Introduction
Hepatocellular carcinoma (HCC) is one of the most
frequently occurring malignant tumors globally (1). In the past decade, patients with HCC
have had to endure a high incidence of recurrence and metastasis; a
5-year recurrence rate of 75–100% is reported (2). Surgical resection, liver
transplantation and radiofrequency ablation may provide a cure for
certain early stage patients however, due to the asymptomatic
nature of HCC, the majority of patients are diagnosed at an
advanced stage (3). The molecular
mechanisms involved in HCC remain poorly understood. The
development of novel strategies to further the understanding of HCC
is required. Alterations in molecular expression in HCC have been
extensively investigated. Dysregulated gene expression has been
observed during the development of HCC (4). Kruppel-like factor 5 (KLF5), a zinc
finger protein that belongs to the KLF family, is a transcription
factor that binds the epidermal growth factor response element
(5). KLF5 was recently
demonstrated to be upregulated in metastatic HCC (6). Additionally, increased expression of
KLF5 promotes cell proliferation and inhibits apoptosis (7). However, the mechanism of aberrant
KLF5 expression remains unclear.
As a family of endogenous, small non-coding RNAs
(20–25 nucleotides in length), microRNAs (miRNAs)
post-transcriptionally regulate the expression of complementary
target mRNA in eukaryotes, which influences several biological
processes, including cell infection, development, immunity and
carcinogenesis (8,9). miRNA expression profiles reveal that
numerous miRNA signatures exist in various different cancer types
(10–13). It is indicated that miRNA
signatures may be predictive for cancer prognosis, classification
and response to therapy (14,15).
Emerging evidence indicates that miRNAs have significant roles in
the development of HCC, and certain tumor suppressive miRNAs in HCC
have already been identified (16). Decreased expression of miRNA
(miR)-145-5p in several cancer types, including prostate (17) and liver cancer (18), has been previously reported. The
present study determined the expression of miR-145-5p in HCC and
miR-145-5p overexpression, through use of miRNA mimics, was
observed to inhibit the proliferation and metastasis of HCC cells.
Furthermore, the present study identified KLF5 as a target of
miR-145-5p in HCC cells. The current study also demonstrated the
association between miR-145-5p and KLF5.
Materials and methods
HCC tissues and cell lines
HCC tissues and paired normal liver tissues were
obtained from 25 patients with primary HCC at The Second Xiangya
Hospital of Central South University (Changsha, China). The Ethics
Committee of The Second Xiangya Hospital of Central South
University approved this study after written informed consent was
obtained from each patient. Details of the patients are presented
in Table I. L02 normal human liver
cell line and HuH-7, HepG2 and SK-Hep-1 HCC cell lines were
obtained from American Type Culture Collection (Manassas, VA, USA).
The cells were cultured in Dulbecco's modified Eagle's medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), which
was supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific) and at 37°C in a 5% CO2
incubator.
 | Table I.Characteristics patients with
hepatocellular carcinoma (n=25). |
Table I.
Characteristics patients with
hepatocellular carcinoma (n=25).
| Characteristic | Number of
patients |
|---|
| Age (years) |
|
|
<50 | 10 |
| ≥50 | 15 |
| Sex |
|
| Male | 17 |
|
Female | 8 |
| Hepatitis
history |
|
| Yes | 11 |
| No | 14 |
| Liver cirrhosis |
|
| Yes | 13 |
| No | 12 |
| Tumor diameter
(cm) |
|
|
<5 | 17 |
| ≥5 | 8 |
| Tumor differentiation
grade |
|
| I+II | 18 |
|
III+IV | 7 |
RNA isolation & reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
By using TRIzol or miRNeasy Mini kit (Qiagen, Inc.,
Valencia, CA, USA), RNA was extracted, followed by reverse
transcription using cDNA Reverse Transcription kit (Qiagen, Inc.).
RT-qPCR was performed using SYBR-Green Master Mix (cat. no. RR420A;
Takara Biotechnology Co., Ltd., Dalian, China) on
LightCycler® 480 System (Roche Diagnostics GmbH,
Mannheim, Germany). miR-145-5p primers were purchased from
Sigma-Aldrich (Merck KGgA, Darmstadt, Germany; cat. no.
MIRAP00180). The reaction system included 2.5 µl cDNA (1:20
dilution by double-distilled water), 10 µl reaction mixture
(Qiagen, Inc.), 2.0 mM forward primer, and 2.0 mM reverse primer,
respectively. The experiment was repeated three times. The primers
of KLF5 were as follows: 5′-CTTCCACAACAGGCCACTTACTT-3′ (forward)
and 5′-AGAAGCAATTGTAGCAGCATAGGA-3′ (reverse). GAPDH primers were as
follows: 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and
5′-GAAGATGGTGATGGGATTTC-3′ (reverse). The cycling conditions were
as follows: Initial denaturation step at 95°C for 5 min, followed
by 40 cycles of 95°C for 30 sec, and 60°C for 30 sec. Relative
expression was calculated using the 2−ΔΔCq method and
levels were normalized to the reference gene, GAPDH.
miRNA mimics and construction of
expression vectors
miR-145-5p mimics (cat. no. miR30000157-1-2), which
mimic endogenous miRNA, and inhibitors (cat. no. miR20000437-1-2)
were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
The control group was transfected with an empty pcDNA3.1 vector
(Invitrogen; Thermo Fisher Scientific, Inc.). The entire coding
region of KLF5 was amplified and restriction enzymes, NotI
and EcoRI (New England BioLabs, Inc., Ipswich, MA, USA),
were employed to insert the KLF5 fragment into the mammalian
expression vector, pcDNA3.1. The primers were as follows:
5′-GGGCGGCCATGGCTACAAGGGTGCTGAGCATGAG-3′ (forward);
5′-GGGAATTCTCAGTTCTGGTGCCTCTTCATATGCAGGGCC-3′ (reverse). The
control involved empty vectors without inserted fragments. For the
luciferase reporter, Dual-Luciferase Reporter Assay System (Promega
Corporation, Madison, WI, USA) was used and the 3′-untranslated
region (3′-UTR) target sequence from KLF5 was amplified and
subsequently inserted into psiCHECK-2 vector (Promega
Corporation).
Cell viability and apoptosis
analysis
Using Cell Counting kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Rockville, MD, USA), cell proliferation rates
were measured. Cells (1×105 cells/well) were seeded
before incubation with mimics/inhibitors (50 nM mimics and 200 nM
inhibitors). The transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The mimics and inhibitors were
combined with 10 µl Lipofectamine® 2000 diluted in 250
µl Opti-MEM I (Invitrogen; Thermo Fisher Scientific, Inc.) and
incubated for 30 min. The mixture was then added to cells which
attained 90% confluence in the 96-well plates. After 24, 48, and 72
h transfection, 10 µl CCK-8 reagent was added to each well. The
optical density at 450 nm was determined using a Multiskan MK3
microplate reader (Thermo Fisher Scientific, Inc.). In order to
analyze apoptosis, cells were harvested and washed in ice-cold PBS
and were then fixed in 70% ice-cold ethanol at 4°C for 1 h. Cells
were incubated with 20 µg/ml propidium iodide and Annexin V-APC
(Sigma-Aldrich; Merck KGgA) for a period of 20 min at room
temperature, and were then analyzed using Fluorescence Activated
Cell Sorter (BD Biosciences, Franklin Lakes, NJ, USA). The cells
were analyzed by CellQuest software (version 3; BD
Biosciences).
Cells migration analysis
A total of 48 h following transfection, cell
migration was evaluated using a Transwell chamber (EMD Millipore,
Billerica, MA, USA). Transwell inserts were pre-coated with
Matrigel (BD Biosciences) and were placed in the upper compartment
prior to cell seeding. Cells (5×104) were seeded in the
upper chamber without FBS. The lower chamber was then filled with
medium that was supplemented with 10% FBS (FBS; Gibco; Thermo
Fisher Scientific, Inc.). Migratory cells on the bottom surface
were then fixed by 4% paraformaldehyde solution for 20 min at room
temperature, and stained with 0.1% crystal violet for 15 min at
room temperature. From each membrane, the number of migrated cells
in four random fields were counted by a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Experiments were conducted
in triplicate.
Western blot analysis
HCC and control tissues were lysed using radio
immunoprecipitation assay buffer (cat. no. P0013B; Beyotime
Institute of Biotechnology, Haimen, China) 48 h following
transfection. Protein concentration was determined using a
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Then, for each sample, 20 µg extracted protein was
separated on a 10% SDS-PAGE gel (cat. no. P0012A, Beyotime
Institute of Biotechnology) at 90 V for 25 min followed by 120 V
for 1.5 h and transferred to polyvinylidene difluoride membranes
(EMD Millipore). The transfer was performed using a Mini Trans-Blot
Transfer Cell (Bio-Rad Laboratories, Inc.) at 4°C for 1.5 h at a
constant voltage setting of 100 V. The membranes were blocked using
4% non-fat dry milk in tris-buffered saline (TBS) with Tween-20
(Sigma-Aldrich; Merck KGgA) at room temperature for 30 min.
Membranes were then incubated with rabbit anti-human KLF5
polyclonal antibody (1:500; cat. no. ab24331) and rabbit anti-human
GAPDH polyclonal antibody (1:1,000; cat. no. ab9485; both Abcam,
Cambridge, MA, USA) overnight at 4°C. Subsequently, horseradish
peroxidase-labeled goat anti-rabbit IgG secondary antibody
(1:2,000; cat. no. ab7090; Abcam) was incubated at 37°C for 1 h
with the membranes and protein bands were visualized with the
enhanced chemiluminescence system (EMD Millipore). Experiments were
conducted in triplicate. The relative expression of protein was
calculated by densitometric analysis using ImageJ software version
2.0.0 (National Institutes of Health, Bethesda, MD, USA).
Luciferase reporter assay
HepG2 cells (1×105 cells/well) were
cultured in a 24-well plate and incubated for 24 h. The cells were
divided into four groups: i) miRNA negative control (cat. no.
B04001; Shanghai GenePharma Co., Ltd., Shanghai, China) and
wild-type 3′UTR of KLF5 group; ii) miR-145-5p mimic and wild-type
3′UTR of KLF5 group; iii) miRNA negative control and mutant 3′UTR
of KLF5 group; and iv) miR-145-5p mimic and mutant 3′UTR of KLF5
group. HepG2 cells were harvested 48 h after transfection. Using
the Dual-Luciferase Reporter System (Promega Corporation), relative
luciferase activity was evaluated and normalized to the activity of
Renilla luciferase.
Statistical analysis
Data are presented as the mean ± standard deviation,
and were analyzed with SPSS 12.0 software (SPSS, Inc., Chicago, IL,
USA). Using Student's t-test or one-way analysis of variance
followed Bonferroni's post-hoc test, quantitative variables were
analyzed. Pearson's correlation coefficient was used to determine
the correlation between KLF5 mRNA expression and miR-145-5p levels
in HCC tissues. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-145-5p expression is decreased in
HCC
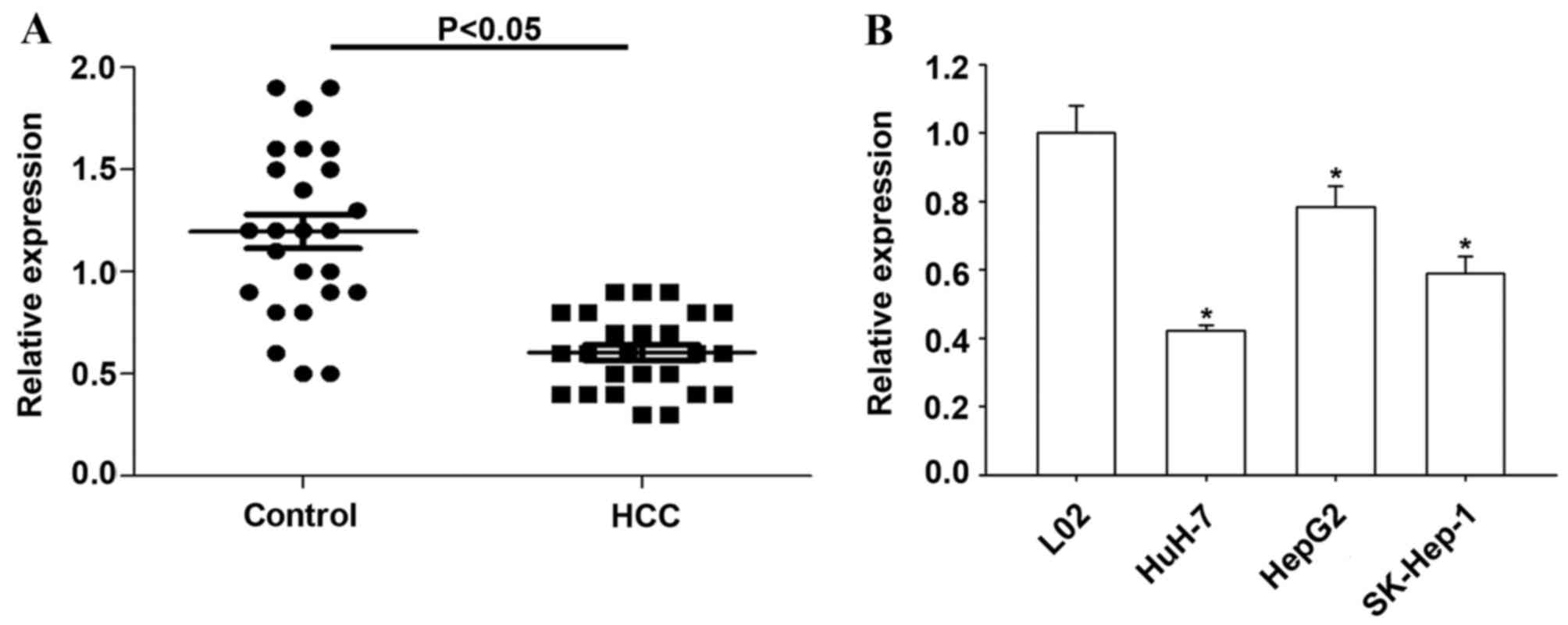
RT-qPCR was used to determine the expression of
miR-145-5p in 25 HCC and matched non-tumor tissues. Compared with
non-tumor tissues, the present study observed significantly
decreased miR-145-5p level in HCC tissues (P<0.05; Fig. 1A). The expression of miR-145-5p was
significantly reduced in all three HCC cell lines compared with L02
normal human liver cells (P<0.05; Fig. 1B).
miR-145-5p inhibits HCC cell
proliferation and migration
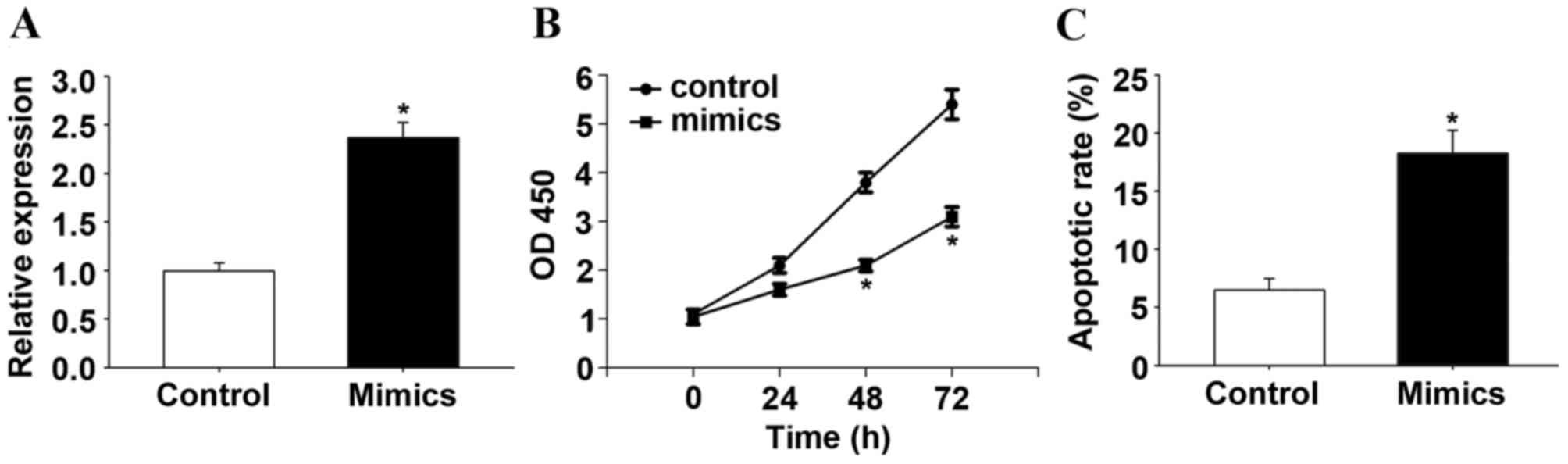
Additionally, the present study observed that
miR-145-5p suppressed the proliferation of HCC cells. RT-qPCR
confirmed that transfection with the miR-145-5p mimic increased the
expression of miR-145-5p (P<0.05; Fig. 2A). Following transfection with
miR-145-5p or control mimics in HepG2 cells, the biological effect
of miR-145-5p on the progression of HCC was determined by CCK-8
assay. The CCK-8 assay indicated that overexpression of miR-145-5p,
induced by miR-145-5p mimics, significantly suppressed the cell
proliferation rate after 48 h (P<0.05; Fig. 2B). A significant increase in the
percentage of apoptotic cells in miR-145-5p-overexpressing HepG2
cells was observed when compared with control HepG2 cells
(P<0.05; Fig. 2C). In addition,
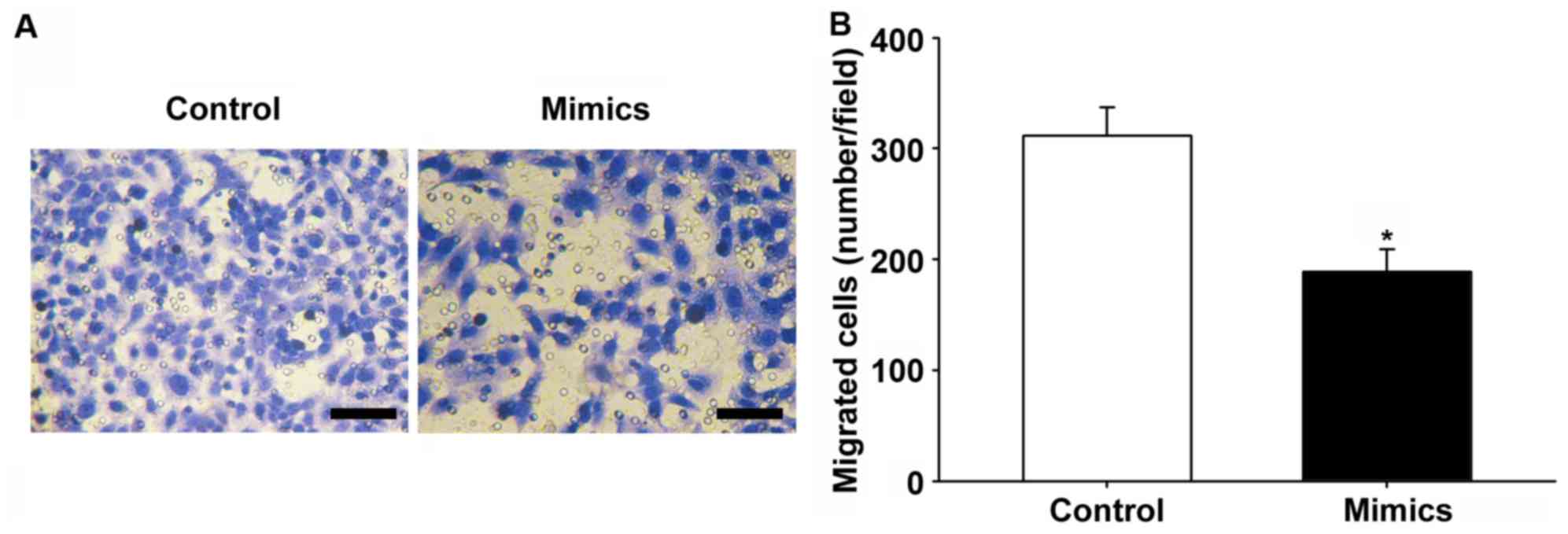
miR-145-5p or control mimics were transfected into HepG2 cells to
determine the effect of miR-145-5p on motility of HCC cells, and
migration assays were conducted. Overexpression of miR-145-5p
significantly inhibited the migratory capabilities of HepG2 cells
(P<0.05; Fig. 3A and B).
KLF5 is a direct target of
miR-145-5p
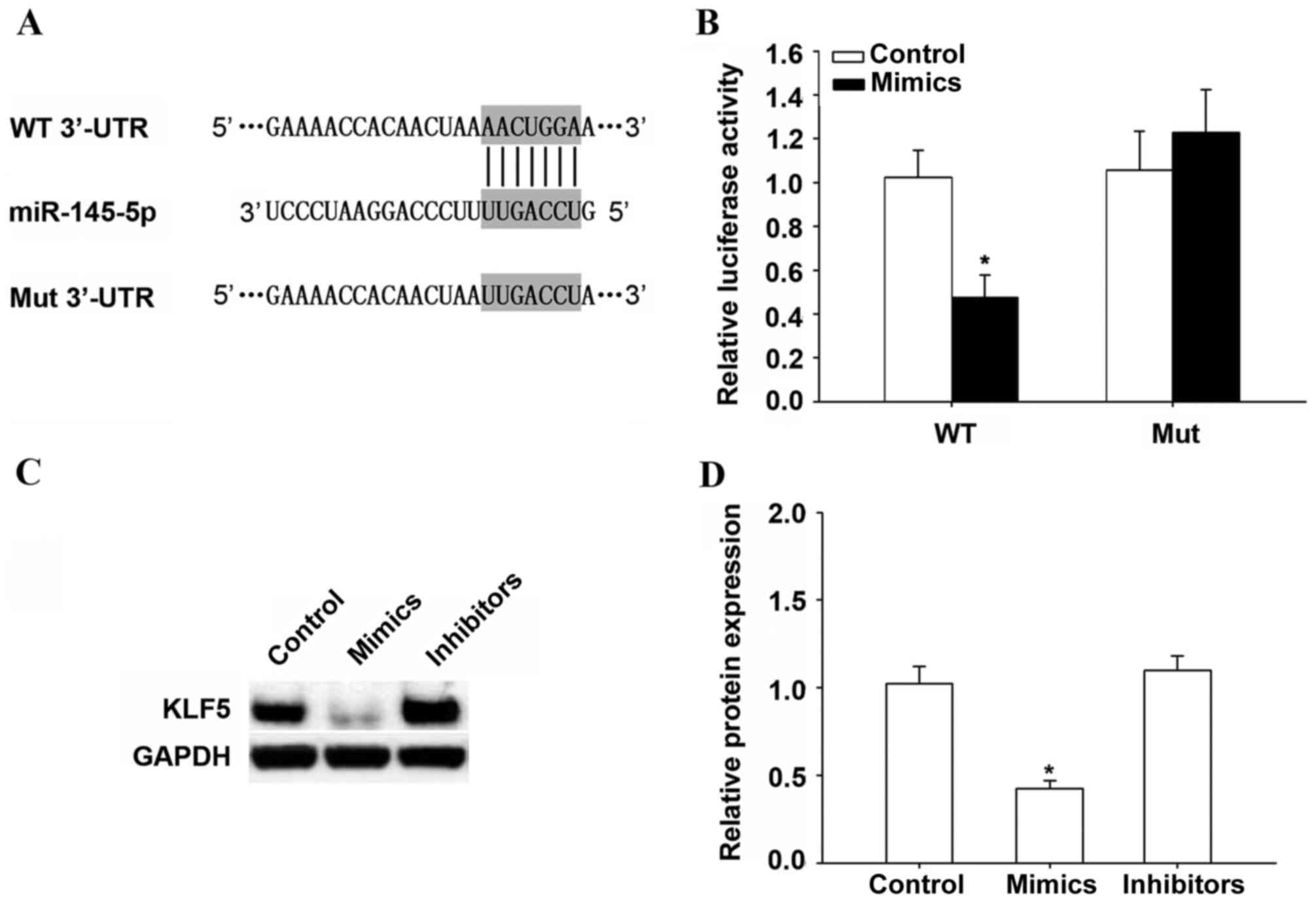
In order to determine the target of miR-145-5p in
HCC, TargetScan 7.0 (www.targetscan.org) was used. It was demonstrated that
KLF5 is a potential target of miR-145-5p (Fig. 4A). Results of the luciferase
activity assay suggested that miR-145-5p overexpression
significantly inhibited the luciferase activity of the WT 3′-UTR of
KLF5 in HepG2 cells compared with the control (P<0.05; Fig. 4B). In addition, miR-145-5p
overexpression significantly suppressed KLF5 protein expression
(P<0.05), while miR-145-5p inhibition did not change KLF5
protein expression compared with the control (Fig. 4C and D).
Overexpression of KLF5 reduces the
tumor suppressive effects of miR-145-5p
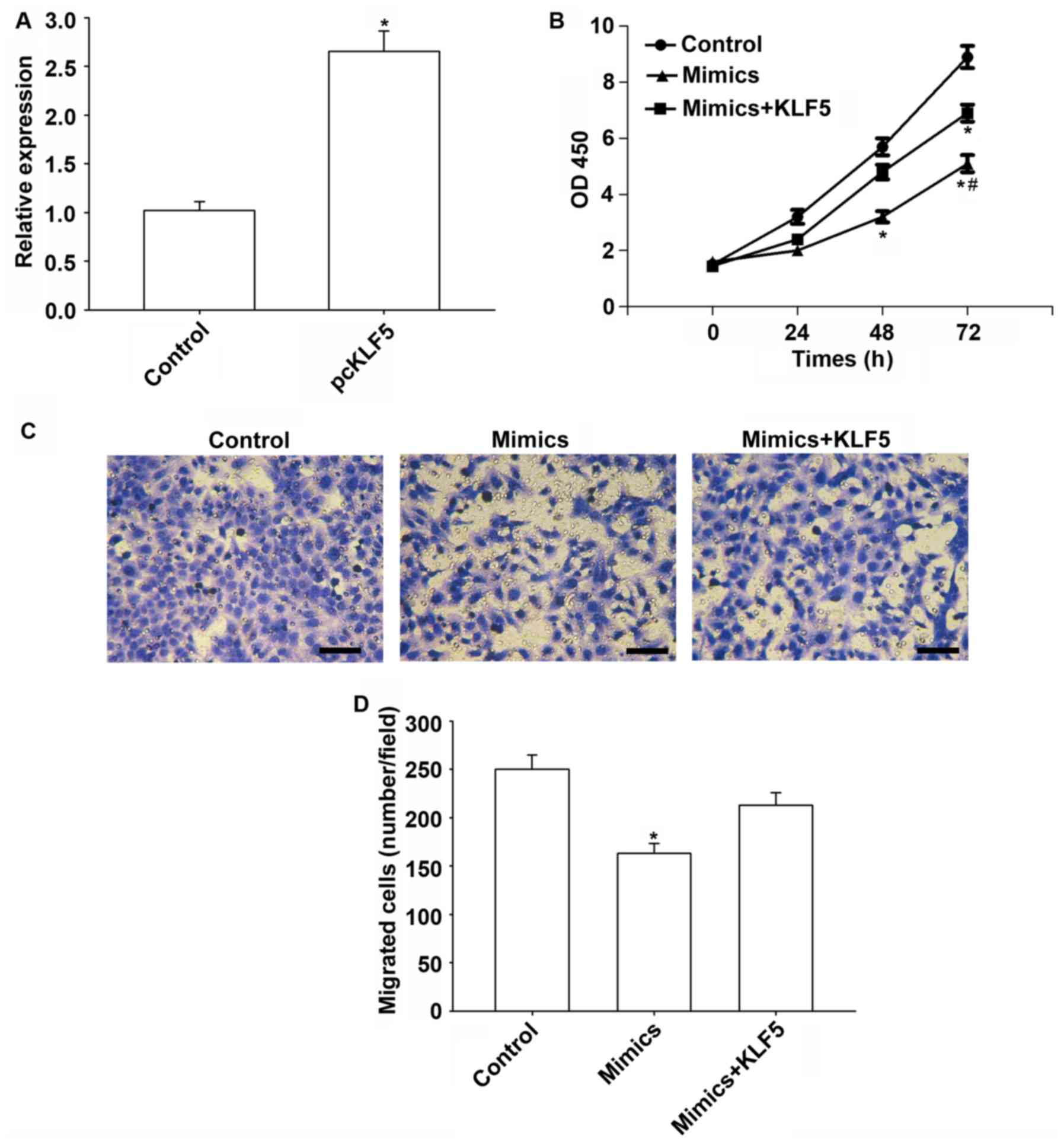
Transfection of the KLF5 vector increased KLF5
expression significantly (P<0.05; Fig. 5A). Furthermore, KLF5 overexpression
weakened the tumor suppressive effect of miR-145-5p in HepG2 cells
(Fig. 5B). Additionally, the
migration of HepG2 cells was significantly reduced by miR-145-5p
compared with the control (P<0.05), while KLF5 overexpression
reversed these suppressive effects (Fig. 5C and D).
miR-145-5p and KLF5 are negatively
correlated in HCC
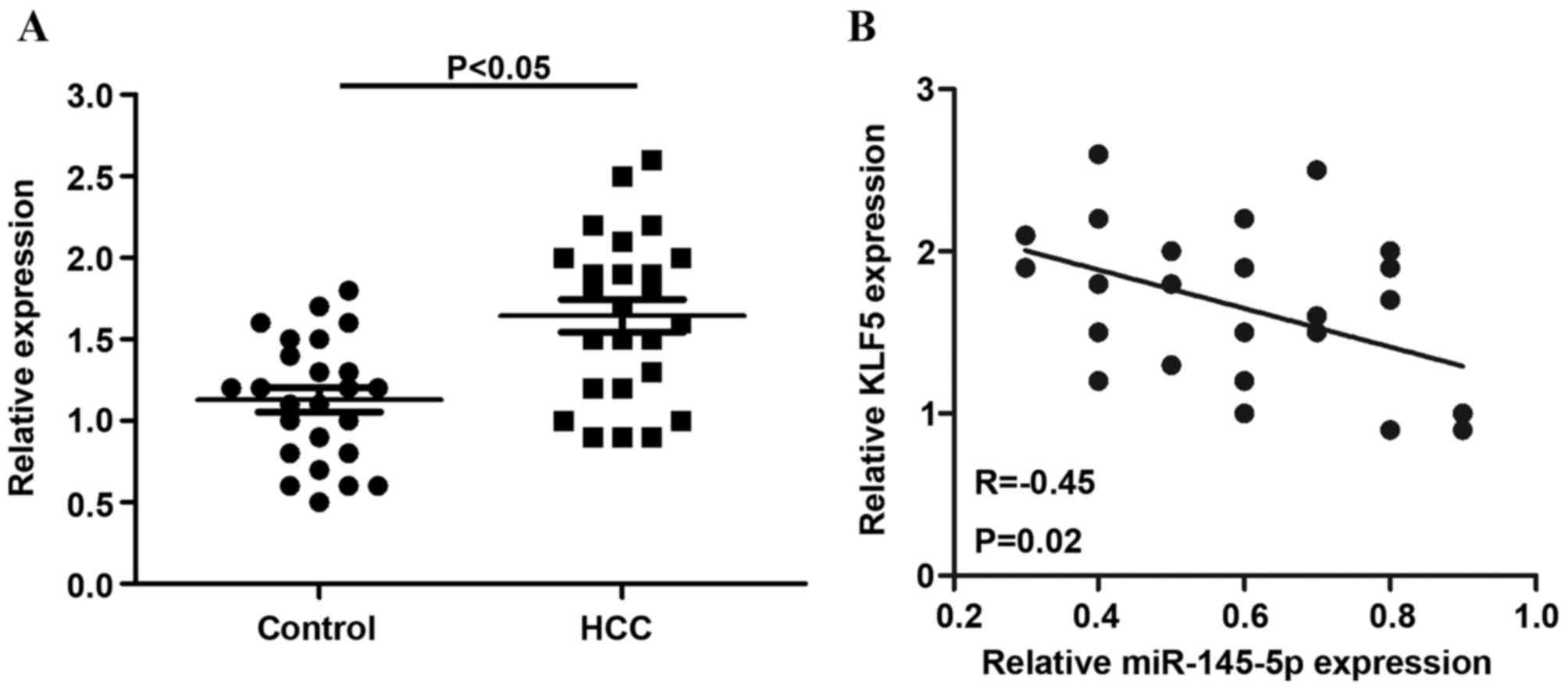
The relative expression of KLF5 was detected in 25
non-tumor tissues and HCC tissues. The results indicated that, in
HCC tissues, KLF5 mRNA levels were significantly increased compared
with non-tumor tissues (P<0.05; Fig. 6A). In addition, the present study
observed that KLF5 mRNA expression was inversely correlated with
miR-145-5p levels in HCC tissues (r=−0.45) (Fig. 6B).
Discussion
HCC has one of the highest mortality rates despite
significant improvements in diagnosis and treatment in recent years
(19). Although the mechanisms
involved in HCC have been previously demonstrated, the involvement
of epigenetic regulation remains predominantly unknown (20). As miRNAs are the most promising
components of the epigenetic pathway in terms of targets for the
development of novel therapeutic approaches, understanding the role
of miRNA in HCC is essential. It is currently known that multiple
miRNAs are involved in HCC development and progression. It has
previously been observed that several miRNAs have aberrant
expression in HCC and it is thought that these miRNAs may be used
as prognostic indicators in HCC (21). miR-145-5p is a novel miRNA that is
suggested to be implicated in cancer treatment and carcinogenesis.
Downregulation of miR-145-5p has been reported in prostate cancer
(17). Additionally, miR-145-5p
was downregulated in prostate cancer (22), lung cancer (23) and colorectal cancer (24), indicating that miR-145-5p may have
tumor suppressive effects. Expression levels of miR-145-5p were
inversely associated with the proliferation of prostate cancer
cells. Furthermore, in embryonic stem cells, by targeting the 3′UTR
of SRY-box 2 mRNA, miR-145-5p suppresses self-renewal of human
embryonic stem cells (17). Thus,
investigation of miR-145-5p provides novel insight into the
molecular mechanisms of cancer progression and carcinogenesis. The
present study aimed to analyze the effects of miR-145-5p on HCC and
to identify the potential target genes of miR-145-5p in order to
study the molecular mechanism behind its role in HCC oncogenesis.
Expression of endogenous miR-145-5p in 25 HCC tissues was
significantly downregulated. Similarly, miR-145-5p was
downregulated in HCC cell lines, including HuH-7, HepG2 and
SK-Hep-1, compared with the L02 normal liver cell line.
Additionally, overexpression of miR-145-5p decreased proliferation
rate and induced apoptosis in HCC cells. Transwell analysis
indicated that miR-145-5p inhibited migration of HCC cells. This
evidence suggested that downregulation of miR-145-5p may lead to
increases in the proliferation, migration and aggression of HCC.
The current study was extended to identify the potential targets of
miR-145-5p in HCC cells. In silico bioinformatics analyses
demonstrated that the 3′UTR of KLF5 is one potential target of
miR-145-5p. In vivo and in vitro results demonstrated
that KLF5 expression levels were significantly decreased in HCC
cells overexpressing miR-145-5p. It is regarded that KLF5 is highly
expressed in HCC tissues compared with healthy control tissues. The
involvement of KLF5 in tumor progression has also been demonstrated
in breast (25), intestinal
(26), esophageal (27) and gastric cancer (28). The current study proposes that
miR-145-5p is an important regulator of KLF5 since miR-145-5p
overexpression in HCC cells reduced KLF5 expression. Decreased
levels of miR-145-5p may be a key step in the pathogenesis of HCC.
It is possible that miRNAs regulate several genes, by targeting the
3′UTR of mRNA, and the subsequent changes in the expression of
these target genes may participate in specific tissue development
or cancer progression. The present study has identified that KLF5,
a target of miR-145-5p, is a key marker gene in cancer. miR-145-5p
may also contribute to the dysregulation of other functional genes
during tumor development. Thus, further studies should focus on the
gene network of miR-145-5p in HCC and the miRNA profiles of
circulating HCC tumor cells, which may demonstrate the roles of
miRNA and improve the understanding of its function in HCC
pathogenesis.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81302328)
and the Science and Technology Department of Hunan province (grant
no. 2013FJ4059).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HL and HS performed the experiments and wrote the
paper. JY and CY designed the study and edited the manuscript. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
The Ethics Committee of The Second Xiangya Hospital
of Central South University approved this study and written
informed consent was obtained from each patient.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bruix J and Sherman M: American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yoo HY, Patt CH, Geschwind JF and
Thuluvath PJ: The outcome of liver transplantation in patients with
hepatocellular carcinoma in the United States between 1988 and
2001: 5-year survival has improved significantly with time. J Clin
Oncol. 21:4329–4335. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sala M, Fuster J, Llovet JM, Navasa M,
Solé M, Varela M, Pons F, Rimola A, García-Valdecasas JC, Brú C, et
al: High pathological risk of recurrence after surgical resection
for hepatocellular carcinoma: an indication for salvage liver
transplantation. Liver Transpl. 10:1294–1300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoshida Y, Villanueva A, Kobayashi M, Peix
J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et
al: Gene expression in fixed tissues and outcome in hepatocellular
carcinoma. New Engl J Med. 359:1995–2004. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parisi S, Passaro F, Aloia L, Manabe I,
Nagai R, Pastore L and Russo T: Klf5 is involved in self-renewal of
mouse embryonic stem cells. J Cell Sci. 121:2629–2634. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saxena NK, Sharma D, Ding X, Lin S, Marra
F, Merlin D and Anania FA: Concomitant activation of the JAK/STAT,
PI3K/AKT, and ERK signaling is involved in leptin-mediated
promotion of invasion and migration of hepatocellular carcinoma
cells. Cancer Res. 67:2497–2507. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dong JT and Chen C: Essential role of KLF5
transcription factor in cell proliferation and differentiation and
its implications for human diseases. Cell Mol Life Sci.
66:2691–2706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang B, Pan X, Cobb GP and Anderson TA:
MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zimmerman AL and Wu S: MicroRNAs, cancer
and cancer stem cells. Cancer Lett. 300:10–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao T, Li H, Hu Y, Ma D and Cai X: miR-144
suppresses the proliferation and metastasis of hepatocellular
carcinoma by targeting E2F3. Tumor Biol. 35:10759–10764. 2014.
View Article : Google Scholar
|
|
14
|
Ludwig JA and Weinstein JN: Biomarkers in
cancer staging, prognosis and treatment selection. Nat Rev Cancer.
5:845–856. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ozen M, Karatas OF, Gullluoglu S, Bayrak
OF, Sevli S, Guzel E, Ekici ID, Caskurlu T, Solak M, Creighton CJ
and Ittmann M: Overexpression of miR-145-5p inhibits proliferation
of prostate cancer cells and reduces SOX2 expression. Cancer
Invest. 33:251–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Noh JH, Chang YG, Kim MG, Jung KH, Kim JK,
Bae HJ, Eun JW, Shen Q, Kim SJ, Kwon SH, et al: MiR-145 functions
as a tumor suppressor by directly targeting histone deacetylase 2
in liver cancer. Cancer Lett. 335:455–462. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yuen MF, Hou JL and Chutaputti A: Asia
Pacific Working Party on Prevention of Hepatocellular Carcinoma:
Hepatocellular carcinoma in the Asia pacific region. J
Gastroenterol. Hepatol. 24:346–353. 2009.
|
|
20
|
Iliopoulos D, Satra M, Drakaki A,
Poultsides GA and Tsezou A: Epigenetic regulation of hTERT promoter
in hepatocellular carcinomas. Int J Oncol. 34:391–399.
2009.PubMed/NCBI
|
|
21
|
Milazzo M, Fornari F and Gramantieri L:
MicroRNA and hepatocellular carcinoma: Biology and prognostic
significance. Minerva Gastroenterol Dietol. 57:257–271.
2011.PubMed/NCBI
|
|
22
|
Avgeris M, Stravodimos K, Fragoulis EG and
Scorilas A: The loss of the tumour-suppressor miR-145 results in
the shorter disease-free survival of prostate cancer patients. Br J
Cancer. 108:2573–2581. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sanfiorenzo C, Ilie MI, Belaid A, Barlési
F, Mouroux J, Marquette CH, Brest P and Hofman P: Two panels of
plasma microRNAs as non-invasive biomarkers for prediction of
recurrence in resectable NSCLC. PLoS One. 8:e545962013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schee K, Lorenz S, Worren MM, Günther CC,
Holden M, Hovig E, Fodstad O, Meza-Zepeda LA and Flatmark K: Deep
sequencing the microRNA transcriptome in colorectal cancer. PLoS
One. 8:e661652013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tong D, Czerwenka K, Heinze G, Ryffel M,
Schuster E, Witt A, Leodolter S and Zeillinger R: Expression of
KLF5 is a prognostic factor for disease-free survival and overall
survival in patients with breast cancer. Clin Cancer Res.
12:2442–2448. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bateman NW, Tan D, Pestell RG, Black JD
and Black AR: Intestinal tumor progression is associated with
altered function of KLF5. J Biol Chem. 279:12093–12101. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Y, Goldstein BG, Chao HH and Katz JP:
KLF4 and KLF5 regulate proliferation, apoptosis and invasion in
esophageal cancer cells. Cancer Biol Ther. 4:1216–1221. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kwak MK, Lee HJ, Hur K, Park DJ, Lee HS,
Kim WH, Lee KU, Choe KJ, Guilford P and Yang HK: Expression of
Krüppel-like factor 5 in human gastric carcinomas. J Cancer Res
Clin Oncol. 134:163–167. 2008. View Article : Google Scholar : PubMed/NCBI
|