Introduction
Renal cell carcinoma (RCC) is the 9th most common
cancer diagnosis and cause of cancer-associated mortality, and
accounts for 3% of all malignancies worldwide (1,2).
Considering that, at present, there are no available biomarkers for
RCC screening, ~30% of patients develop metastasis at the time of
diagnosis, and 20–40% patients develop recurrence or metastases
following initial surgical resection (3–5).
Furthermore, RCC is insensitive to conventional chemotherapy and
radiotherapy (6). In total,
approximately a third (~71,300) of patients diagnosed with RCC each
year worldwide will ultimately succumb to metastatic disease
(7). Thus, identification of novel
biomarkers is important for the early diagnosis and treatment of
RCC, and therapeutic strategies based on novel molecular targets
are urgently required in RCC diagnosis and therapeutics.
Recently, microRNAs (miRNAs/miRs) have been revealed
to have important roles in numerous carcinomas, including kidney
cancer (8–12). miRNAs are short non-coding single
stranded RNAs containing 20–22 nucleotides and are able to regulate
gene expression at the post-transcriptional level (13). Increasing evidence has suggested
that miRNAs function as oncogenes or tumor suppressors in RCC
(3,14,15).
miR-181a-5p is located on chromosome 9, and its
activity is dysregulated in numerous tumor types, including gastric
cancer (16), pituitary adenoma
(17) and hepatocellular carcinoma
(18). Thus, it may be suggested
that miR-181a-5p may function as either a tumor suppressor gene or
an oncogene. However, the effect of miR-181a-5p on RCC remains
largely undetermined. In the present study, the expression levels
of miR-181a-5p in RCC tissues and cell lines were investigated, in
addition to the effect of miR-181a-5p on cell function.
Materials and methods
Ethics statement
All human paired RCC samples and adjacent normal
tissue samples were obtained at the Peking University Shenzhen
Hospital (Shenzhen, China), between July 2015 and July 2016. All
patients provided written informed consent, and the present study
was approved by the Ethical Review Committee of Peking University
Shenzhen Hospital (Shenzhen, China) and complied with the
Declaration of Helsinki. Patient characteristics are presented in
Table I.
 | Table I.Clinicopathological characteristics
of patients with renal cell carcinoma. |
Table I.
Clinicopathological characteristics
of patients with renal cell carcinoma.
| Characteristic | Number |
|---|
| Age range
(years) | 27–72 |
| Gender
(male/female) | 16/5 |
| Histological type
(clear cell/papillary) | 18/3 |
| Fuhrman grade
(I/II/III/IV) | 13/6/1/1 |
| AJCC clinical stage
(I/II/III+IV) | 13/7/1 |
Cell culture
786-O, Caki-1 and ACHN are renal cell carcinoma cell
lines. 293T is a normal renal cell line. Cell lines were purchased
from the Guangdong and Shenzhen Key Laboratory of Male Reproductive
Medicine and Genetics (Shenzhen, China). ACHN, Caki-1 and 293T
cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10%
fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc.), 1% antibiotics (100 µl/ml penicillin and 100 mg/ml
streptomycin sulfate) and 1% glutamine, and maintained in a
humidified atmosphere with 5% CO2 at 37°C. 786-O cells
were cultured in RPMI 1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from excised tumor specimens
using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and
purified with the RNeasy Maxi kit (Qiagen GmbH, Hilden, Germany),
following the manufacturer's protocol. The concentration of RNA was
determined using a NanoDrop 2000/2000c (Thermo Fisher Scientific,
Inc.). Following this, RNA (1 µg) was reverse-transcribed to cDNA
using a miScript Reverse Transcription kit (Qiagen GmbH). Total RNA
was converted into cDNA using the miScript II RT kit (Qiagen GmbH)
The reaction was performed at 37°C for 1 h, followed by RT
inactivation at 95°C. qPCR was subsequently performed to determine
the expression level of miR-181a-5p using a miScript
SYBR® Green PCR kit (Qiagen GmbH) on the Roche
Lightcycler 480 Real Time PCR system (Roche Diagnostics, Basel,
Switzerland), according to the manufacturer's protocol. The miRNA
expression was normalized to U6 expression. The thermocycling
conditions of qPCR were as follows: Initial denaturation of 95°C
for 2 min; followed by 40 cycles of denaturation at 95°C for 10
sec, annealing and elongation at 55°C for 30 sec, and final
extension at 72°C for 30 sec. The sequences of the primers used
were as follows: miR-181a-5p forward, 5′-AACAUUCAACGCUGUCGGUGAGU-3′
and reverse, miScript universal primer (miScript SYBR-Green PCR
kit; sequence unavailable); U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-ACGCTTCACGAATTTGCGT-3′. The expression levels of
miR-181a-5p in tissues and cells were determined using the
2−ΔΔCq method (19).
Cell transfection
786-O and ACHN cells were seeded into a 6-well plate
(3×105 cells/well). Following culture for 24 h, cells
were transfected with 5 ml miR-181a-5p mimics (forward,
5′-AACAUUCAACGCUGUCGGUGAGU-3′ and reverse,
5′-UCACCGACAGCGUUGAAUGUUUU-3′) or negative control (NC; forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) purchased from Guangzhou RiboBio, Co.,
Ltd. (Guangzhou, China) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), which was mixed with
Opti-MEM® I Reduced Serum Medium (Gibco; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. In
order to confirm the efficiency of transfection, RT-qPCR was
performed to determine the levels of miR-18a-5p expression 24 h
post-transfection.
Wound healing assay
786-O and ACHN cells were seeded into a 6-well plate
(3×105 cells/well) and incubated in a humidified
atmosphere with 5% CO2 at 37°C for 24 h. Following this,
cells were transfected with miR-181a-5p mimics or negative control
(NC) using Lipofectamine® 2000. A wound was created in a
monolayer of 786-O cells or ACHN cells using a sterile 200-µl
pipette tip 6 h post-transfection. PBS was used to wash away the
cell debris. Images of cells were recorded at 0 and 12 h time
intervals following the initial creation of the wound using a light
microscope (magnification, ×100; Olympus Corporation, Tokyo,
Japan).
Transwell assay
The cellular migratory and invasive ability of 786O
and ACHN cells was determined using a Transwell assay. Transwell
chambers (BD Biosciences, Franklin Lakes, NJ, USA) with Matrigel
were used to analyze the invasive capacity of cells, whereas
Transwell chambers without Matrigel were used to analyze the
migratory ability of cells. At 24 h post-transfection,
~2×104 cells were added to the upper chamber with
serum-free DMEM, and DMEM supplemented with 10% FBS was plated in
the lower chamber. Following this, the chambers were incubated for
48 h in a 5% CO2 incubator at 37°C. Cells adhering to
the upper side of the inserts were gently scraped off, and invasive
cells on the lower surface were stained with 0.1% crystal violet at
room temperature for 25 min and counted using a light microscope at
a magnification of ×100 (Olympus Corporation).
TT assay
The cytoactivity of the 786O and ACHN cells was
determined using a MTT assay (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). Renal cancer cells (~5,000 cells) were inoculated in a
96-well plate (5×103 cells/well) and transfected with
miR-181a-5p mimics and NC using Lipofectamine® 2000. A
total of 96 h post-transfection, 20 µl MTT (5 mg/ml; Sigma-Aldrich;
Merck KGaA) was added to each well of the 96-well plate. After 4 h,
a total of 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA)
was added to the 96-well plate, which was shaken on a reciprocating
decolorization shaking table (TSB-108; Haimen LinBair Instruments
Manufacturing Co., Ltd., Haimen, China) for 10 min in the dark at
room temperature. Finally, the optical density (OD) values of each
well were determined using an ELISA microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) at a wavelength of 595 nm
(620 nm as the reference wavelength).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was determined using a CCK-8
assay (Beyotime Institute of Biotechnology, Haimen, China)
according to the manufacturer's protocol. A total of
5×103 cells/well were seeded in 96-well plates and
incubated in a humidified atmosphere with 5% CO2 at 37°C
for 24 h until 30–50% confluence was reached. Cells were
subsequently transfected with miR-181a-5p mimics−, in
addition to NC−. The OD values of experimental wells
were investigated at 450 nm at 0, 24, 48 and 72 h time intervals
post-transfection using an ELISA microplate reader (Bio-Rad
Laboratories, Inc.).
Flow cytometry assay
The apoptosis rate of 786-O and ACHN cells was
determined via flow cytometry assays. Cells were added to a 6-well
plate (3×105 cells/well) and subsequently transfected
with-miR-181a-5p mimics or NC. Cells were collected 48 h
post-transfection and washed with cold PBS (4°C). Subsequently, the
cells were resuspended in 100 µl 1X binding buffer, and 5 µl
Annexin V-fluorescein isothiocyanate (Invitrogen; Thermo Fisher
Scientific, Inc.) and 5 µl propidium iodide (Invitrogen; Thermo
Fisher Scientific, Inc.) was added to each cell suspension. Cells
were subsequently incubated at room temperature for 15 min in the
dark, and 400 µl binding buffer was added to each tube. The
apoptosis rate was determined using flow cytometry (EPICS, Xl-4;
Beckman Coulter, Inc., Brea, CA, USA) and was analyzed with FlowJo
software (version 10; Flow Jo LLC, Ashland, OR, USA).
Statistical analysis
Data are presented as mean ± standard error of the
mean. All assays were repeated at least three times. Significance
of differential expression was analyzed using one way analysis of
variance followed by Tukey's post-hoc test. The SPSS 23.0
statistical software package (IBM Corp., Armonk, NY, USA) was used
to perform statistical analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-181a-5p is upregulated in RCC
tissues and cell lines
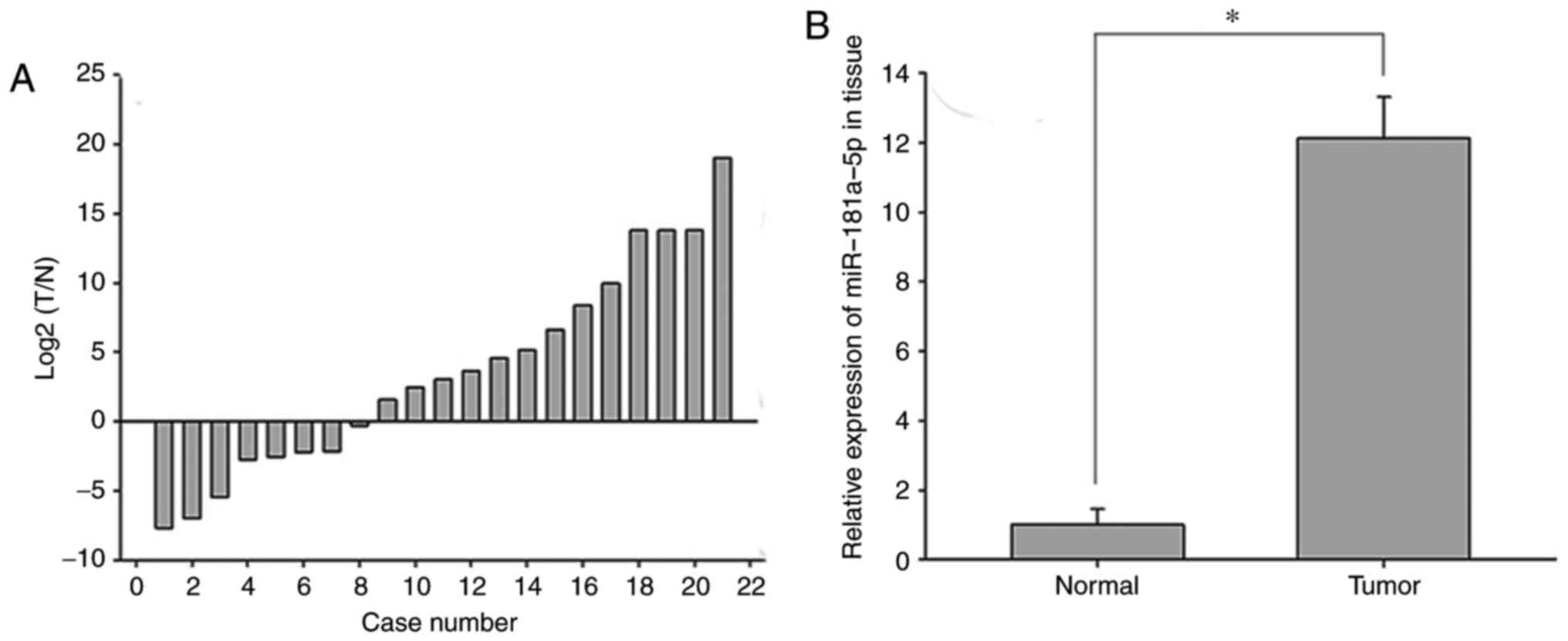
To investigate the role of miR-181a-5p in RCC,
RT-qPCR was performed to determine the expression levels of
miR-181a-5p in 21 paired RCC tissues and adjacent normal renal
tissues (Fig. 1A). miR-181a-5p was
revealed to be upregulated in RCC tissues compared with normal
renal tissues (P<0.05; Fig.
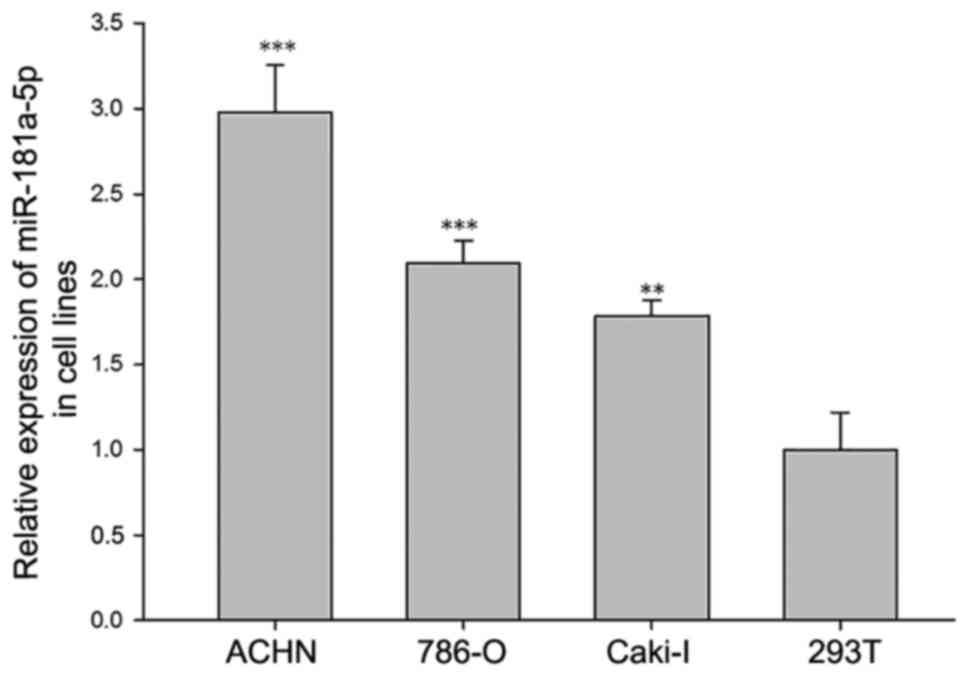
1B). Furthermore, miR-181a-5p was demonstrated to be expressed
in RCC cell lines and normal renal cells; however, it was revealed
that the 786-O, Caki-1 and ACHN cell lines exhibited significantly
increased expression levels of miR181a-5p compared with the 293T
normal kidney cell line (P<0.05; Fig. 2).
Cell transfection efficiency
validation
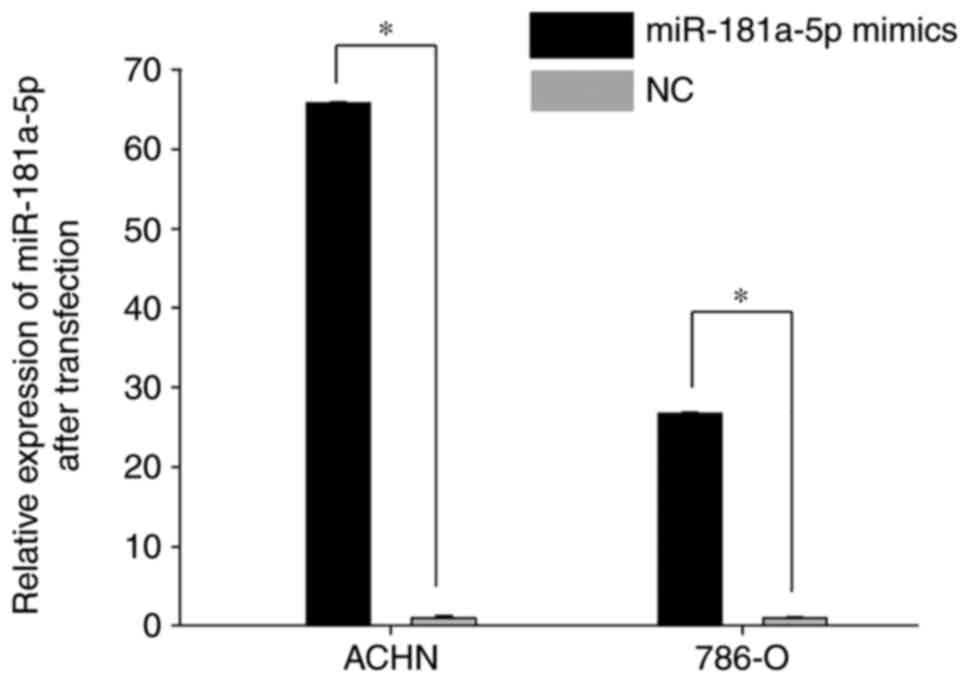
RT-qPCR was performed to determine the transfection
efficiency of miR-181a-5p mimics-compared with the NC. As revealed
in Fig. 3, the expression level of
miR-181a-5p was significantly enhanced in cells transfected with
miR-181a-5p mimics 24 h post-transfection compared with the NC
(Fig. 3; P<0.05).
Effect of miR-181a-5p on RCC cell
proliferation
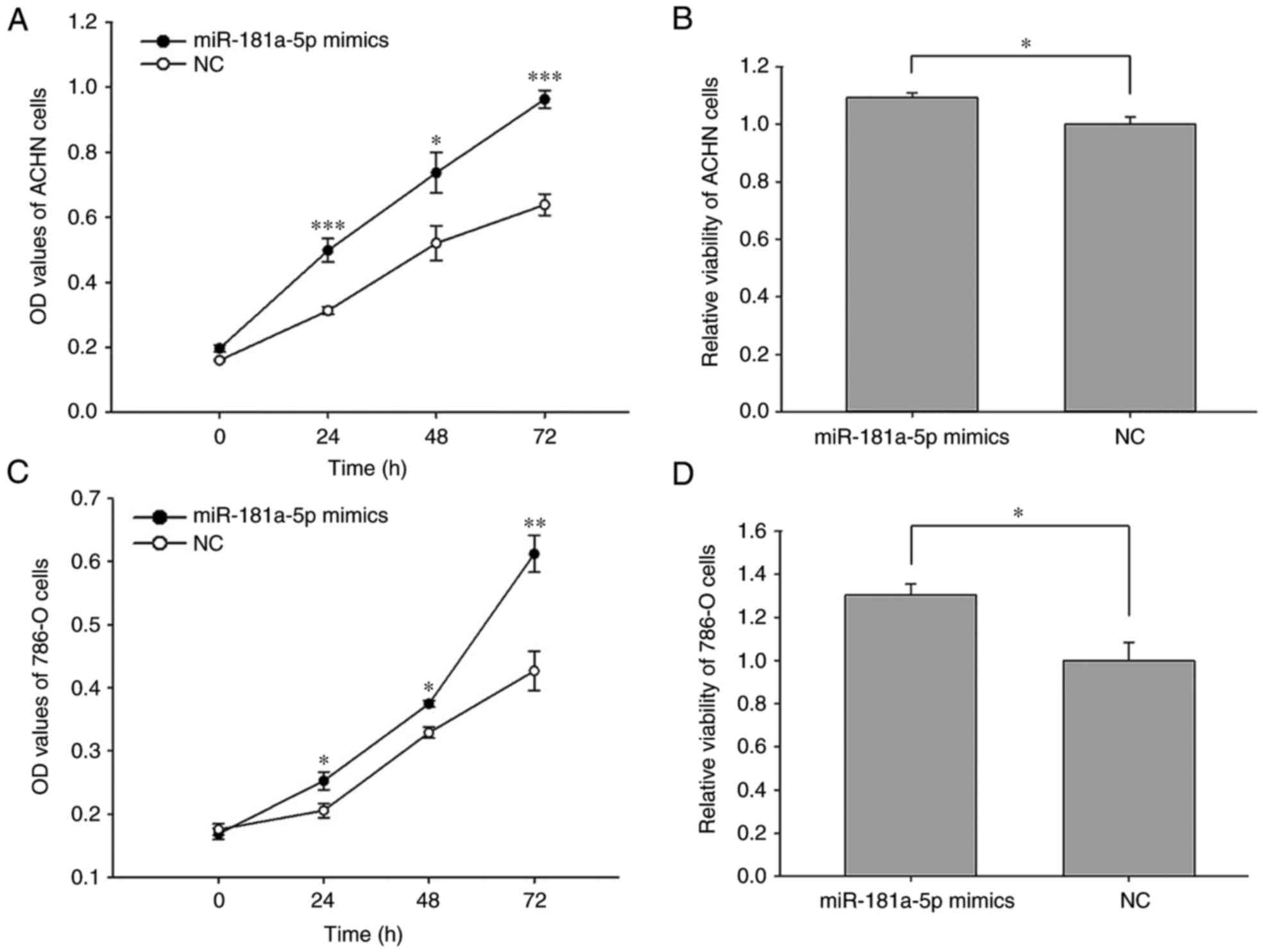
The results of the MTT and CCK-8 assays revealed
that the upregulation of miR-181a-5p enhanced cell proliferation-.
The proliferation of ACHN cells transfected with miR-181a-5p mimics
was significantly enhanced at 24 (59.15%), 48 (41.73%) and 72
(50.81%) h time intervals post-transfection, respectively, compared
with cells transfected with the NC (Fig. 4A; P<0.05, P<0.001). The
proliferation of 786-O cells transfected with miR-181a-5p mimics
significantly increased by 22.82% (P<0.05), 13.98% (P<0.05)
and 43.33% (P<0.01) at 24, 48 and 72 h time intervals
post-transfection, respectively, when compared with cells
transfected with the NC (Fig. 4B).
The results of the MTT assay demonstrated that the viability of
ACHN and 786-O cells increased by 9.20% (P<0.05) and 30.3%
(P<0.05), respectively following transfection with miR-181a-5p
compared with cells transfected with the NC (Fig. 4C and D).
Effect of miR-181a-5p on RCC cell
motility
A Transwell assay and wound scratch assay were
performed to investigate the effect of miR-181a-5p on the motility
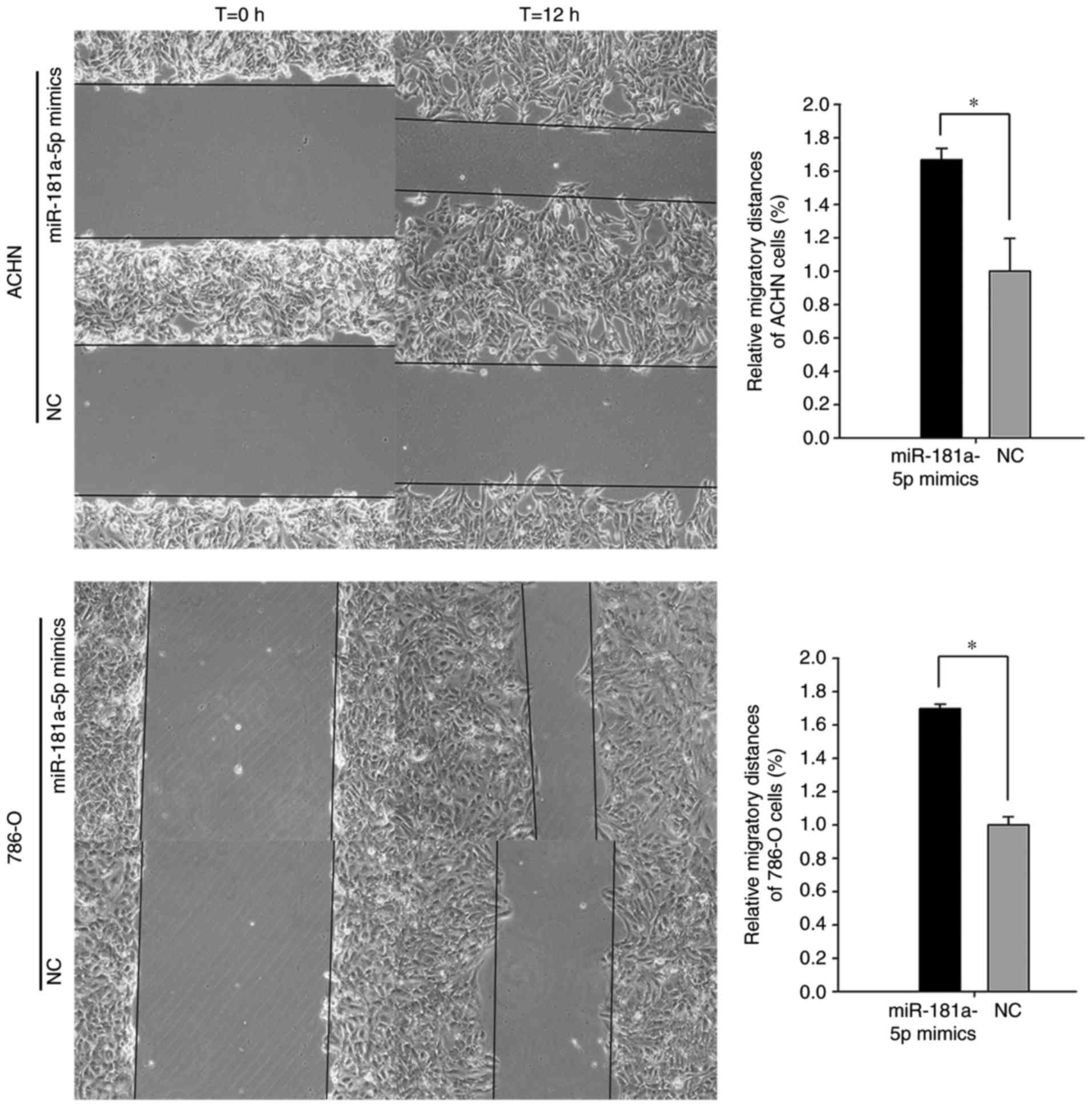
of the 786-O and ACHN cell lines. As revealed in Fig. 5, the results of the wound scratch
assay demonstrated that the distance migrated by 786-O cells
transfected with miR-181a-5p mimics was significantly increased at
the 12 h time interval post-transfection compared with cells
transfected with the NC (69.90%; P<0.01). Furthermore, the
distance migrated by ACHN cells transfected with miR-181a-5p mimics
was significantly increased at the 12 h time interval
post-transfection compared with cells transfected with the NC
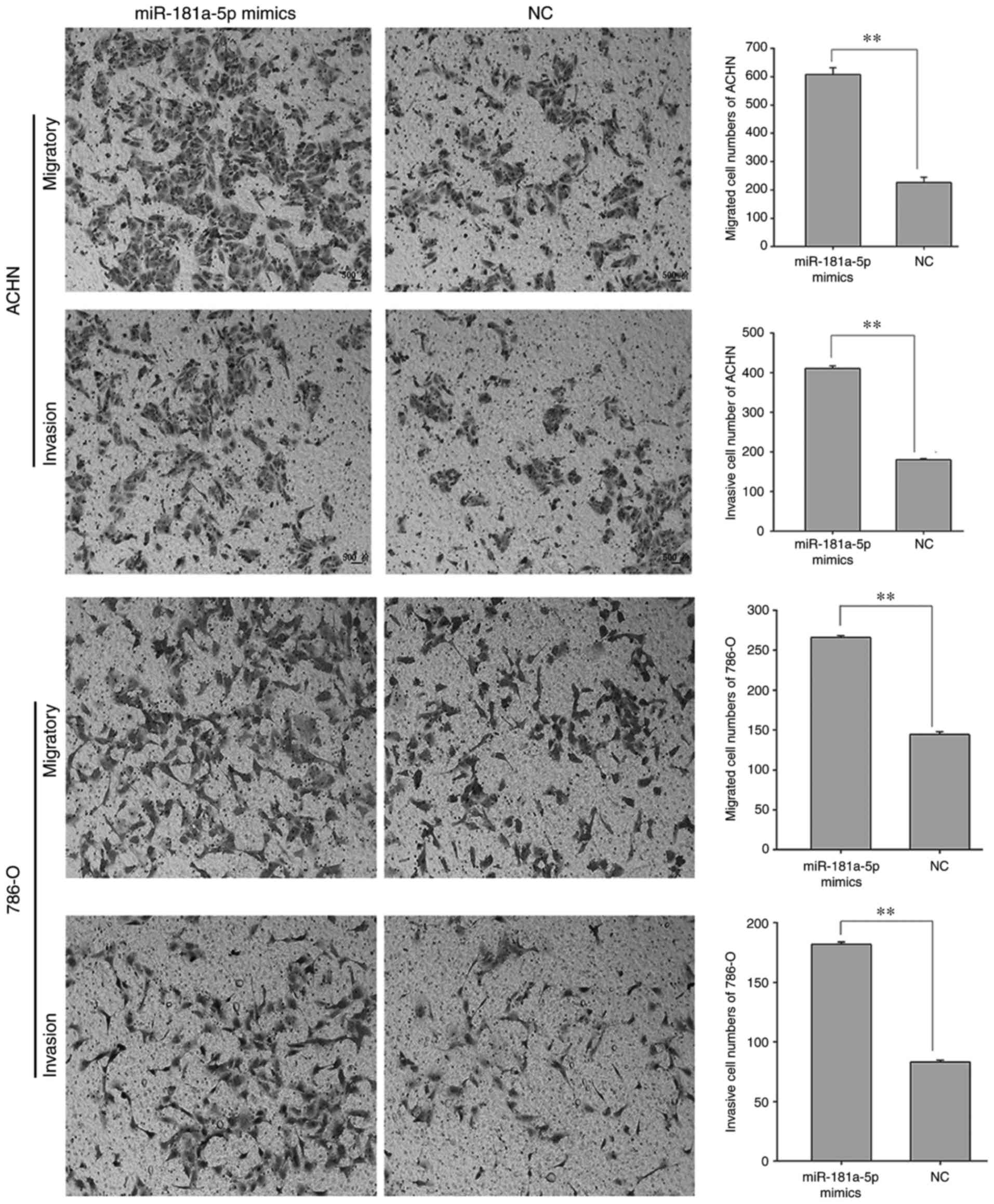
(66.80%; P<0.01). Furthermore, the results of the Transwell
assay revealed that 786-O cells transfected with miR-181a-5p mimics
exhibited a significantly increased migratory capacity compared
with cells transfected with the NC (84.40%; Fig. 6; P<0.01). In addition, ACHN cell
migration was significantly increased in cells transfected with
miR-181a-5p mimics compared with cells transfected with the NC
(168.43%; Fig. 6; P<0.01).
Furthermore, the invasive ability of 786-O cells transfected with
miR-181a-5p mimics was significantly increased compared with cells
transfected with the NC (118.32%; P<0.01; Fig. 6). In addition, the invasive ability
of ACHN cells was significantly increased in cells transfected with
miR-181a-5p mimics compared with cells transfected with the NC
(127.78%; P<0.01; Fig. 6).
These results suggested that miR-181a-5p may enhance the motility
of RCC cells.
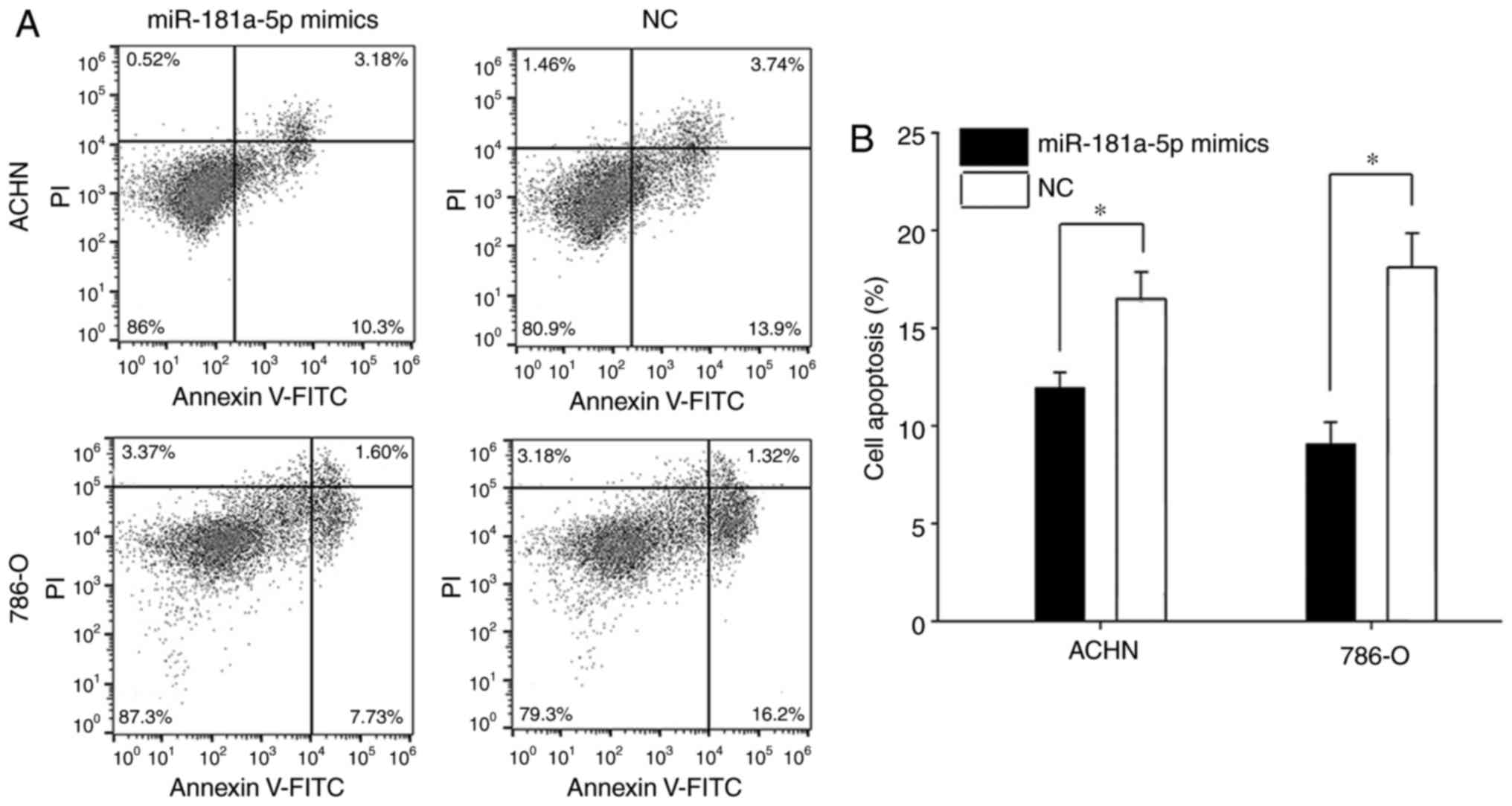
Upregulation of miR-181a-5p reduces
the apoptosis of ACHN and 876-O cells
The apoptosis rate was investigated by flow
cytometry, and the results demonstrated that miR-181a-5p
overexpression significantly suppressed cellular apoptosis in 786-O
and ACHN cell lines (Fig. 7). The
apoptosis rate of 786-O and ACHN cells transfected with miR-181a-5p
significantly decreased by 49.96 and 27.82%, respectively, when
compared with cells transfected with the NC (Fig. 7B; P<0.05)−.
Discussion
miRNAs have important roles in numerous biological
processes, including cell development, differentiation, metabolism,
proliferation, the cell cycle and apoptosis (1). Aberrant miRNA expression has been
associated with a number of chronic illnesses, including heart
disease, diabetes and cancer (20). Deregulated expression of miRNAs is
involved in the initiation and progression of tumors, metastasis
and therapeutic resistance (21).
Previous studies have demonstrated that miR-181a-5p
serves as an oncogene or a tumor suppressor via one or more
signaling pathways in certain types of tumors. In colorectal
cancer, downregulation of miR-181a-5p has been demonstrated to
enhance cell proliferation and chemoresistance by targeting protein
Wnt/β-catenin and transcription factor 4 (22). A previous study that investigated
hepatocarcinogenesis revealed that the downregulation of
miR-181a-5p activated hepatocyte growth factor receptor-mediated
oncogenic signaling (18).
Furthermore, miR-181a-5p has been demonstrated to be downregulated
in aggressive human breast and colon cancer, which promotes cancer
cell migration and angiogenesis (23). miR-181a-5p has been revealed to
function as a tumor suppressor by targeting GTPase KRas in
non-small cell lung cancer (24).
Mi et al (25) demonstrated
that miR-181a-5p may function as an onco-miRNA via activation of
Ras association domain-containing protein 6-mediated
mitogen-activated protein kinase signaling in gastric cancer. In a
further study, the transcription factor SOX2/miR-181a-5p/tumor
suppressor candidate 3 axis was revealed to have an important role
in the proliferation, migration and invasiveness of breast cancer
cells (26). Petrillo et al
(27) demonstrated that
concomitant expression of phosphorylated mothers against
decapentaplegic homolog 2 and miR-181a-5p represents a biomarker
for poor prognosis in patients with ovarian cancer. Furthermore,
Boguslawska et al (28)
revealed that serine/arginine-rich splicing factor 7 and
miR-181a-5p form a regulatory feedback loop in renal cancer cells,
which affects cell proliferation.
The results of the present study demonstrated that
the expression of miR-181a-5p was upregulated in 786-O and ACHN
cell lines compared with normal renal tissues. In addition, the
results of the present study revealed that expression of
miR-181a-5p promoted cell proliferation, invasion and migration,
and suppressed cellular apoptosis.
In conclusion, the results of the present study
demonstrated that miR-181a-5p was upregulated in RCC tissues and
cell lines, and that miR-181a-5p was associated with cell
migration, proliferation and apoptosis in RCC. The results of the
present study additionally suggested that miR-181a-5p may function
as an onco-miRNA in RCC. However, Brodaczewska et al
(29) revealed that RCC cell lines
used in in vitro experiments may be unable represent the
full pathological features of RCC, which remains a limitation that
merits further study. In addition, 293T cells have been
demonstrated to exhibit many features of neuronal cells (29); therefore, caution is required when
interpreting the results (30,31).
Furthermore, considering that renal cancer cells are adherent
cells, a small number of surviving cells may have undergone
mechanical death rather than apoptosis in the apoptosis assays.
Therefore, these limitations require consideration in future
studies aiming to investigate the effect of miR-181a-5p on RCC.
Future studies investigating the effect of miR-181a-5p on RCC may
focus on determining the underlying role of miR-181a-5p, including
its potential application and use in early diagnosis and prognostic
prediction in RCC. Further studies are required to determine the
cellular mechanism of miR-181a-5p in RCC tumorigenesis, and its
potential use in targeted therapy for RCC.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81101922), Science and
Technology Development Fund Project of Shenzhen (grant nos.
JCYJ20150403091443329 and JCYJ20170307111334308), the fund of
‘San-ming’ Project of Medicine in Shenzhen and the fund of
Guangdong Key Medical Subject.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LN and YoL conceptualized and refined the study
design. YuL, LZ and JH collected the literature data. JX, XG, JQ
and PC performed the experiments. YuL evaluated and selected the
data. YuL and LZ drafted the manuscript. YuL edited the manuscript.
JX and XG designed the study. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent, and
the present study was approved by the Ethical Review Committee of
Peking University Shenzhen Hospital (Shenzhen, China) and complied
with the Declaration of Helsinki.
Consent for publication
All patients provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gandellini P, Doldi V and Zaffaroni N:
microRNAs as players and signals in the metastatic cascade:
Implications for the development of novel anti-metastatic
therapies. Semin Cancer Biol. 44:132–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Trevisani F, Ghidini M, Larcher A, Lampis
A, Lote H, Manunta P, Alibrandi MT, Zagato L, Citterio L,
Dell'Antonio G, et al: MicroRNA 193b-3p as a predictive biomarker
of chronic kidney disease in patients undergoing radical
nephrectomy for renal cell carcinoma. Br J Cancer. 115:1343–1350.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao L, Zhu J, Zhou H, Zhao Z, Zou Z, Liu
X, Lin X, Zhang X, Deng X, Wang R, et al: Identification of
cellular microRNA-136 as a dual regulator of RIG-I-mediated innate
immunity that antagonizes H5N1 IAV replication in A549 cells. Sci
Rep. 5:149912015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Teixeira AL, Dias F, Gomes M, Fernandes M
and Medeiros R: Circulating biomarkers in renal cell carcinoma: The
link between microRNAs and extracellular vesicles, where are we
now? J Kidney Cancer VHL. 1:84–98. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou J, Yun EJ, Chen W, Ding Y, Wu K, Wang
B, Ding C, Hernandez E, Santoyo J, Pong RC, et al: Targeting
3-phosphoinositide-dependent protein kinase 1 associated with
drug-resistant renal cell carcinoma using new oridonin analogs.
Cell Death Dis. 8:e27012017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haas NB, Manola J, Uzzo RG, Flaherty KT,
Wood CG, Kane C, Jewett M, Dutcher JP, Atkins MB, Pins M, et al:
Adjuvant sunitinib or sorafenib for high-risk, non-metastatic
renal-cell carcinoma (ECOG-ACRIN E2805): A double-blind,
placebo-controlled, randomised, phase 3 trial. Lancet.
387:2008–2016. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Visone R and Croce CM: MiRNAs and cancer.
Am J Pathol. 174:1131–1138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Voinnet O: Origin, biogenesis, and
activity of plant microRNAs. Cell. 136:669–687. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mikhaylova O, Stratton Y, Hall D, Kellner
E, Ehmer B, Drew AF, Gallo CA, Plas DR, Biesiada J, Meller J, et
al: VHL-regulated MiR-204 suppresses tumor growth through
inhibition of LC3B-mediated autophagy in renal clear cell
carcinoma. Cancer Cell. 21:532–546. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu H, Brannon AR, Reddy AR, Alexe G,
Seiler MW, Arreola A, Oza JH, Yao M, Juan D, Liou LS, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: With
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aguiari G: MicroRNAs in clear cell renal
cell carcinoma: Biological functions and applications. J Kidney
Cancer VHL. 2:140–152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Motzer RJ, Figlin RA, Martini JF,
Hariharan S, Agarwal N, Li CX, Williams JA and Hutson TE: Germline
genetic biomarkers of sunitinib efficacy in advanced renal cell
carcinoma: Results from the RENAL EFFECT trial. Clin Genitourin
Cancer. 15:526–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu F, Chen N, Xiao R, Wang W and Pan Z:
miR-144-3p serves as a tumor suppressor for renal cell carcinoma
and inhibits its invasion and metastasis by targeting MAP3K8.
Biochem Biophys Res Commun. 480:87–93. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen G, Shen ZL, Wang L, Lv CY, Huang XE
and Zhou RP: Hsa-miR-181a-5p expression and effects on cell
proliferation in gastric cancer. Asian Pac J Cancer Prev.
14:3871–3875. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu S, Gu Y, Huang Y, Wong TC, Ding H, Liu
T, Zhang Y and Zhang X: Novel biomarkers for non-functioning
invasive pituitary adenomas were identified by using analysis of
micrornas expression profile. Biochem Genet. 55:253–267. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Korhan P, Erdal E and Atabey N:
MiR-181a-5p is downregulated in hepatocellular carcinoma and
suppresses motility, invasion and branching-morphogenesis by
directly targeting c-Met. Biochem Biophys Res Commun.
450:1304–1312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eichmüller SB, Osen W, Mandelboim O and
Seliger B: Immune modulatory micrornas involved in tumor attack and
tumor immune escape. J Natl Cancer Inst. 109:2017. View Article : Google Scholar
|
|
22
|
Han P, Li JW, Zhang BM, Lv JC, Li YM, Gu
XY, Yu ZW, Jia YH, Bai XF, Li L, et al: The lncRNA CRNDE promotes
colorectal cancer cell proliferation and chemoresistance via
miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Kuscu C, Banach A, Zhang Q,
Pulkoski-Gross A, Kim D, Liu J, Roth E, Li E, Shroyer KR, et al:
miR-181a-5p inhibits cancer cell migration and angiogenesis via
downregulation of matrix metalloproteinase-14. Cancer Res.
75:2674–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma Z, Qiu X, Wang D, Li Y, Zhang B, Yuan
T, Wei J, Zhao B, Zhao X, Lou J, et al: MiR-181a-5p inhibits cell
proliferation and migration by targeting Kras in non-small cell
lung cancer A549 cells. Acta Biochim Biophys Sin (Shanghai).
47:630–638. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mi Y, Zhang D, Jiang W, Weng J, Zhou C,
Huang K, Tang H, Yu Y, Liu X, Cui W, et al: miR-181a-5p promotes
the progression of gastric cancer via RASSF6-mediated MAPK
signalling activation. Cancer Lett. 389:11–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu K, Xie F, Gao A, Zhang R, Zhang L,
Xiao Z, Hu Q, Huang W, Huang Q, Lin B, et al: SOX2 regulates
multiple malignant processes of breast cancer development through
the SOX2/miR-181a-5p, miR-30e-5p/TUSC3 axis. Mol Cancer. 16:622017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Petrillo M, Zannoni GF, Beltrame L,
Martinelli E, DiFeo A, Paracchini L, Craparotta I, Mannarino L,
Vizzielli G, Scambia G, et al: Identification of high-grade serous
ovarian cancer miRNA species associated with survival and drug
response in patients receiving neo-adjuvant chemotherapy: A
retrospective longitudinal analysis using matched tumor biopsies.
Ann Oncol. 27:625–634. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boguslawska J, Sokol E, Rybicka B, Czubaty
A, Rodzik K and Piekielko-Witkowska A: microRNAs target SRSF7
splicing factor to modulate the expression of osteopontin splice
variants in renal cancer cells. Gene. 595:142–149. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brodaczewska KK, Szczylik C, Fiedorowicz
M, Porta C and Czarnecka AM: Choosing the right cell line for renal
cell cancer research. Mol Cancer. 15:832016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zaravinos A, Pieri M, Mourmouras N,
Anastasiadou N, Zouvani I, Delakas D and Deltas C: Altered
metabolic pathways in clear cell renal cell carcinoma: A
meta-analysis and validation study focused on the deregulated genes
and their associated networks. Oncoscience. 1:117–131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de Araújo Júnior RF, Oliveira Leitão AL,
de Melo Silveira RF, de Oliveira Rocha HA, de França Cavalcanti P
and de Araújo AA: Telmisartan induces apoptosis and regulates Bcl-2
in human renal cancer cells. Exp Biol Med (Maywood). 240:34–44.
2015. View Article : Google Scholar : PubMed/NCBI
|