Introduction
Today we are witnessing a notable increase of
bacterial resistance to a wide range of antibiotics, reported
worldwide. This has stimulated intensive efforts to search for new
antibiotics, as well as for valued antibacterial agents, that can
be utilized to treat infectious diseases (1,2).
Severe infections caused by bacteria that have become resistant to
regularly used antibiotics have become a major global healthcare
problem in the twenty-first century (3–5).
Antibiotic resistance, connected with Iatrogenesis, and an
increasing number of hospital-acquired infections, mainly in
critically ill and immunosuppressed patients, has now become
established in the community, causing severe infections that are
difficult to diagnose and treat (6). Bacteria have developed resistance to
most classes of antibiotics that have been discovered so far
(7). Several genes, many of which
can be transferred between bacteria, encode antibiotic resistance.
New resistance mechanisms are continually being described, and new
genes and vectors of transmission are identified on a regular
basis.
The molecular mechanisms by which bacteria have
become resistant to antibiotics are varied and complicated
(8,9). The most common type of bacterial
resistance is acquired and transmitted via horizontal gene transfer
(the antibiotic resistance genes are loaded on plasmids, which can
act as vectors that transfer these genes to other members of a
bacterial species or genus) (10).
New mechanisms of resistance have affected several classes of
antibiotics, leading to the aberrance of multidrug-resistant
bacterial strains, some known as superbugs (7). The overuse and/or misuse of
antimicrobial agents in patient clinics, in hospitalized patients,
and in the food industry are the principal factors leading to
antibiotic resistance (7). In
recent years, the number of new antibiotics licensed for human use
in different parts of the world has decreased. This development of
drug resistance to frequently used antibiotics by human pathogens
has driven the search for new antimicrobial chemicals,
chemotherapeutic agents, and agrochemicals that may combine higher
antimicrobial efficacy with lower toxicity, and minimize a negative
impact on the environment.
For ages, various cultures around the world have
used medicinal plants to treat or cure all sorts of diseases. The
natural products (NPs) of plants, mostly responsible for plant
pigmentation and flavor, are produced as secondary metabolites and
serve as defense mechanisms against bacteria, insects, and
herbivores. NPs have been adjusted to interact with biological
systems via a long natural selection process (11,12).
Consequently, they have long been a basis of therapeutics (13), and most of today's marketed drugs
are natural-based products or their derivatives (14). This supports the claim that
natural-based products are essentially better accepted by the body
than synthetic chemicals and have a better chance to be successful
drugs. (15) During the 1980s and
1990s, following the introduction of combinatorial chemistry and
high throughput synthesis, nature became a less important source of
drug candidates in drug discovery projects. However, even though
drug research global expenditures have more than doubled since
1991, the number of new drug entities that are approved annually by
the Food and Drug Administration in the U.S. (FDA) is dropping off;
in 2016 only 23 therapeutic new chemical entities were approved,
the fewest in almost last five decades and below statistical
expectations. (16) To remedy this
situation, the main players in the field of drug discovery and
development (the pharmaceutical industry and academic researchers)
have returned to searching for new drugs in Nature's pantry
(17,18).
Since the discovery and development of a new drug is
a long and costly process, we use computer methodologies to
facilitate the identification of new lead compounds and to optimize
drugs in clinical use (19,20).
Structural-based (21–23) and ligand-based (19,24–27)
computerized methods are used increasingly for the construction of
models that can predict the bioactivity of molecules and for the
in silico screening of chemical databases. For modeling
process, it is necessary to have sets of active and inactive
chemicals and an optimization technique. We assume that active
ligands have common features that are not easily detectable if only
a small number of active ligands are used (28). For this reason, usage a larger
number of active and inactive ligands in the modeling process
ensure that more significant and robust conclusions can be obtained
regarding the properties of these ligands. As well, it is worth
noting that including compounds in the sample of inactive chemicals
that possess properties similar to those of the compounds in the
screened chemical database increases the applicability of the
prediction model to virtual screening. Since a large number of
physicochemical properties should be considered during the modeling
process, we need extraordinary optimization techniques that are
capable of overcoming the limitations of the combinatorial nature
of the molecular bioactivity-indexing problem. During the last
decade, we developed a new optimization algorithm, termed
iterative stochastic elimination (ISE), that is able to scan
multi-dimensional space and detect the best solutions (the global
minimum and the best set of local minima) (29–31).
We have applied this novel algorithm to several ligand-based
problems (28,32). In this research, we used the ISE
algorithm to build the filters, and the MBI equation to construct
the model for indexing natural products for their potential
antibacterial activity. Analysis of the filters enabled us to map
physicochemical properties/descriptors that might contribute
significantly to antibacterial activity.
Materials and methods
We used a set of 628 anti-bacterial drugs (collected
from the Comprehensive Medicinal Chemistry Database and the
literature) to represent the active domain for modeling and
bioactivity-indexing purposes. The list of antibacterial drugs
(documented in SMILES format and/or by their common names) could be
supplied upon request from the corresponding author. Another set,
composed of 2,892 NPs, was selected to represent the inactive
domain. The database of NPs was prepared by collecting
phytochemicals isolated from more than 800 diverse plants, spread
worldwide, that can be obtained from AnalytiCon Discovery GmbH
(Potsdam, Germany; www.ac-discovery.com). To construct an accurate
predictive model, it is necessary to use sets of molecules that
cover the space of the properties of the molecules in the screened
database. As well, we had to select, as the inactive set, molecules
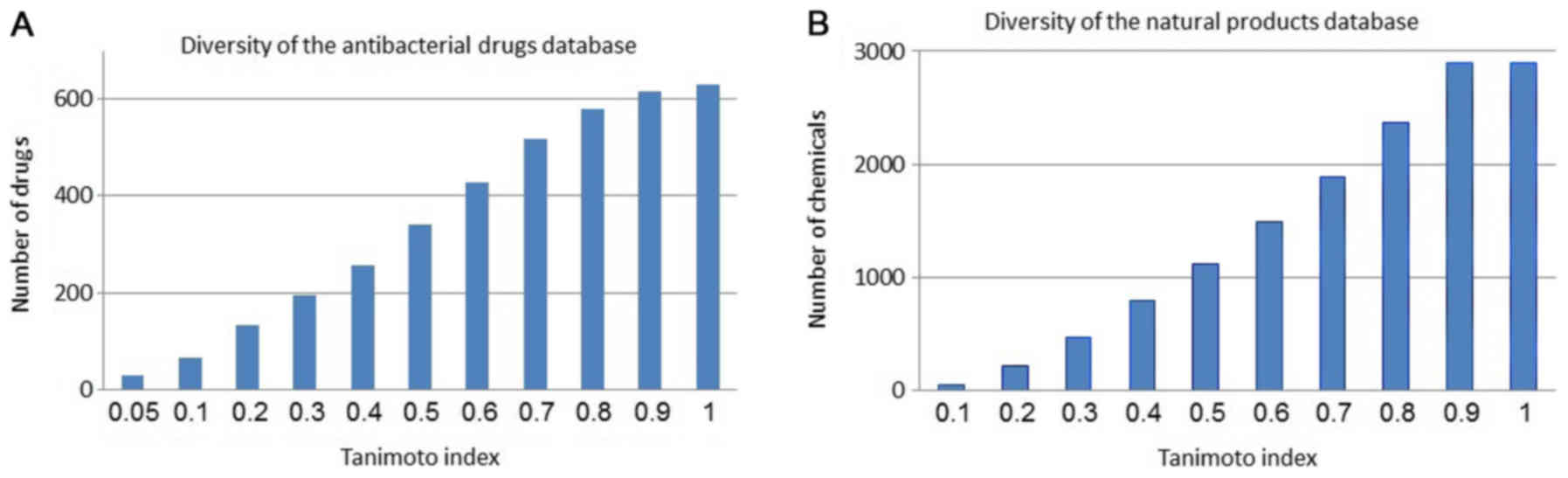
with the same ‘property space’ as the screened molecules. Fig. 1A and B show the diversity within
the antibacterial drugs and the natural products database,
respectively.
Categorizing the 628 antibacterial drugs based on
their own mechanism of action might enable us to construct
different models, depending on the category of active molecules.
However, we prefer to use the entire set of the 628 antibacterial
drugs as active set and not to categorize them based on their
mechanism of action in order to obtain more robust model (due to
utilizing big and diverse set of active ligands) and to be more
focused on the indication (antibacterial) and not on the biological
target. From our past experience, by using the ISE-based indexing
technique, we were successful in constructing discriminative
filters and in proposing highly predictive models when applied for
general properties such as drug likeness (28), antidiabetic (25), anti-inflammatory (33), anticancer (27).
MOE software, v2009.10 (www.chemcomp.com) was used to calculate the
physicochemical properties (termed descriptors) of all the
chemicals in the two databases. The calculated descriptors included
molecular weight, logP, H-bond donors/acceptors, solubility, total
charge and charge distribution, the types and number of atoms, and
so forth (www.chemcomp.com/journal/descr.htm). Both databases of
active/inactive ligands were divided into two-thirds for the
training set and one-third for the test set. An in-house random
picking module performed the split.
The cheminformatics version of the ISE algorithm
(28) was used to construct models
tailored to index phytochemicals for their potential antibacterial
activity. Through efficient searching of the multivariable space,
we constructed a large set of filters tailored to distinguish
between antibacterial and inactive ligands. Each filter is composed
of a certain sets of descriptors, and each is limited to an
assigned range. The process of filter selection and construction is
highly complex and requires the use of a highly efficient
optimization algorithm, since the descriptors generally interact
with each other, and changes in the range of one descriptor can
have an effect on the best range of another descriptor. In order to
arrive at the best set of filters, the optimization process ought
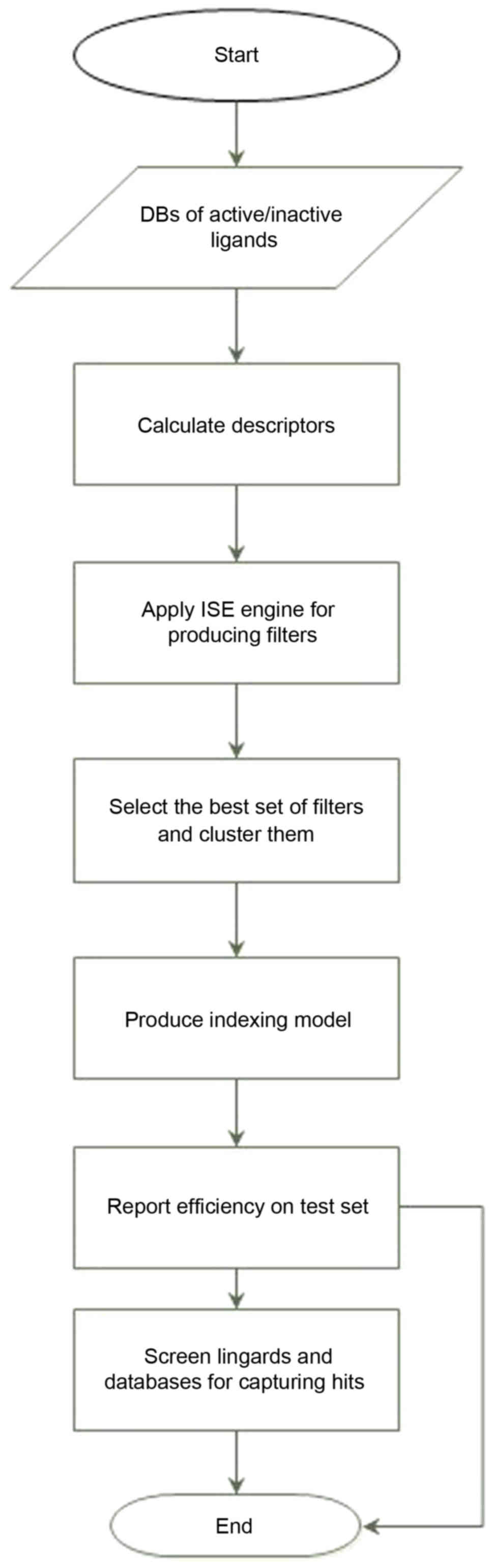
to consider all descriptors in the set simultaneously. Fig. 2 describes the main items in the
modeling process. For detailed descriptions of the ISE optimization
technique and its utility in choosing sets of descriptors and
optimizing their ranges, see our previously reported research
studies (32).
Results and Discussion
Structural similarity analysis was conducted to
assure that both sets of active/inactive chemicals were not biased
and displayed adequate diversity. As shown in Fig. 1, both sets of chemicals are
diverse. The 341 antibacterial drugs and 1,119 natural products had
a diversity of less than 0.5 in terms of the structural Tanimoto
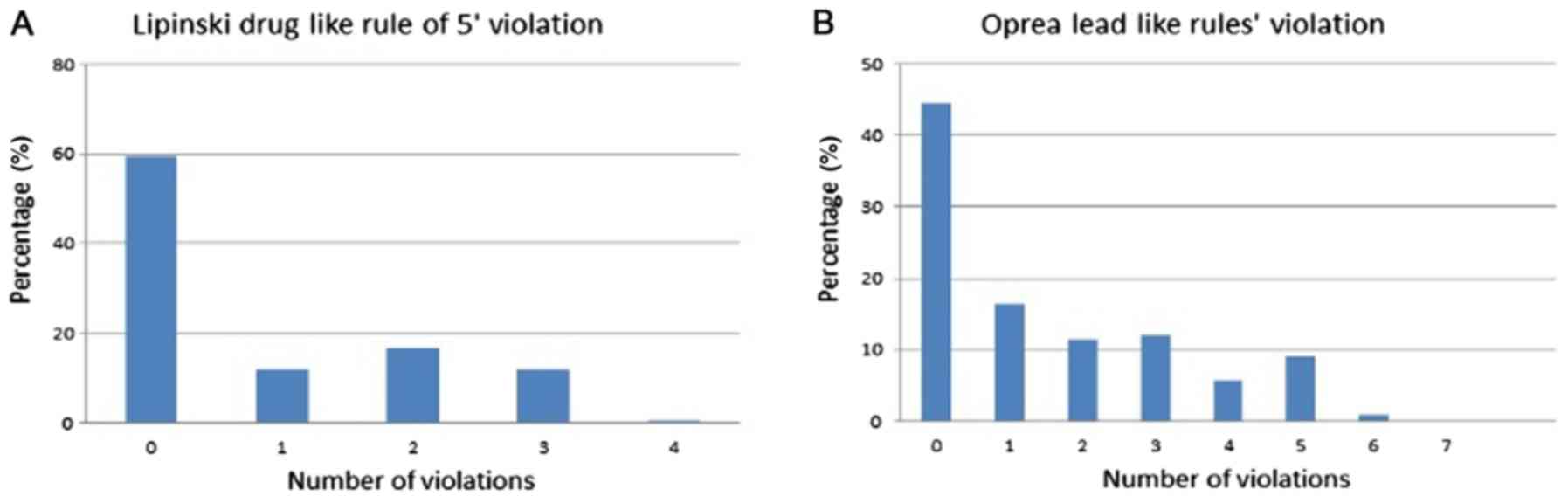
index. As well, analysis of the physicochemical properties noted
that 71.5% of the antibacterial drugs conformed to Lipinski's rule
of 5 (ROF), and 61% conformed to Oprea's rule for lead-likeness
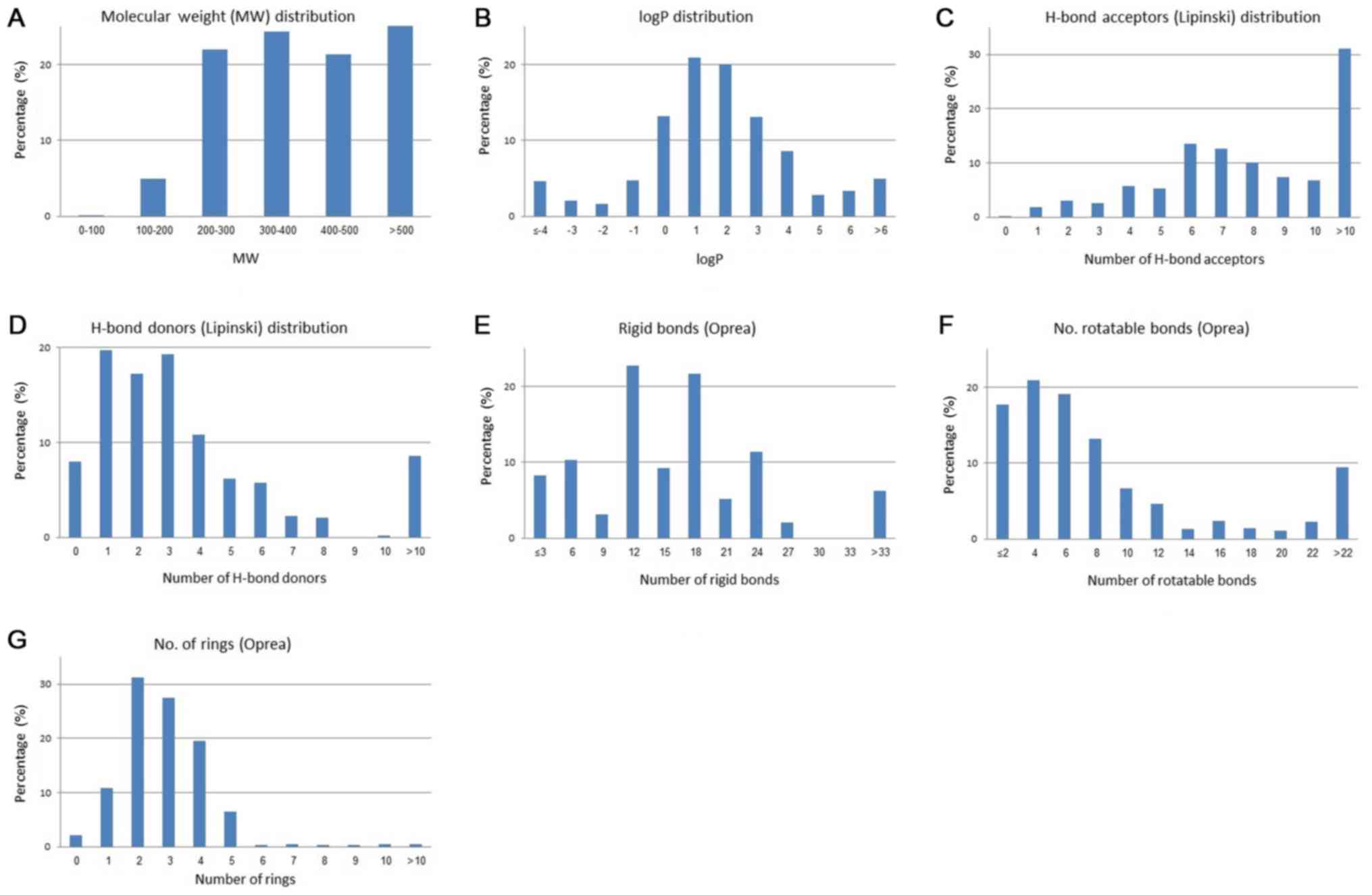
(34) (Fig. 3). Fig.
3 displays the distribution plots for the physicochemical
properties of the antibacterial drugs related to Lipinski's ROF and
Oprea's rule for lead-likeness. The median is around 396 for the
molecular weight; 1.2 for logP; 7–8 and 2–3 for the hydrogen bond
acceptors and hydrogen bond donors, respectively (Fig. 4).
The aforementioned filter-based indexing technique
was utilized to launch an in silico prediction model capable
of discovering novel antibacterial drug candidates. It was built
using a set of 628 antibacterial drugs to represent the active
domain, and a set of 2,892 natural products to represent the
inactive domain. It is worth noting that a few chemicals out of the
2,892 natural products might have had antibacterial activity, but
the effect of that on the quality of the prediction model was
anticipated to be negligible (28,32).
The optimization technique used to construct the filters was the
ISE algorithm. Thirty-six unique filters were produced; each was
composed of either different sets of four descriptors or different
ranges of the same set of descriptors. The best three filters are
described in Table I. Their
efficiencies in terms of MCC are relatively very high. Filter
number 1, shown in Table I, has a
MCC of 0.879, and nearly 94% of the antibacterial drugs (true
positives) were successfully identified using this
four-descriptor-based filter, while less than 6% of the natural
products database was ‘misclassified’ (passed the filter as
positives, but are yet unproven).
 | Table I.Detailed information regarding the
efficiencies and ranges of descriptors for the best 3 filters out
of the 36 used to construct the antibacterial indexing model. |
Table I.
Detailed information regarding the
efficiencies and ranges of descriptors for the best 3 filters out
of the 36 used to construct the antibacterial indexing model.
| A, Parameters for
filter efficiency |
|---|
|
|---|
| Variable | Filter 1 | Filter 2 | Filter 3 |
|---|
| MCC | 0.879 |
0.809 | 0.782 |
| TP (%) | 93.79 | 81.85 | 79.8 |
| TN (%) | 94.09 | 98.03 | 97.2 |
|
| B, Descriptors,
(range) |
|
| 1 | BCUT_SMR_3,
(0–3.33) | PEOE_PC+,
(0–29.795) | PEOE_PC+, 95
(0–29.795) |
| 2 | logS,
(−19.42–2.64) | a_nN, (2–32) | PEOE_VSA_NEG,
(0–1,045.34) |
| 3 | a_nN, (1–32) | Q_VSA_FNEG,
(0–0.808) | a_ICM,
(1.55–2.33) |
| 4 | GCUT_PEOE_3,
(0–3.754) | SMR_SVA0,
(0–658.98) | PEOE_VSA_FNEG, (0
−0.834) |
The content of the thirty-six filters was
investigated; Table II shows the
number of appearances of the most dominant descriptors. The third
column shows how many times each descriptor actually appeared in
the set of best filters, vs. random distribution. Fig. 5 was built using WORDLE module; it
shows the frequency of dominant descriptors in a graphical way. The
most dominant descriptors can be valued more highly than the less
dominant descriptors for differentiating between antibacterial
chemicals and inactive ones.
 | Table II.Number of appearances of the most
dominant descriptors within the set of 36 filters that were
utilized in the construction of the antibacterial model. |
Table II.
Number of appearances of the most
dominant descriptors within the set of 36 filters that were
utilized in the construction of the antibacterial model.
| Descriptor
name | Redundancy | Redundant more
times than random |
|---|
| GCUT_SLOGP_0 | 22 | 36.7 |
| a_ICM | 11 | 18.4 |
| PEOE_VSA+4 | 7 | 11.7 |
| SMR_VSA1 | 7 | 11.7 |
| logS | 6 | 10.0 |
| Nmol | 6 | 10.0 |
| lip_druglike | 5 | 8.3 |
| chi1_C | 4 | 6.7 |
| GCUT_PEOE_0 | 4 | 6.7 |
| opr-leadlike | 4 | 6.7 |
| Q_VSA_FPOS | 4 | 6.7 |
| SMR_VSA3 | 4 | 6.7 |
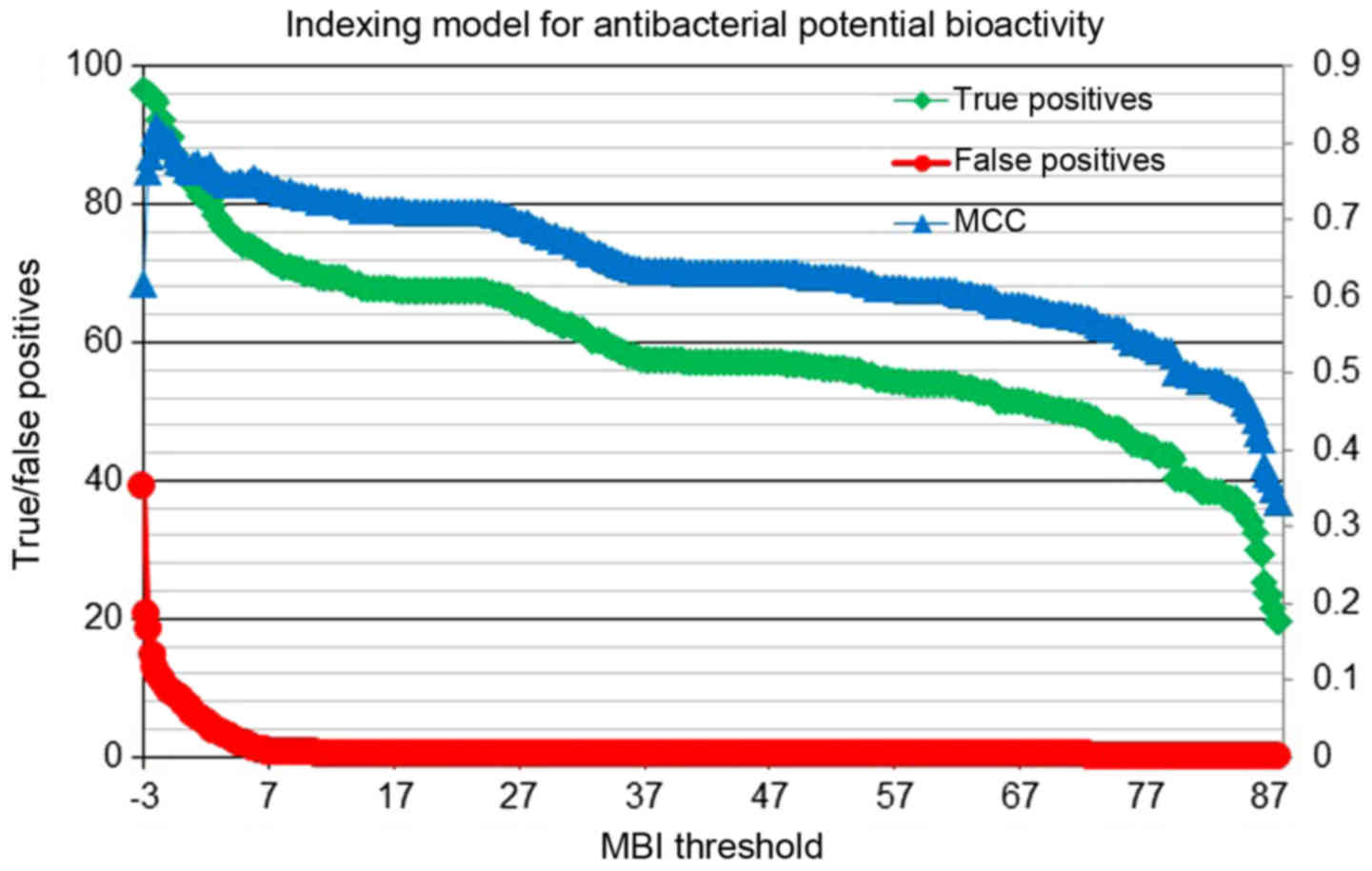
Fig. 6 describes
the antibacterial activity-indexing model, showing changes in
percentage of true positives, true negatives and Matthews'
correlation coefficient (MCC) connected with discriminative
efficiencies along with the index values. The percentages of
true/false positives (left x-axis) and the MCCs (right
y-axis) are plotted against the molecular bioactivity index
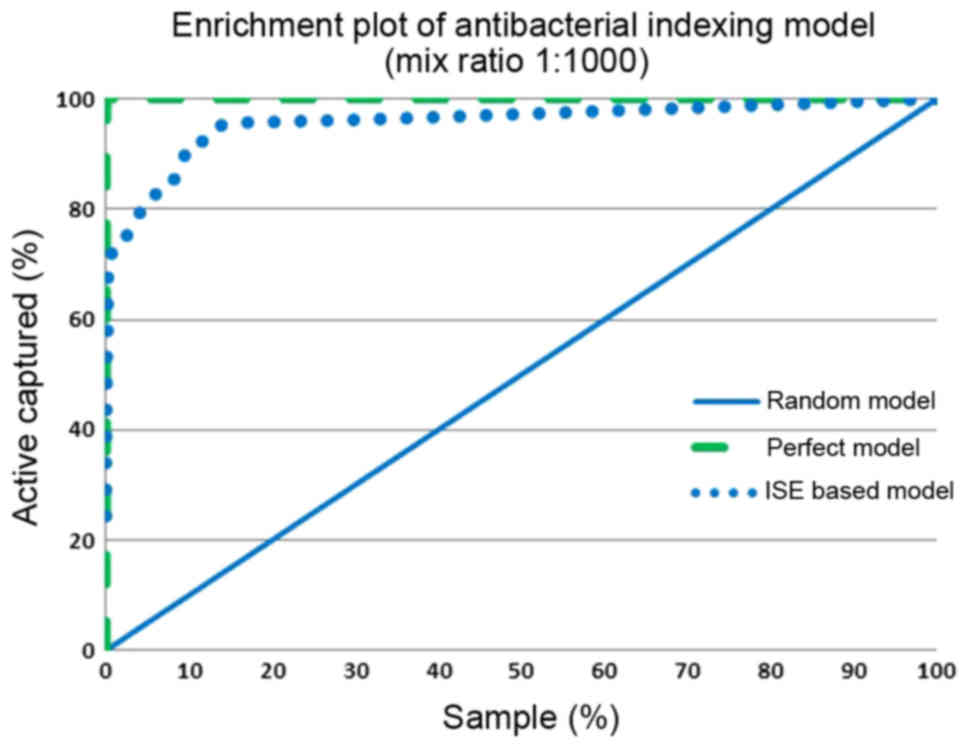
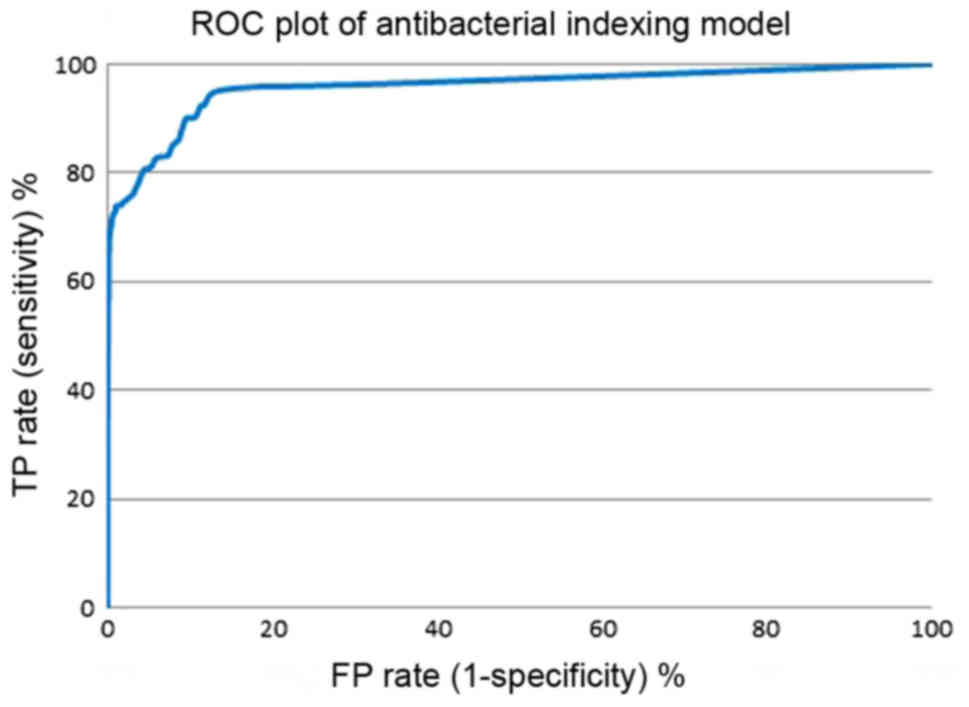
(MBI threshold, x-axis). Figs.
7 and 8 show the enrichment
plot and the receiver-operating characteristic (ROC) plot of our
antibacterial activity-indexing model. The enrichment plot
(Fig. 7) shows how many times
antibacterial drug candidates can be detected if natural products
are ranked according to the ISE-based prediction model rather than
random selection.
If we pick molecules with an MBI above 10.0, our
predictive model and the perfect model overlay to large extent.
Therefore, it seems that the indexing model is highly accurate and
bears high prioritization power. With the use of this antibacterial
activity-indexing model and a mixed set of active and inactive
chemicals (with a ratio of 1:1,000), 72% of the antibacterial drugs
were detected in the top 1% of the screened molecules, compared to
100% in the perfect model and 1% in the random model, yielding an
enrichment factor of 72. The ISE-based model and the perfect model
overlay to some extent in the range of MBI above 7.0. The area
under the curve (AUC) attained was 0.957, which indicates that the
model is excellent and highly efficient in distinguishing
antibacterial drugs from inactive natural products. The natural
products database, composed of 2,892 phytochemicals, was virtually
screened using this filter-based activity-indexing model. The MBI
scores, as shown in Fig. 6, range
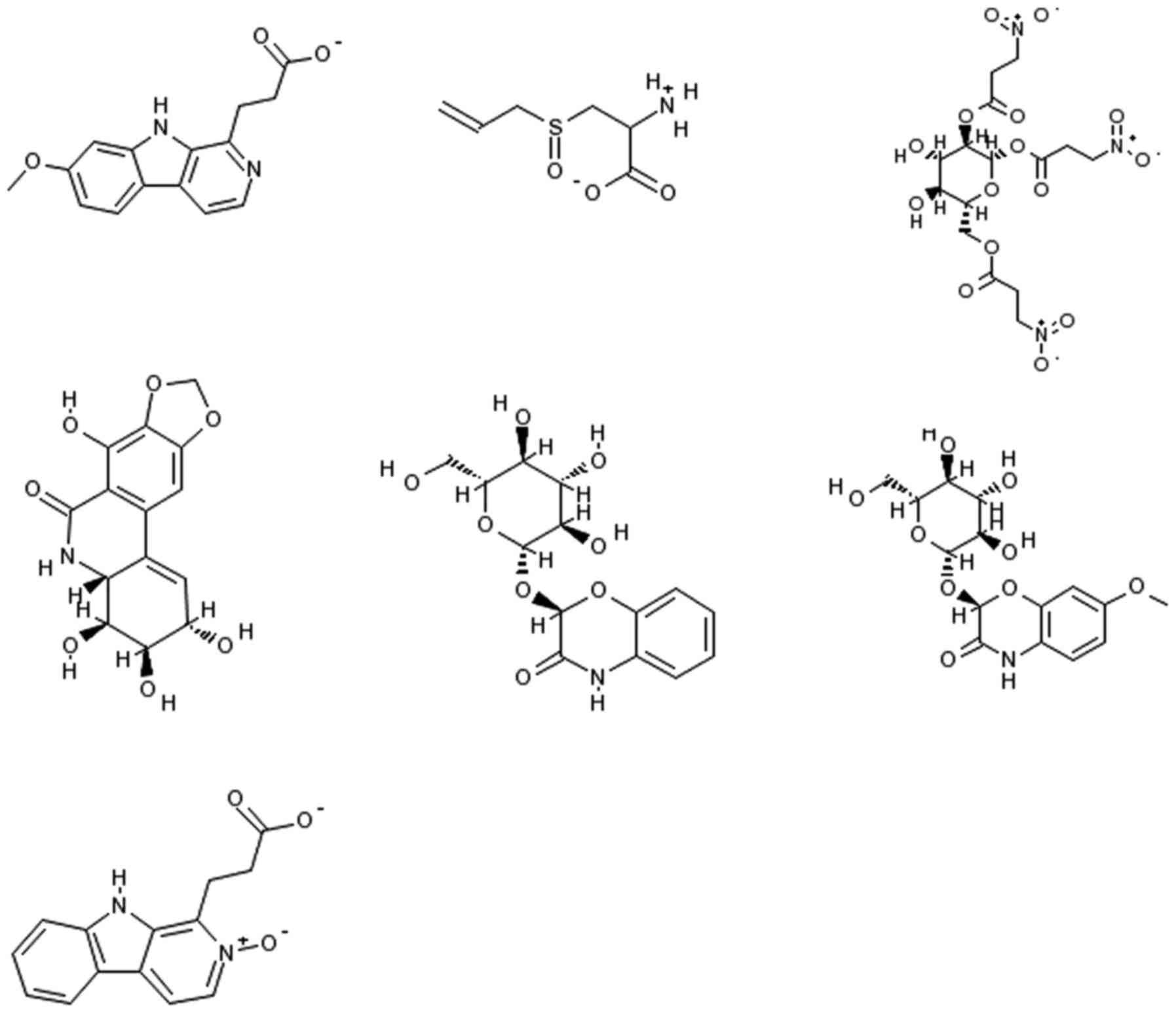
from-3.0 (the lowest score) to 87.0 (the highest score). Figs. 9 and 10 disclose 10 natural products that
scored high as potential antibacterial drug candidates (with MBI
scores above 10.0). When choosing an MBI threshold score of 10.0,
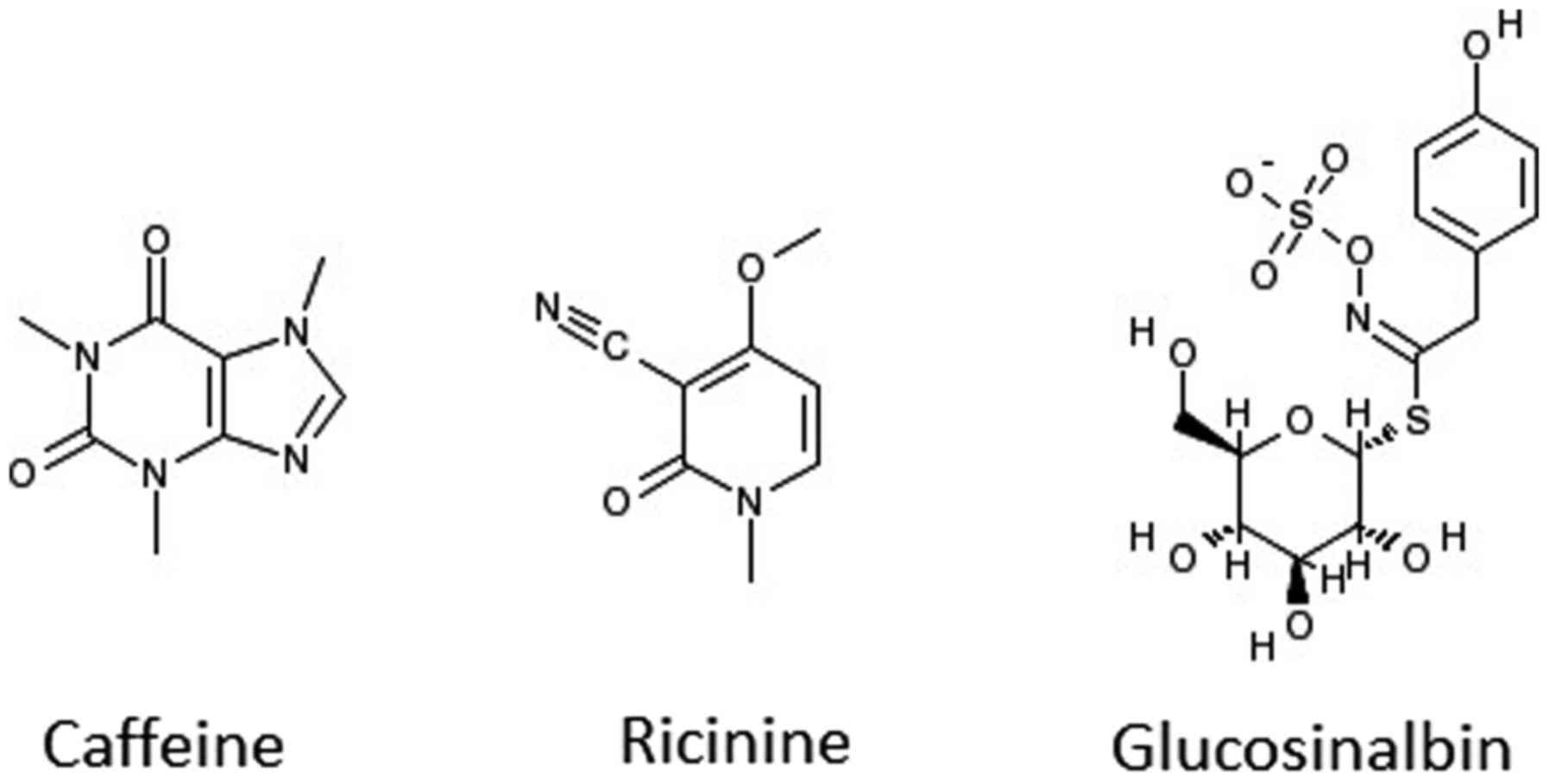
the ratio of TP: FP is 168:1. A search on PubMed revealed that two
of the highly indexed phytochemicals (caffeine and ricinine) have
already been tested experimentally and confirmed as antibacterial
agents (35). Caffeine has been
reported to exert physiological effects on various organisms at µM
concentrations and to act as an antimicrobial agent (36,37).
Ricinine had high nematocidal activity (38) and notable activity against ants
(39). The other eight
phytochemicals await evaluation in the wet lab to ascertain their
potential antibacterial activity. It is worth mentioning that one
of the volatile isolates from Cardaria draba (L.) Desv. that
contained glucosinalbin has shown a wide range of growth inhibition
activity against both Gram-positive and Gram-negative bacteria
(40). It is worth testing to
establish whether the phytochemical glucosinalbin is one of the
main contributors to the extract's antibacterial activity. The
chemical structures of caffeine, ricinine, and glucosinalbin are
shown in Fig. 9. Fig. 10 displays the chemical structures
of the other seven phytochemicals that scored high as potential
antibacterial drug candidates with our model and await validation
in the wet lab.
The current study provides vital insights into the
discriminative properties of antibacterial natural products and
this information might be supportive to medicinal chemists in their
search for novel natural antibacterial products. As well, we think
the one of the aims behind publication of such theoretical work is
to recruit experimental groups that are not in contact with us to
test the disclosed molecules/natural products. This paper should be
cited following their antibacterial activity evaluation.
It is worth to note that application of
structural-based approaches in drug discovery fails to deliver
better results than application of ligand-based approaches due to
low efficient scoring functions and high number of false positive.
The number of false positives is very high, mainly in virtual
high-throughput screening, and we are waiting for development of
novel scoring functions that capable to reduce the number of false
positives. From previous experience in other projects, we have seen
that structural-based approaches, which uses docking, could be
helpful for re-ranking highly scored ligands that are output from
ligand-based approaches (26).
Successful story was published last year describing the utility of
the ISE algorithm and physicochemical properties in discovery of
novel ligands (19). As well,
these days we have submitted a manuscript under the title
‘Accelerating Drug Discovery Process by the Iterative Stochastic
Elimination Algorithm: Discovering Novel Selective Agonists of
PPAR-δ’. In this manuscript, we describe the discovery of novel
molecular hits and leads for PPAR-δ by applying our combinatorial
optimizing algorithm, ISE.
Using the ISE algorithm, we identified 36 unique
filters that enabled us to construct a highly discriminative and
robust model tailored to index natural products for their
antibacterial bioactivity. For modeling purposes, we utilized a set
of 628 antibacterial drugs, representing the active domain, and
2,892 natural products, representing the inactive domain. The area
attained under the curve (AUC) was 0.957, indicating a highly
discriminative and robust model. In this paper, we disclose ten
natural products that scored high as antibacterial drug candidates
with the proposed indexing model. A search on PubMed revealed that
two phytochemicals (caffeine and ricinine) out of the ten highly
indexed molecules have already been tested experimentally and
confirmed as antibacterial agents. The other eight phytochemicals
await experimental evaluation. Due to its high efficiency and
rapidity, this model might be used to virtually screen large
chemical databases and to index natural products for potential
antibacterial bioactivity.
Acknowledgements
Not applicable.
Funding
The present study was supported by unrestricted
grants from Al-Qasemi Academic College (grant no. 898002) and The
Ministry of Science, Space and Technology (Israel), which was
awarded through The Institute of Applied Research-Galilee Society
(Annual Budget of 2017).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AR was involved in study conception and curation,
formal analysis, funding acquisition, the methodology, study
supervision, and reviewing and editing the manuscript. MM
contributed to the formal analysis, performed the investigations,
validated the study and assisted with the original draft of the
manuscript. MR performed the investigation and validation. AA and
ZA contributed to the formal analysis and completed the original
draft of the manuscript. MM, MR, AA, ZA and AR gave final approval
for the version to be published.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interest.
Glossary
Abbreviations
Abbreviations:
|
ISE
|
iterative stochastic elimination
|
|
MCC
|
Matthews correlation coefficient
|
|
EF
|
enrichment factor
|
|
vHTS
|
virtual high-throughput screening
|
|
TP
|
true positive
|
|
FP
|
false positive
|
|
MBI
|
molecular bioactivity index
|
References
|
1
|
Alanis AJ: Resistance to antibiotics: Are
we in the post-antibiotic era? Arch Med Res. 36:697–705. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Klein E, Smith DL and Laxminarayan R:
Hospitalizations and deaths caused by methicillin-resistant
Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis.
13:1840–1846. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Appelbaum PC: The emergence of
vancomycin-intermediate and vancomycin-resistant Staphylococcus
aureus. Clin Microbiol Infect. 12 Suppl 1:S16–S23. 2006. View Article : Google Scholar
|
|
4
|
Núñez-Núñez M, Navarro MD, Gkolia P,
Rajendran Babu N, Del Toro MD, Voss A, Sharland M, Sifakis F,
Tacconelli E, Rodríguez-Baño J, et al: Surveillance Systems from
Public Health Institutions and Scientific Societies for
Antimicrobial Resistance and Healthcare-Associated Infections in
Europe (SUSPIRE): Protocol for a systematic review. BMJ Open.
7:e0145382017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kalesinskas P, Kačergius T, Ambrozaitis A,
Pečiulienė V and Ericson D: Reducing dental plaque formation and
caries development. A review of current methods and implications
for novel pharmaceuticals. Stomatologija. 16:44–52. 2014.PubMed/NCBI
|
|
6
|
Pitiriga V, Dimitroulia E, Saroglou G and
Tsakris A: The challenge of curbing aminoglycoside resistance: Can
antimicrobial stewardship programs play a critical role? Expert Rev
Anti Infect Ther. 15:947–954. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cantón R, Horcajada JP, Oliver A,
Garbajosa PR and Vila J: Inappropriate use of antibiotics in
hospitals: The complex relationship between antibiotic use and
antimicrobial resistance. Enferm Infecc Microbiol Clin. 31 Suppl
4:S3–S11. 2013. View Article : Google Scholar
|
|
8
|
Abachi S, Lee S and Rupasinghe HP:
Molecular mechanisms of inhibition of streptococcus species by
phytochemicals. Molecules. 21:pii: E215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mestrovic T and Ljubin-Sternak S:
Molecular mechanisms of Chlamydia trachomatis resistance to
antimicrobial drugs. Front Biosci (Landmark Ed). 23:656–670. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vaidya VK: Horizontal transfer of
antimicrobial resistance by extended-spectrum β lactamase-producing
enterobacteriaceae. J Lab Physicians. 3:37–42. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wijesekera RO: Is there an industrial
future for phytopharmaceutical drugs? An outline of UNIDO
programmes in the sector. J Ethnopharmacol. 32:217–224. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kacergius T, Abu-Lafi S, Kirkliauskiene A,
Gabe V, Adawi A, Rayan M, Qutob M, Stukas R, Utkus A, Zeidan M and
Rayan A: Inhibitory capacity of Rhus coriaria L. extract and its
major component methyl gallate on Streptococcus mutans biofilm
formation by optical profilometry: Potential applications for oral
health. Mol Med Rep. 16:949–956. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cragg GM and Newman DJ: Medicinals for the
millennia: The historical record. Ann N Y Acad Sci. 953:3–25. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Andrade-Carrera B, Clares B, Noé V,
Mallandrich M, Calpena AC, García ML and Garduño-Ramírez ML:
Cytotoxic evaluation of (2S)-5,7-dihydroxy-6-prenylflavanone
derivatives loaded PLGA nanoparticles against MiaPaCa-2 cells.
Molecules. 22:pii: E1553. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zaid H, Raiyn J, Nasser A, Saad B and
Rayan A: Physicochemical properties of natural based products
versus synthetic chemicals. Open Nutraceut J. 3:194–202. 2010.
View Article : Google Scholar
|
|
16
|
Griesenauer RH and Kinch MS: 2016 in
review: FDA approvals of new molecular entities. Drug Discov Today.
22:1593–1597. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Harvey AL and Cree IA: High-throughput
screening of natural products for cancer therapy. Planta Med.
76:1080–1086. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frank A, Abu-Lafi S, Adawi A, Schwed JS,
Stark H and Rayan A: From medicinal plant extracts to defined
chemical compounds targeting the histamine H4 receptor: Curcuma
longa in the treatment of inflammation. Inflamm Res. 66:923–929.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zatsepin M, Mattes A, Rupp S, Finkelmeier
D, Basu A, Burger-Kentischer A and Goldblum A: Computational
discovery and experimental confirmation of TLR9 receptor antagonist
leads. J Chem Inf Model. 56:1835–1846. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zaid H, Raiyn J, Osman M, Falah M, Srouji
S and Rayan A: In silico modeling techniques for predicting the
tertiary structure of human H4 receptor. Front Biosci (Landmark
Ed). 21:597–619. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pappalardo M, Rayan M, Abu-Lafi S,
Leonardi ME, Milardi D, Guccione S and Rayan A: Homology-based
modeling of rhodopsin-like family members in the inactive State:
Structural analysis and deduction of tips for modeling and
optimization. Mol Inform. 36:2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shahaf N, Pappalardo M, Basile L, Guccione
S and Rayan A: How to choose the suitable template for homology
modelling of GPCRs: 5-HT7 receptor as a test case. Mol Inform.
35:414–423. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Michaeli A and Rayan A: Modeling ensembles
of loop conformations by iterative stochastic elimination. Lett
Drug Design Discov. 13:1–6. 2016. View Article : Google Scholar
|
|
24
|
Fradera X and Babaoglu K: Overview of
methods and strategies for conducting virtual small molecule
screening. Curr Protoc Chem Biol. 9:196–212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zeidan M, Rayan M, Zeidan N, Falah M and
Rayan A: Indexing natural products for their potential
anti-diabetic activity: Filtering and mapping discriminative
physicochemical properties. Molecules. 22:pii: E1563. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pappalardo M, Shachaf N, Basile L, Milardi
D, Zeidan M, Raiyn J, Guccione S and Rayan A: Sequential
application of ligand and structure based modeling approaches to
index chemicals for their hH4R antagonism. PLoS One. 9:e1093402014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rayan A, Raiyn J and Falah M: Nature is
the best source of anticancer drugs: Indexing natural products for
their anticancer bioactivity. PLoS One. 12:e01879252017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rayan A, Marcus D and Goldblum A:
Predicting oral druglikeness by iterative stochastic elimination. J
Chem Inf Model. 50:437–445. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Glick M, Rayan A and Goldblum A: A
stochastic algorithm for global optimization and for best
populations: A test case of side chains in proteins. Proc Natl Acad
Sci USA. 99:703–708. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rayan A, Noy E, Chema D, Levitzki A and
Goldblum A: Stochastic algorithm for kinase homology model
construction. Curr Med Chem. 11:675–692. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rayan A, Senderowitz H and Goldblum A:
Exploring the conformational space of cyclic peptides by a
stochastic search method. J Mol Graph Model. 22:319–333. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rayan A, Falah M, Raiyn J, Da'adoosh B,
Kadan S, Zaid H and Goldblum A: Indexing molecules for their hERG
liability. Eur J Med Chem. 65:304–314. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aswad M, Rayan M, Abu-Lafi S, Falah M,
Raiyn J, Abdallah Z and Rayan A: Nature is the best source of
anti-inflammatory drugs: Indexing natural products for their
anti-inflammatory bioactivity. Inflamm Res. 67:67–75. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hann MM and Oprea TI: Pursuing the
leadlikeness concept in pharmaceutical research. Curr Opin Chem
Biol. 8:255–263. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dash SS and Gummadi SN: Inhibitory effect
of caffeine on growth of various bacterial strains. Res J
Microbiol. 3:457–465. 2008. View Article : Google Scholar
|
|
36
|
Sandlie I, Solberg K and Kleppe K: The
effect of caffeine on cell growth and metabolism of thymidine in
Escherichia coli. Mutat Res. 73:29–41. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Whitney AK and Weir TL: Interaction of
caffeine with the SOS response pathway in Escherichia coli. Gut
Pathog. 7:212015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao QY, Hu FL, Zhu HH, Liu MQ, Li HX and
Hu F: Control effects of Ricinus communis extracts on Meloidogyne
incognita. Ying Yong Sheng Tai Xue Bao. 22:3033–3038. 2011.(In
Chinese). PubMed/NCBI
|
|
39
|
Bigi MF, Torkomian VL, de Groote ST,
Hebling MJ, Bueno OC, Pagnocca FC, Fernandes JB, Vieira PC and da
Silva MF: Activity of Ricinus communis (Euphorbiaceae) and ricinine
against the leaf-cutting ant Atta sexdens rubropilosa (Hymenoptera:
Formicidae) and the symbiotic fungus Leucoagaricus gongylophorus.
Pest Manag Sci. 60:933–938. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Radonić A, Blažević I, Mastelić J, Zekić
M, Skočibušić M and Maravić A: Phytochemical analysis and
antimicrobial activity of Cardaria draba (L.) Desv. volatiles. Chem
Biodivers. 8:1170–1181. 2011. View Article : Google Scholar : PubMed/NCBI
|