentially expressed in nicotine-treated osteoblasts
compared with control cells. |
Table III.
The top 10 downregulated genes that
were differentially expressed in nicotine-treated osteoblasts
compared with control cells.
| Gene name | Fold change | P-value |
|---|
| Notch1 | 5.466 | 0.0001 |
| Hey2 | 4.727 | 0.0448 |
| Ccdc40 | 4.000 | 0.0019 |
| Cdhr4 | 3.893 | 0.0015 |
| Adora2a | 3.862 | 0.0001 |
| Scin | 3.754 | 0.0015 |
| Smad3 | 3.715 | 0.0006 |
| Slc25a47 | 3.646 | 0.0009 |
| Sema3g | 3.540 | 0.0205 |
| Filip1 | 3.483 | 0.0001 |
 | Table IV.Osteogenesis-associated genes that
were differentially expressed in nicotine-treated osteoblasts
compared with control cells. |
Table IV.
Osteogenesis-associated genes that
were differentially expressed in nicotine-treated osteoblasts
compared with control cells.
| Gene name | Fold change | P-value | Regulation |
|---|
| Smad3 | 3.716 | 0.0007 | Down |
| MMP-3 | 2.645 | 0.0001 | Down |
| Ltbp2 | 2.11 | 0.0006 | Down |
 | Table V.Kyoto Encyclopedia of Genes and
Genomes pathway analysis of the top 10 significantly upregulated
genes following nicotine treatment. |
Table V.
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of the top 10 significantly upregulated
genes following nicotine treatment.
| Pathway ID | Definition | Count | P-value | Genes |
|---|
| rno04640 | Hematopoietic cell
lineage (rat) | 4 | 0.0005 | CD34, CD38, IL2RA,
ITGA2 |
| rno04921 | Oxytocin signaling
pathway (rat) | 5 | 0.0008 | CCND1, CD38, FOS,
JUN, MYLK3 |
| rno04510 | Focal adhesion
(rat) | 5 | 0.0028 | ACTN3, CCND1,
ITGA2, JUN, MYLK3 |
| rno04380 | Osteoclast
differentiation (rat) | 4 | 0.0029 | FCGR2B, FOS, JUN,
LILRA5 |
| rno05210 | Colorectal cancer
(rat) | 3 | 0.0033 | CCND1, FOS,
JUN |
| rno04630 | Jak-STAT signaling
pathway (rat) | 4 | 0.0044 | BCL2L1, CCND1,
IL2RA, TSLP |
| rno04662 | B cell receptor
signaling pathway (rat) | 3 | 0.0046 | FCGR2B, FOS,
JUN |
| rno05133 | Pertussis
(rat) | 3 | 0.0046 | C1QC, FOS, JUN |
| rno05412 | Arrhythmogenic
right ventricular cardiomyopathy (ARVC) (rat) | 3 | 0.0050 | ACTN3, ITGA2,
SGCG |
| rno05222 | Small cell lung
cancer (rat) | 3 | 0.0080 | BCL2L1, CCND1,
ITGA2 |
 | Table VI.Kyoto Encyclopedia of Genes and
Genomes pathway analysis of the top 10 significantly downregulated
genes following nicotine treatment. |
Table VI.
Kyoto Encyclopedia of Genes and
Genomes pathway analysis of the top 10 significantly downregulated
genes following nicotine treatment.
| Pathway ID | Definition | Count | P-value | Genes |
|---|
| rno05150 | Staphylococcus
aureus infection (rat) | 4 | 0.0002 | C3, C4A, CFB,
SELP |
| rno05133 | Pertussis
(rat) | 4 | 0.0007 | C3, C4A, C4BPA,
IL23A |
| rno04610 | Complement and
coagulation cascades (rat) | 4 | 0.0007 | C3, C4A, C4BPA,
CFB |
| rno05322 | Systemic lupus
erythematosus (rat) | 4 | 0.0058 | C3, C4A, HIST1H2AN,
HIST1H3A |
| rno05323 | Rheumatoid
arthritis (rat) | 3 | 0.0128 | IL23A, MMP3,
TNFSF11 |
| rno04514 | CAMs (rat) | 4 | 0.0156 | CLDN4, NEGR1,
NTNG1, SELP |
| rno05033 | Nicotine addiction
(rat) | 2 | 0.0212 | CACNA1B,
GABRA1 |
| rno04668 | TNF signaling
pathway (rat) | 3 | 0.0227 | BIRC3, CX3CL1,
MMP3 |
| rno04340 | Hedgehog signaling
pathway (rat) | 2 | 0.0298 | IHH, LRP2 |
| rno04330 | Notch signaling
pathway (rat) | 2 | 0.0345 | DLL3, NOTCH1 |
 | Table VII.Differentially expressed genes
associated with Hedgehog and Notch signaling pathways following
nicotine treatment. |
Table VII.
Differentially expressed genes
associated with Hedgehog and Notch signaling pathways following
nicotine treatment.
| Gene name | Fold change | P-value | Regulation | Pathway |
|---|
| IHH | 2.619 | 0.0016 | Down | Hedgehog |
| LRP2 | 2.406 | 0.0058 | Down | Hedgehog |
| DLL3 | 2.056 | 0.0087 | Up | Notch |
| NOTCH1 | 5.466 | 0.0001 | Down | Notch |
Discussion
A substantial amount of clinical and experimental
research has demonstrated that tobacco smoking has deleterious
effects on numerous biological systems, including an association
with oral diseases (1). As the
major toxic component, nicotine appears to serve a major role in
the negative effects of tobacco smoke on bone metabolism, primarily
by causing the absorption of alveolar bone. Various studies have
investigated the effects of nicotine on the osteogenic activity by
analyzing proliferation, differentiation, and specific protein and
gene expression. However, the results remain controversial. Fang
et al (22) reported that
nicotine suppressed cellular proliferation and stimulated ALP
activity in a dose-dependent fashion in UMR 106-01 rat osteoblastic
osteosarcoma cells. Furthermore, nicotine was reported to inhibit
the proliferation of osteoblasts as well as the expression of
certain important osteogenic mediators in rabbit osteoblasts
(23). Yuhara et al
(11) further identified that in
ROB-C26 cells, nicotine increased ALP activity in a dose-dependent
manner and increased the deposition of Ca2+ similarly.
However, nicotine was reported to reduce these two activities in
MC3T3-E1 cells. A previous study has also indicated that nicotine
may affect bone metabolism in a biphasic manner, stimulating
proliferation and gene expression at low doses analogous to
concentrations acquired by light smokers, and suppressing
proliferation at high doses analogous to concentrations acquired by
heavy smokers (13). Although
these studies used nicotine in similar concentrations to treat
osteoblasts, the results differed from each other. These
differences may have occurred as a result of differences in cell
culture conditions, species differences, the type of osteoblast
model selected, including differentiation stage, and differences in
experimental designs. In previous studies, cell lines such as
MC3T3-E1 and MG-63 were often used, whereas few studies have been
performed on primary osteoblasts. After several passages of cell
lines, apparent phenotypic heterogeneity can develop and cells may
arrest at a certain stage of differentiation, therefore no longer
fully reflecting the normal phenotype of osteoblasts (13,24).
Compared with cell lines, primary osteoblast cells are more similar
to human osteoblasts. In 1995, the U.S. Food and Drug
Administration approved the use of the primary rat osteoblasts as
osteoporosis and other bone metabolic disease models to evaluate
preclinical and clinical drug efficacy (25). Consequently, in the present study,
experiments were conducted on SD rat primary osteoblasts using
nicotine at concentrations of 1×10−6, 1×10−5,
1×10−4 and 1×10−3 mol/l. These concentrations
were selected based on nicotine levels of 0.06–1.2 mM/l detected in
the blood of smokers of varying regularity; these concentrations
were significantly lower compared with the 0.6–9.6 mM/l identified
in the saliva (26).
In a previous study, 10−6 and
10−4 mol/l nicotine were reported to promote cell cycle
progression during proliferation by downregulating p53 expression
and upregulating cyclin D1 in the mouse pre-osteoblastic cell line
MC3T3-E1 (27). However, the
results of the present study revealed an inhibitory effect of
nicotine on the proliferation of primary osteoblasts. The
difference in the results may be due to the use of different cell
models.
Apoptosis is an active programmed cell death process
that, together with cell proliferation and differentiation,
maintains the homeostasis of the organism (28). To the best of our knowledge, the
effects of nicotine on osteoblast apoptosis have rarely been
reported. In the present study, the early apoptosis of cells was
significantly promoted by nicotine concentrations of
10−4 and 10−3 mol/l. However, the mechanism
underlying this effect requires further investigation. Previous
studies have demonstrated that members of the Bcl-2 family,
including Bcl-2, Bcl-2-like 2 and Bcl-2-associated X, members of
the caspase family, including caspase 1 and caspase 3, and the
oncogene c-Myc may be involved in osteoblast apoptosis (29,30).
As an indicator of the differentiation potential of
osteoblasts in the initial stage, ALP is able to hydrolyze the
phosphate ester bonds on substrate molecules to generate phosphate
ions, which have a strong affinity for Ca2+ (31). The quality and quantity of ALP is
important in affecting the viability of osteoblasts. In previous
studies, the effect of nicotine on the ALP activity of osteoblasts
has been controversial. Yuhara et al (11) reported that nicotine reduced ALP
activity in a dose-dependent manner in MC3T3-E1 cells and increased
ALP activity in ROB-C26 cells during days 3–12. By contrast,
Gullihorn et al (32)
demonstrated that nicotine promoted ALP activity on day 3 in
MC3T3-E1 cells. Sato et al (27) reported that following a transient
increase in ALP activity at day 3, nicotine led to a marked
reduction in the activity of ALP at day 7 in MC3T3-E1 cells, which
is similar to the results of the present study. The results of the
present study demonstrated that ALP activity was significantly
higher in nicotine-treated groups compared with the control group
at day 4, which may occur due to the stress response of the cells
following nicotine stimulation and subsequent activation of the
regulatory mechanism of ALP, potentially through the bone
morphogenetic protein/Smads signaling pathway (33). In addition, studies have also
demonstrated that nitric oxide affected the differentiation of
osteoblast-like cells (34,35).
Therefore, differences in the effects of nicotine on osteoblasts
may be associated with differences in nitric oxide-associated
pathways. Thus, it is noted that the regulatory effect of nicotine
on the ALP activity of osteoblasts requires further confirmation
and the regulatory mechanism, particularly the molecular pathways
involved, also requires investigation.
During in vitro osteogenesis, Col1 functions
as the scaffold for the nucleation of hydroxyapatite crystals and
acts on osteoblast cells in an autocrine manner to promote the
expression of ALP, OCN, OPN and osteonectin by activating the
protein kinase C signal transduction pathway (36). OCN is usually expressed in the
early stage of calcification and regulates the speed and direction
of bone matrix mineral formation (37). OPN is a phosphorylated sialoprotein
with a highly conserved RGD motif that interacts with various
receptors, including αvβ3, αvβ1 and αvβ5, on the surface of some
cell types, such as osteoclast and endothelial cells (38). A previous study has reported that
the phosphorylation of OPN may inhibit the formation of
hydroxyapatite crystals in vitro in a dose-dependent manner.
The mechanism of this inhibition may involve OPN interacting with
osteoclast surface integrins (primarily αvβ3) through the RGD motif
to mediate osteoclast adhesion in bone tissue and promote bone
resorption (39). OPN has also
been reported to inhibit osteoblast differentiation (40). BSP is a highly sulfated and
phosphorylated glycoprotein that nucleates hydroxyapatite crystal
formation. Nakayama et al (41) identified that nicotine suppressed
the transcription of BSP mediated through CRE, FRE and HOX elements
within the proximal promoter of the BSP gene in rats. According to
the results of the present study, it may be concluded that the
inhibition of mineralized nodule formation may depend on the effect
of nicotine on ALP activity at 1×10−6 mol/l, on the
decrease of ALP and BSP protein at1×10−5 mol/l and on
the decrease of Col1-α1 and OCN protein, and the increase of OPN
protein, at 1×10−4 and 1×10−3 mol/l nicotine.
The inhibitory effect of nicotine on the expression levels of
osteoblast metabolism associated mRNA and proteins was
dose-dependent. Notably, in the presents study, the gene expression
of each osteogenesis-associated factor was not completely
consistent with the protein expression, which may be due to
multiple regulatory steps that occur between transcription and
translation.
The effects of nicotine on osteoblasts involve
complex intracellular signal transduction pathways; however, the
specific molecular mechanism of regulation remains to be
elucidated. The whole genome expression microarray is a
high-throughput gene detection method that can provide a reliable
basis for investigating the molecular regulatory mechanism of
biological effects by utilizing bioinformatics analysis to evaluate
the expression of genes in cells rapidly and comprehensively. GO
analysis indicated that nicotine altered several functions of the
SD rat osteoblasts, including organic metabolism, intercellular
biological process regulation, cytokine receptor activity,
inflammatory response, ion transport, calcium ion adhesion and
transcription factor adhesion. KEGG pathway analysis demonstrated
that these differentially expressed genes were primarily involved
in pathways concerning proliferation, differentiation, apoptosis,
adhesion and carcinogenesis. Among the pathways identified, the
Hedgehog and Notch pathways have been reported to directly regulate
osteoblast differentiation (42,43).
In the Hedgehog pathway, Sonic hedgehog protein acts
as an important signal molecule in the early stage of osteoblast
differentiation, while Indian hedgehog protein (Ihh) is involved in
the later stage to promote the differentiation and maturation of
osteoblasts (44). The results of
the present study demonstrated that the expression of Ihh was
significantly decreased following nicotine treatment, indicating
that nicotine may inhibit the proliferation, differentiation and
function of SD rat osteoblasts by disturbing the Hedgehog signaling
pathway.
Concerning the role of the Notch pathway in the
regulation of osteoblast differentiation, in vitro studies
have reported contrasting results. Certain studies demonstrated
that the overexpression of Notch1, one of the notch receptors,
inhibited osteoblastic differentiation (45,46).
Conversely, Tezuka et al (47) demonstrated that osteoblastic cell
differentiation was regulated positively by Notch1. In the present
study, the results demonstrated that the expression of Notch1 was
significantly decreased following nicotine treatment, indicating
that nicotine may inhibit the activation of the Notch signaling
pathway and thereby decrease osteoblastic proliferation. However,
this hypothesis requires confirmation.
In conclusion, the present study demonstrated that
nicotine exhibited an inhibitory effect on SD rat osteoblast
metabolism. Microarray analysis revealed that the Notch and
Hedgehog signaling pathways were enriched with differentially
expressed genes in osteoblasts following nicotine treatment and
that these pathways were strongly associated with osteoblast
differentiation, which may result in improved future understanding
of nicotine-induced bone diseases. These results may provide a
theoretical basis for the prevention and treatment of
smoking-related diseases in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Sichuan
science and technology support project, Chengdu, China (grant no.
2014SZ0019).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW conceived and designed the present study. DL and
KW performed cell culture, western blot analyses, reverse
transcription-quantitative polymerase chain reaction and microarray
analysis, and were also the predominant contributors in the writing
of the manuscript. ZT and RL performed experiments for the
determination of cellular proliferation, apoptosis, alkaline
phosphatase activity and formation of mineralized nodules. FZ and
MC analyzed and interpreted the data. QL made contributions to
interpretation of data and critically revised the manuscript for
important intellectual content. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of West China Hospital of Stomatology of Sichuan
University (Chengdu, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vellappally S, Fiala Z, Smejkalová J,
Jacob V and Somanathan R: Smoking related systemic and oral
diseases. Acta Medica (Hradec Kralove). 50:161–166. 2007.
View Article : Google Scholar
|
|
2
|
Johnson GK and Guthmiller JM: The impact
of cigarette smoking on periodontal disease and treatment.
Periodontol. 44:178–194. 2007. View Article : Google Scholar
|
|
3
|
Tymkiw KD, Thunell DH, Johnson GK, Joly S,
Burnell KK, Cavanaugh JE, Brogden KA and Guthmiller JM: Influence
of smoking on gingival crevicular fluid cytokines in severe chronic
periodontitis. J Clin Periodontol. 38:219–228. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Heitz-Mayfield LJ and Huynh-Ba G: History
of treated periodontitis and smoking as risks for implant therapy.
Int J Oral Maxillofac Implants. 24 Suppl:S39–S68. 2009.
|
|
5
|
Larrazábal C, García B and Peñarrocha M
and Peñarrocha M: Influence of oral hygiene and smoking on pain and
swelling after surgical extraction of impacted mandibular third
molars. J Oral Maxillofac Surg. 68:43–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
García B, Penarrocha M, Martí E,
Gay-Escodad C and Von Arx T: Pain and swelling after periapical
surgery related to oral hygiene and smoking. Oral Surg Oral Med
Oral Pathol Oral Radiol Endod. 104:271–276. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Krall EA, Sosa Abreu C, Garcia C, Nunn ME,
Caplan DJ and Garcia RI: Cigarette smoking increases the risk of
root canal treatment. J Dent Res. 85:313–317. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Henningfield JE and Goldberg SR: Progress
in understanding the relationship between the pharmacological
effects of nicotine and human tobacco dependence. Pharmacol Biochem
Behav. 30:217–220. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
En-Nosse M, Hartmann S, Trinkaus K, Alt V,
Stigler B, Heiss C, Kilian O, Schnettler R and Lips KS: Expression
of non-neuronal cholinergic system in osteoblast-like cells and its
involvement in osteogenesis. Cell Tissue Res. 338:203–215. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alford AI and Hankenson KD: Matricellular
proteins: Extracellular modulators of bone development, remodeling,
and regeneration. Bone. 38:749–757. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuhara S, Kasagi S, Inoue A, Otsuka E,
Hirose S and Hagiwara H: Effects of nicotine on cultured cells
suggest that it can influence the formation and resorption of bone.
Eur J Pharmacol. 383:387–393. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramp WK, Lenz LG and Galvin RJ: Nicotine
inhibits collagen synthesis and alkaline phosphatase activity, but
stimulates DNA synthesis in osteoblast-like cells. Proc Soc Exp
Biol Med. 197:36–43. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rothem DE, Rothem L, Soudry M, Dahan A and
Eliakim R: Nicotine modulates bone metabolism-associated gene
expression in osteoblast cells. J Bone Miner Metab. 27:555–561.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rothem DE, Rothem L, Dahan A, Eliakim R
and Soudry M: Nicotinic modulation of gene expression in osteoblast
cells, MG-63. Bone. 48:903–909. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Torshabi M, Esfahrood ZR, Gholamin P and
Karami E: Effects of nicotine in the presence and absence of
vitamin E on morphology, viability and osteogenic gene expression
in MG-63 osteoblast-like cells. J Basic Clin Physiol Pharmacol.
27:595–602. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The Gene Ontology (GO) database and informatics resource.
Nucleic Acids Res. 32:(Database Issue). D258–D261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zanotti S, Smerdel-Ramoya A, Stadmeyer L,
Durant D, Radtke F and Canalis E: Notch inhibits osteoblast
differentiation and causes osteopenia. Endocrinology.
149:3890–3899. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Canalis E: Role of Notch signaling in
osteoblast differentiation and bone remodeling. Bone. 45 Suppl
3:S120–S121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Chen J, Karner CM and Long F:
Hedgehog signaling activates a positive feedback mechanism
involving insulin-like growth factors to induce osteoblast
differentiation. Proc Natl Acad Sci USA. 112:4678–4683. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang MA, Frost PJ, Iida-Klein A and Hahn
TJ: Effects of nicotine on cellular function in UMR 106-01
osteoblast-like cells. Bone. 12:283–286. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma L, Zwahlen RA, Zheng LW and Sham MH:
Influence of nicotine on the biological activity of rabbit
osteoblasts. Clin Oral Implants Res. 22:338–342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang D, Christensen K, Chawla K, Xiao G,
Krebsbach PH and Franceschi RT: Isolation and characterization of
MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo
differentiation/mineralization potential. J Bone Miner Res.
14:893–903. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thompson DD, Simmons HA, Pirie CM and Ke
HZ: FDA Guidelines and animal models for osteoporosis. Bone. 17 4
Suppl:125S–133S. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benowitz NL: Drug therapy. Pharmacologic
aspects of cigarette smoking and nicotine addition. N Engl J Med.
319:1318–1330. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sato T, Abe T, Nakamoto N, Tomaru Y,
Koshikiya N, Nojima J, Kokabu S, Sakata Y, Kobayashi A and Yoda T:
Nicotine induces cell proliferation in association with cyclin D1
up-regulation and inhibits cell differentiation in association with
p53 regulation in a murine pre-osteoblastic cell line. Biochem
Biophys Res Commun. 377:126–130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peter ME: Programmed cell death: Apoptosis
meets necrosis. Nature. 471:310–312. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Youle RJ and Strasser A: The BCL-2 protein
family: Opposing activities that mediate cell death. Nat Rev Mol
Cell Biol. 9:47–59. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ola MS, Nawaz M and Ahsan H: Role of Bcl-2
family proteins and caspases in the regulation of apoptosis. Mol
Cell Biochem. 351:41–58. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Golub EE, Harrison G, Taylor AG, Camper S
and Shapiro IM: The role of alkaline phosphatase in cartilage
mineralization. Bone Miner. 17:273–278. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gullihorn L, Karpman R and Lippiello L:
Differential effects of nicotine and smoke condensate on bone cell
metabolic activity. J Orthop Trauma. 19:17–22. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kuo PL, Hsu YL, Chang CH and Chang JK:
Osthole-mediated cell differentiation through bone morphogenetic
protein-2/p38 and extracellular signal-regulated kinase 1/2 pathway
in human osteoblast cells. J Pharmacol Exp Ther. 314:1290–1299.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Koyama A, Otsuka E, Inoue A, Hirose S and
Hagiwara H: Nitric oxide accelerates the ascorbic acid-induced
osteoblastic differentiation of mouse stromal ST2 cells by
stimulating the production of prostaglandin E(2). Eur J Pharmacol.
391:225–231. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
MacPherson H, Noble BS and Ralston SH:
Expression and functional role of nitric oxide synthase isoforms in
human osteoblast-like cells. Bone. 24:179–185. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Green J, Schotland S, Stauber DJ, Kleeman
CR and Clemens TL: Cell-matrix interaction in bone: Type I collagen
modulates signal transduction in osteoblast-like cells. Am J
Physiol. 268:1090–1103. 1995. View Article : Google Scholar
|
|
37
|
Boskey AL, Gadaleta S, Gundberg C, Doty
SB, Ducy P and Karsenty G: Fourier transform infrared
microspectroscopic analysis of bones of osteocalcin-deficient mice
provides insight into the function of osteocalcin. Bone.
23:187–196. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giachelli CM, Schwartz SM and Liaw L:
Molecular and cellular biology of osteopontin Potential role in
cardiovascular disease. Trends Cardiovasc Med. 5:88–95. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chellaiah MA, Kizer N, Biswas R, Alvarez
U, Strauss-Schoenberger J, Rifas L, Rittling SR, Denhardt DT and
Hruska KA: Osteopontin deficiency produces osteoclast dysfunction
due to reduced CD44 surface expression. Mol Biol Cell. 14:173–189.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kojima H, Uede T and Uemura T: In vitro
and in vivo effects of the overexpression of osteopontin on
osteoblast differentiation using a recombinant adenoviral vector. J
Biochem. 136:377–386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nakayama Y, Mezawa M, Araki S, Sasaki Y,
Wang S, Han J, Li X, Takai H and Ogata Y: Nicotine suppresses bone
sialoprotein gene expression. J Periodontal Res. 44:657–663. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zanotti S and Canalis E: Notch and the
skeleton. Mol Cell Biol. 30:886–896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Day TF and Yang Y: Wnt and hedgehog
signaling pathways in bone development. J Bone Joint Surg Am. 90
Suppl 1:S19–S24. 2008. View Article : Google Scholar
|
|
44
|
Tu X, Joeng KS and Long F: Indian hedgehog
requires additional effectors besides Runx2 to induce osteoblast
differentiation. Dev Biol. 362:76–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deregowski V, Gazzerro E, Priest L,
Rydziel S and Canalis E: Notch 1 overexpression inhibits
osteoblastogenesis by suppressing Wnt/beta-catenin but not bone
morphogenetic protein signaling. J Biol Chem. 281:6203–6210. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sciaudone M, Gazzerro E, Priest L, Delany
AM and Canalis E: Notch 1 impairs osteoblastic cell
differentiation. Endocrinology. 144:5631–5639. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tezuka K, Yasuda M, Watanabe N, Morimura
N, Kuroda K, Miyatani S and Hozumi N: Stimulation of osteoblastic
cell differentiation by notch. J Bone Miner Res. 17:231–239. 2002.
View Article : Google Scholar : PubMed/NCBI
|