Introduction
In Traditional Chinese Medicine, the
‘exterior-interior relationship between the lung and the large
intestine’ is a classical basic theory, which was first postulated
in the Inner Canon of Huangdi ancient Chinese medical text
(1,2). This theory serves as guidance for
treatment of certain pulmonary diseases combined with disorders of
the large intestine (3). In
addition, an increasing amount of clinical evidence demonstrated
that there is a relationship between the physiology and pathology
of the lung and large intestine (4,5).
However, at present, systemic reports regarding molecular
mechanisms underlying mutual interactions between the lung and the
large intestine are lacking.
Allergic asthma is a complex chronic airway
inflammatory reaction mediated by mastocytes, eosinophils and T
lymphocytes (6,7). The prevalence of asthma in
industrialized countries has been increasing and asthma is now the
most common chronic disease of children in the United States
(8). It is believed that an
additional 100 million people will be suffering with asthma by 2025
(9). Therefore, allergic asthma
has become a public health issue. Increasing amount of research
indicated that allergic asthma is closely associated with the
intestinal flora disorder and is considered a typical disease model
for investigating the mutual interactions between the lung and the
large intestine (1,10). Therefore, in the present study, an
animal model of allergic asthma complicated with intestinal flora
disorder was established in rats to elucidate the molecular
mechanism of mutual interactions between the lung and the large
intestine.
Materials and methods
Animals
A total of 30 male Sprague-Dawley rats (3–4 weeks
old, 200±20 g) were purchased from the Dashuo Laboratory Animal
Co., Ltd. (Chengdu, China; http://www.jianyang.ccoo.cn/post/zhaopin/minqi/index462587.html).
The animals were housed in a temperature and humidity controlled
room (temperature 22±2°C, atmosphere 40–60% CO2, and
10–12-h light/dark cycle) with food and water ad libitum.
All animal experimental protocols were approved by the Ethics
Committee for Laboratory Animal Experimentation of Chengdu
University of Traditional Chinese Medicine.
Chemicals and reagents
Cefoperazone was purchased from the North China
Pharmaceutical Co., Ltd. (Shijiazhuang, China). Culture media (the
selective culture media for Enterococcus, the selective
culture media for enteric Bacilli, the BS culture media for
Bifidobacterium, the selective culture media for
Lactobacillus and the SDA culture media for
Candida) for Enterococcus, enteric Bacilli,
Bifidobacterium, Lactobacillus and Candida were
purchased from the Qingdao Haibo Biotechnology Co., Ltd. (Qingdao,
China). Ovalbumin (OVA) was purchased from Sigma-Aldrich (Merck
KGaA, Darmstadt, Germany). Aluminum hydroxide was purchased from
Chengdu Chron Chemicals Co., Ltd., (Chengdu, China). Live combined
Bacillus subtilis and Enterococcus faecium granules
(BEG) were purchased from the Hanmi Pharmaceutical Co., Ltd.
(Beijing, China). Aminophylline (ANP) was purchased from the
Southwest Pharmaceutical Co., Ltd. (Chongqing, China). Candida
albicans was purchased from the Guangdong Huankai Microbial
Technology Co., Ltd. (Guangzhou, China; http://huankaiye.bioon.com.cn/). TRIzol reagent was
purchased from Invitrogen (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Rat ELISA kits for secretory(s) immunoglobulin (Ig) A
(cat. no. 201411), IgE (cat. no. R141126-117a) interleukin (IL)-4
(cat. no. R141126-002a) and interferon (IFN)-γ (cat. no.
R141126-101a) were purchased from the Neobioscience Technology Co.,
Ltd. (Shenzhen, China). HiScript 1st Strand cDNA Synthesis kit and
SYBR-Green Master Mix were purchased from the Vazyme Biotech Co.,
Ltd. (Nanjing, China). Hematoxylin and eosin (H&E) and
Wright-Giemsa kits were purchased from the Baso Biotech Co., Ltd.
(Taiwan, China). All primers used in the present study were
designed by Primer-Express version 3.0 (Thermo Fisher Scientific,
Inc.) and synthetized by Sangon Biotech Co., Ltd. (Shanghai, China;
Table I).
 | Table I.Primers used in this research. |
Table I.
Primers used in this research.
|
| Primer sequence
(5′→3′) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| TLR-2 |
5′-GTTGCGTTACATCTTGGA-3′ |
5′-GGAATACACAGTGCTCAG-3′ |
| TLR-4 |
5′-CAGCTCGTTTCTCACCCAGT-3′ |
5′-TGTATCGGTGGTCAGTGTGC-3′ |
| MyD88 |
5′-CGACGCCTTCATCTGCTA-3′ |
5′-GCCGATAGTCTGTCTGTTCT-3′ |
| TRAF6 |
5′-CAGTCCCCTGCACATTCAGT-3′ |
5′-CTGGGCCAACAGTCTCATGT-3′ |
| β-arrestin |
5′-GGGCATTTGTACTGAGCTGT-3′ |
5′-TGCACCTTGAGGCATCTCTG-3′ |
| β-actin |
5′-AGGGAAATCGTGCGTGACAT-3′ |
5′-GAACCGCTCATTGCCGATAG-3′ |
Experimental protocols and animal
model preparation
A total of 30 rats were randomly divided into 3
groups (n=10): i) Normal group; ii) model group; and iii) positive
treatment group (treated with live combined BEG and ANP). With the
exception of the normal rats, all animals in the model and positive
groups were freely administered orally Cefoperazone aqueous
solution (0.5 g/l) for consecutive 5 days. Subsequently, rats of
the model and positive treatment groups were orally administered 50
µl Candida albicans [109 colony forming unit
(CFU)/ml] on the sixth day. OVA was used to induce allergic asthma
in rats according to the previous methods (11,12)
with minor modifications. Briefly, rats were immunized via
intraperitoneal (IP) injection of 1 ml OVA-aluminum hydroxide
mixture on days 0 and 7 (1 mg OVA and 200 mg aluminum hydroxide
were dissolved in 1 ml saline). Subsequently, allergic asthma in
rats was induced with 1% OVA-saline solution by aerosol inhalation
in a glass box (10×10×20 cm) from day 14 to day 21 (30 min/day).
Rats in the normal group were treated using the same protocol, with
saline instead of OVA-aluminum hydroxide mixture. In the positive
treatment group, rats were orally administered BEG (500 mg/kg) and
ANP (300 mg/kg) from day 14 to day 21. On the day 21, the pulmonary
functions were determined, then rats were sacrificed under
pentobarbital sodium anesthesia (45 mg/kg; IP) after the blood
samples and bronchoalveolar lavage fluid (BALF) were collected.
Subsequently, the lung tissues, the large intestine tissues and
intestinal mucous were harvested for the following biochemical
assays.
Determination of the rat pulmonary
function
Rat pulmonary function including respiratory rate
and airway responsiveness were determined using a Buxco®
animal pulmonary function analysis system (FinePointe Non-Invasive
Airway Mechanics; DSI; Harvard Bioscience, Inc., Holliston, MA,
USA; https://www.datasci.com/products/buxco-respiratory-products/finepointe-non-invasive-airway-mechanics).
Airway responsiveness was evaluated using the enhanced pause values
(Penh value) according to a previously reported method (13).
Blood cell counting
Blood smears were prepared with blood taken from the
heart of the rats and fixed with formalin for 2–3 min at 25°C.
Subsequently, Wright-Giemsa staining was performed for 15 min at
25°C and cell counting was carried out under an optical light
microscope (CH20BIMF200; Olympus Corporation, Tokyo, Japan) at
magnification, ×100 and ×400.
Bacterial colony counting
All experiments were carried out under sterile
conditions. Briefly, 0.1 g rat feces were dissolved in saline at a
dilution of 1:1010. Subsequently, the diluted feces
samples were cultured in an anaerobic incubator for 48–72 h at 37°C
for detection of Bifidobacterium, Lactobacillus and
Candida albicans. Diluted feces samples were also cultured
in an aerobic incubator for 24–48 h at 37°C for detection of
Enterococcus and Enterobacterium. The colony counting
was expressed as logCFU/g.
Examination of pathological
alterations of the lung tissues
The histopathological examination was performed as
previously described (14).
Briefly, the lung tissues were collected and fixed with 4%
paraformaldehyde for 24 h. The tissues were subsequently embedded
in paraffin and cut into 5-µm-thick sections. The samples were
deparaffinized and stained with H&E. Finally, pathological
alterations of lung tissues were examined under an optical
microscope (CH20BIMF200; Olympus Corporation).
ELISA assays for detection of sIgA,
IgE, IL-4 and IFN-γ
The levels of sIgA in BALF and intestinal mucosa,
and levels of IgE, IL-4 and IFN-γ in serum were determined using
commercial ELISA kits according to the manufacturers' protocols and
determined using a microplate reader at a wavelength of 450 nm
(Multiskan Ascent 413MBY042078; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) assays
Lung and intestine tissues were collected and
homogenized. Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, total
RNA was used for cDNA synthesis using HiScript 1st Strand cDNA
Synthesis kit, according to the manufacturer's protocol. qPCR was
performed using SYBR-Green Master Mix, according to the
manufacturer's protocol with primers specific to TRL-2, TRL-4,
myeloid differentiation primary response 88 (MyD88), TRAF6,
β-arrestin and β-actin (Table I).
The thermocycling conditions for qPCR: Pre-denaturation at 95°C for
5 min and 95°C for 10 sec at 53.5°C for 30 sec, the data were
recorded by fluorescent reading board, and 39 cycles were recorded.
The PCR reactions were performed using CFX96TM™
Real-Time system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
The relative mRNA expressions were determined by 2−ΔΔCq
relative quantitative analysis according to the previous reported
method (15).
Rat intestine 16S ribosomal DNA (16S
rDNA) assay
For the determination of rat intestine 16S rDNA, 6
rats were selected from each group (n=6). Subsequently, the DNA was
collected from the feces of rats. The DNA samples used in this
study were isolated and purified using the QIAamp® DNA
Stool isolation and purification kit (Qiagen China Co., Ltd.,
Shanghai, China) and the protocol is adopted following the
manufacturer's protocol. Further analysis was performed using a DNA
sequencer (Illumina HiSeq™ 2000; Illumina, Inc., San
Diego, CA, USA) and BGI Tech Solutions Co., Ltd., (Shenzhen, China)
analyzed and interpreted the sequencing data and the parameters
were investigated as described previously (16,17).
Statistical analysis
Data are presented as the mean ± standard deviation,
and each experiment was repeated at least 3 times. Differences
between groups were determined using one-way analysis of variance
followed by Dunnett's multiple comparisons test. The statistical
significance of differences was analyzed using SPSS software
(version 13.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Results of the determination of
pulmonary function
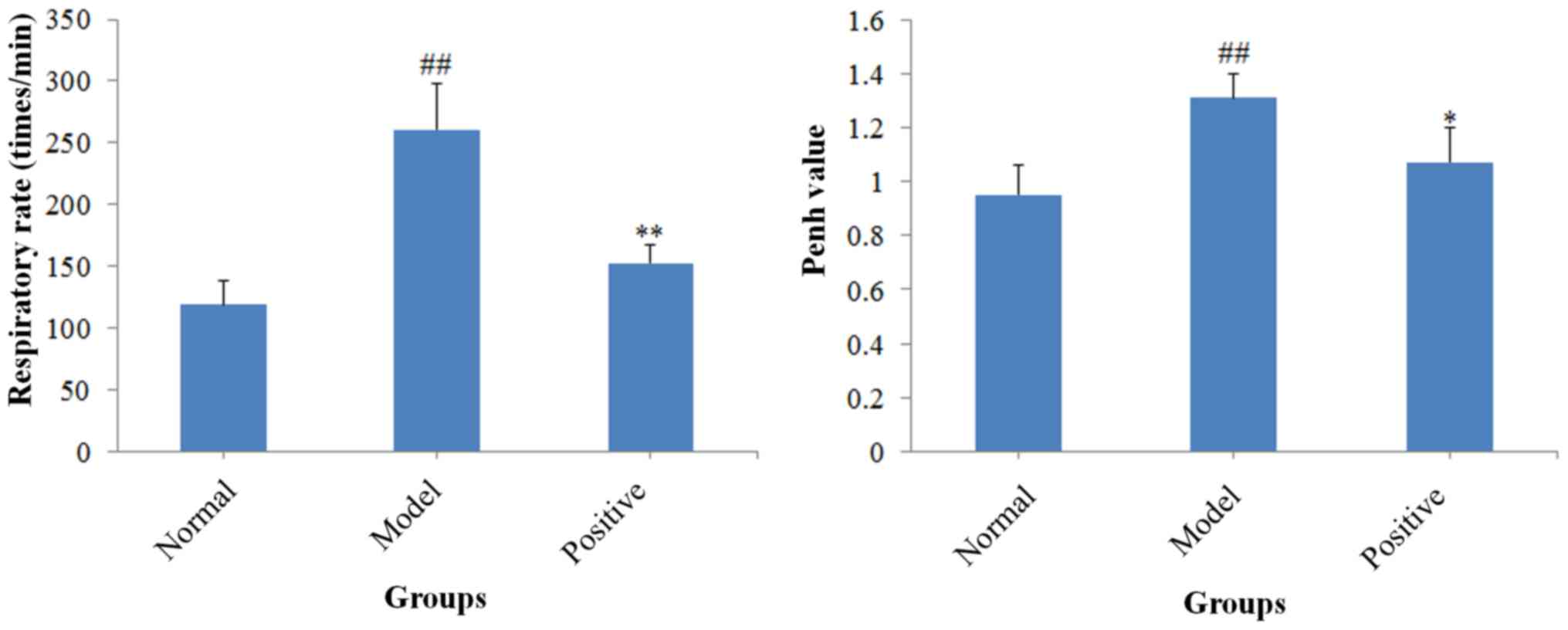
The respiratory rate and Penh value were determined
(Fig. 1). Compared with the normal
rats, the respiratory rate (P<0.01) and Penh value (P<0.01)
of the model rats increased significantly. However, the positive
drug treatment significantly decreased the elevated respiratory
rate (P<0.01) and Penh value (P<0.05) compared with the model
rats. The present results revealed that a rat model of allergic
asthma was successfully established.
Results of cell counting in blood
samples
The results of cell counting (Table II) indicated that the numbers of
the four types of inflammatory cells (eosinophils, neutrophils,
lymphocytes and monocytes) increased in the model rats compared
with the normal rats (all P<0.01). By contrast, the positive
treatment decreased the number of inflammatory cells compared with
the model group (P<0.01, P<0.05, P<0.05 and P<0.01,
respectively for eosinophils, neutrophils, lymphocytes and
monocytes).
 | Table II.Blood cell count
(×106/l). |
Table II.
Blood cell count
(×106/l).
|
| Inflammatory cell
type |
|---|
|
|
|
|---|
| Group | Eosinophil | Neutrophil | Lymphocyte | Monocyte |
|---|
| Normal | 2.66±1.2 | 22.50±5.5 | 37.00±5.0 | 2.00±0.36 |
| Model |
27.75±2.19c |
43.00±1.82c |
66.80±3.21c |
5.33±0.88c |
| Positive |
4.66±0.55b |
32.00±2.08a |
50.40±4.93a |
2.88±0.26b |
Results of bacterial colony counting
in rat feces
The number of colonies of three bacterial strains
including Enterococcus, Bifidobacterium and
Lactobacillus significantly decreased in model rats compared
with normal rats (all P<0.01; Table III), whereas the numbers of
colonies of enteric Bacilli (P<0.05) and Candida albicans
(P<0.01) significantly increased. By contrast, in the positive
drug treatment group, the number of Enterococcus
(P<0.01), Bifidobacterium (P<0.01), enteric Bacilli
(P<0.05) and Lactobacillus (P<0.01) colonies
significantly increased, whereas the colony numbers of Candida
albicans decreased (P<0.01), compared with the model rats.
The above results indicated that the model rats exhibited
significant alterations of the intestinal flora.
 | Table III.Intestinal flora determination (log
colony forming unit/g). |
Table III.
Intestinal flora determination (log
colony forming unit/g).
|
| Gut
microorganism |
|---|
|
|
|
|---|
| Group |
Enterococcus | Enteric
bacilli |
Bifidobacterium |
Lactobacillus | Candida
albicans |
|---|
| Normal | 6.88±0.67 | 6.78±0.69 | 10.66±1.63 | 9.28±0.64 | 0.00±000 |
| Model |
4.70±0.46c |
7.99±0.96b |
0.00±0.00c |
0.00±0.00c |
7.30±0.50c |
| Positive |
5.78±0.69a | 6.78±0.67 |
1.68±0.59a |
8.86±0.55a |
3.04±0.59a |
Examination of pathological
alterations of the lungs
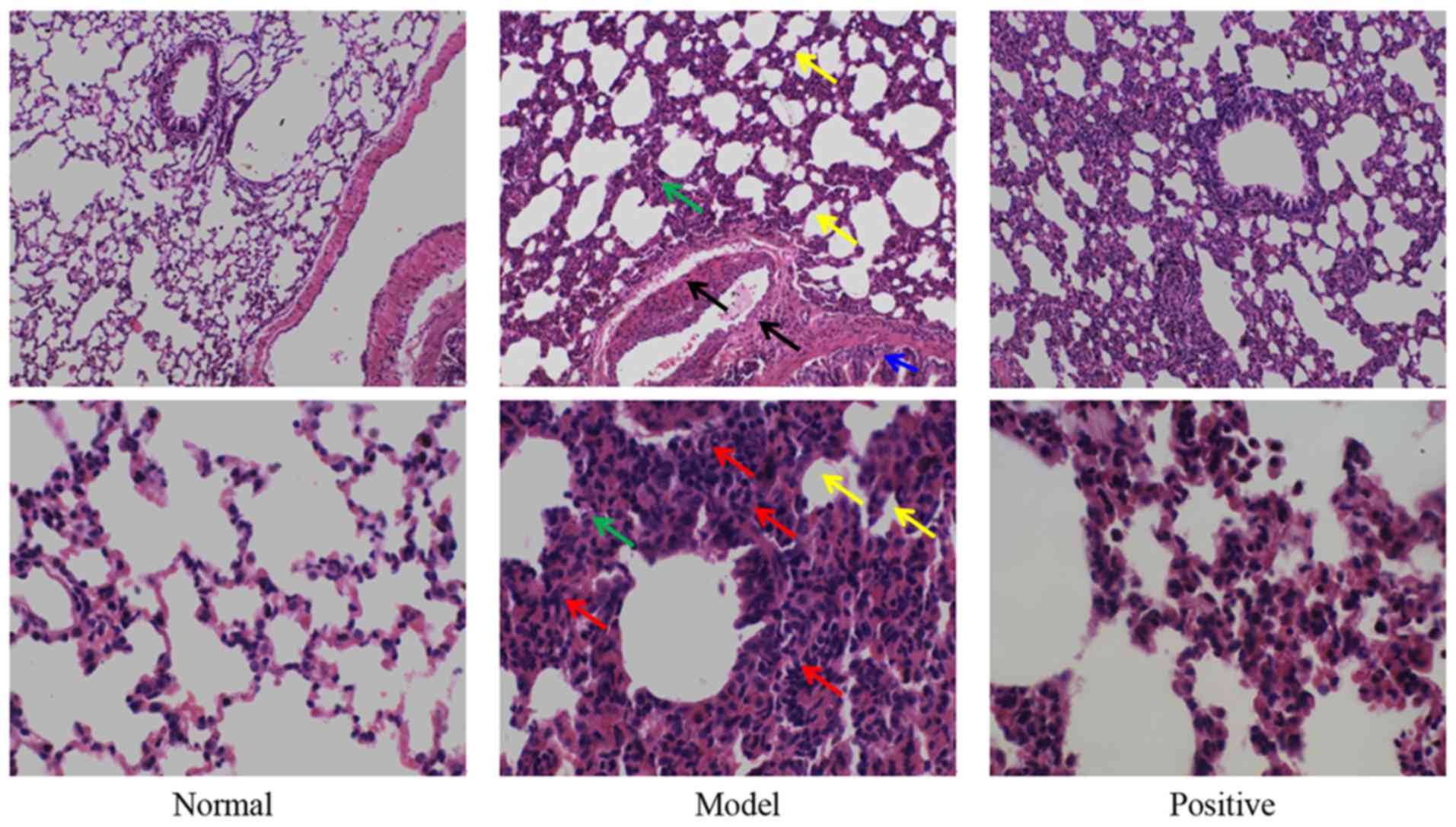
Results of the pathological examination indicated
that, in the normal group, no obvious pathological alterations were
observed (Fig. 2). Compared with
the normal rats, marked inflammatory cell infiltration was observed
in the lung tissues of the model rats, and the pulmonary septum
became thick and alveolar space became narrow (Fig. 2). In addition, edema could be also
observed in the blood vessel and tracheal wall. However, in the
positive group, the aforementioned abnormal alterations were
markedly alleviated.
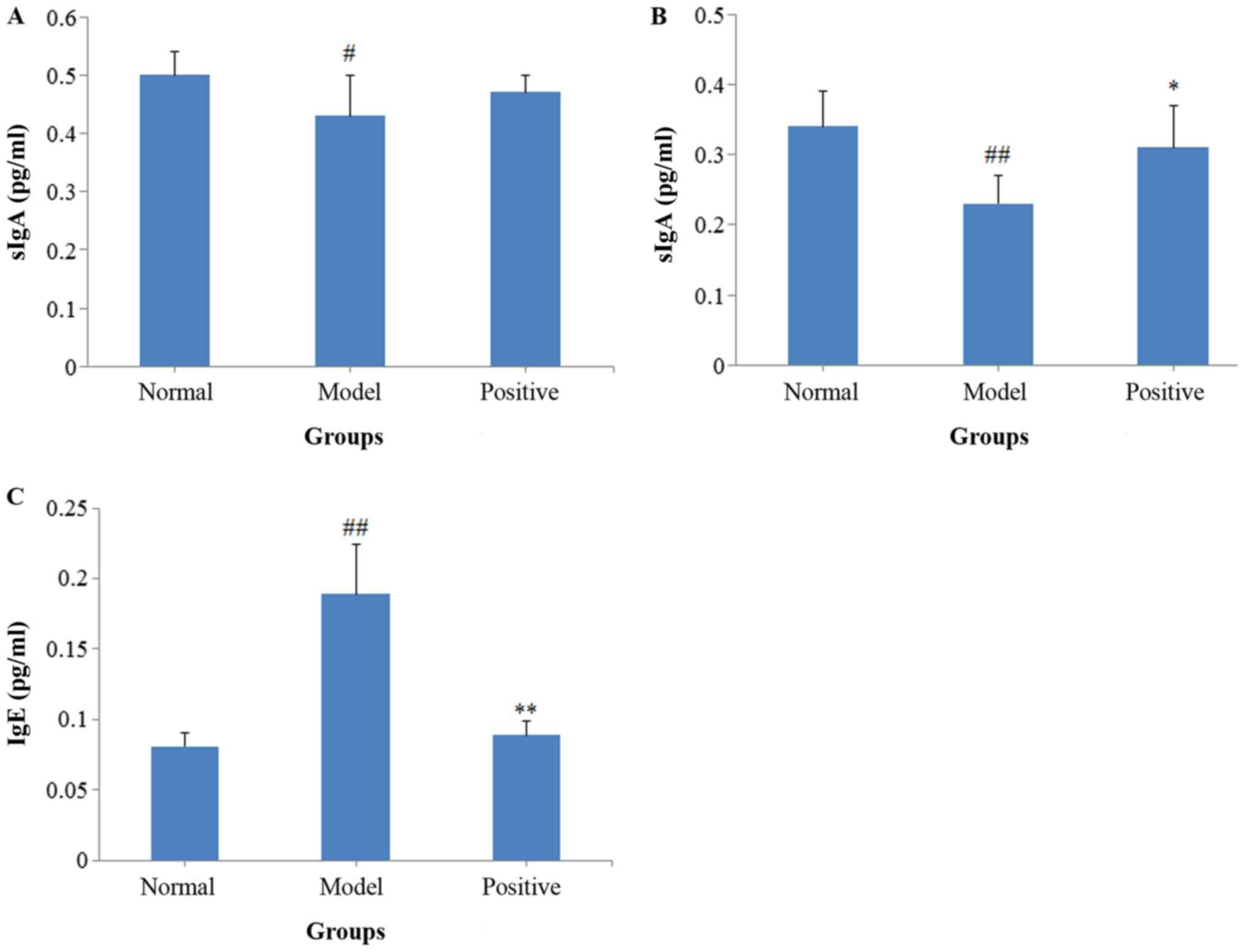
Results of the ELISA assays
sIgA levels in both BALF (P<0.05; Fig. 3A) and intestinal mucosa (P<0.01;
Fig. 3B) decreased in the model
rats compared with the normal rats, and the serum IgE levels
significantly increased (P<0.01; Fig. 3C). Following treatment with BEG and
ANP, the sIgA levels in the intestinal mucosa (P<0.05)
increased, whereas the serum IgE levels decreased (P<0.01),
compared with the model rats.
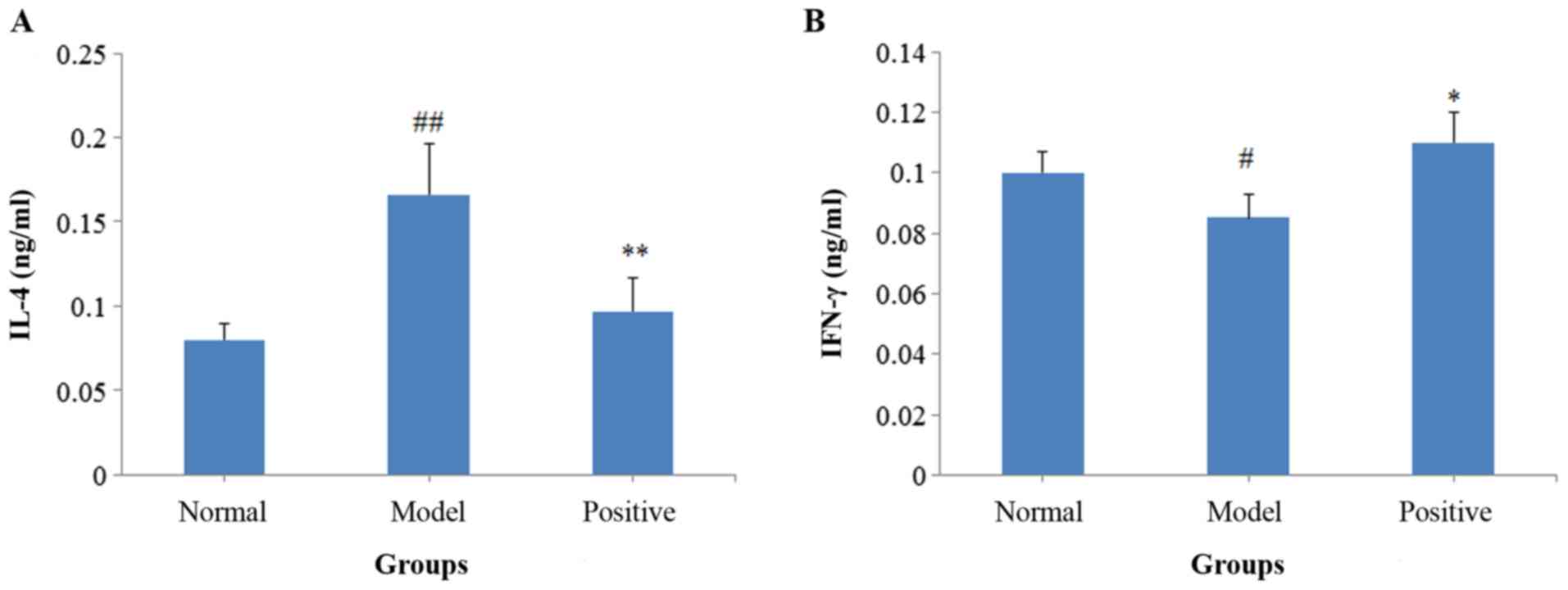
Following activation with OVA, the IL-4 levels
(P<0.01) of the model rats significantly increased compared with
the normal group, whereas the IFN-γ levels significantly decreased
(P<0.05; Fig. 4). By contrast,
in the positive treatment group, the levels of IL-4 significantly
decreased (P<0.01; Fig. 4A) and
the levels of IFN-γ (P<0.05; Fig.
4B) significantly increased compared with the model rats.
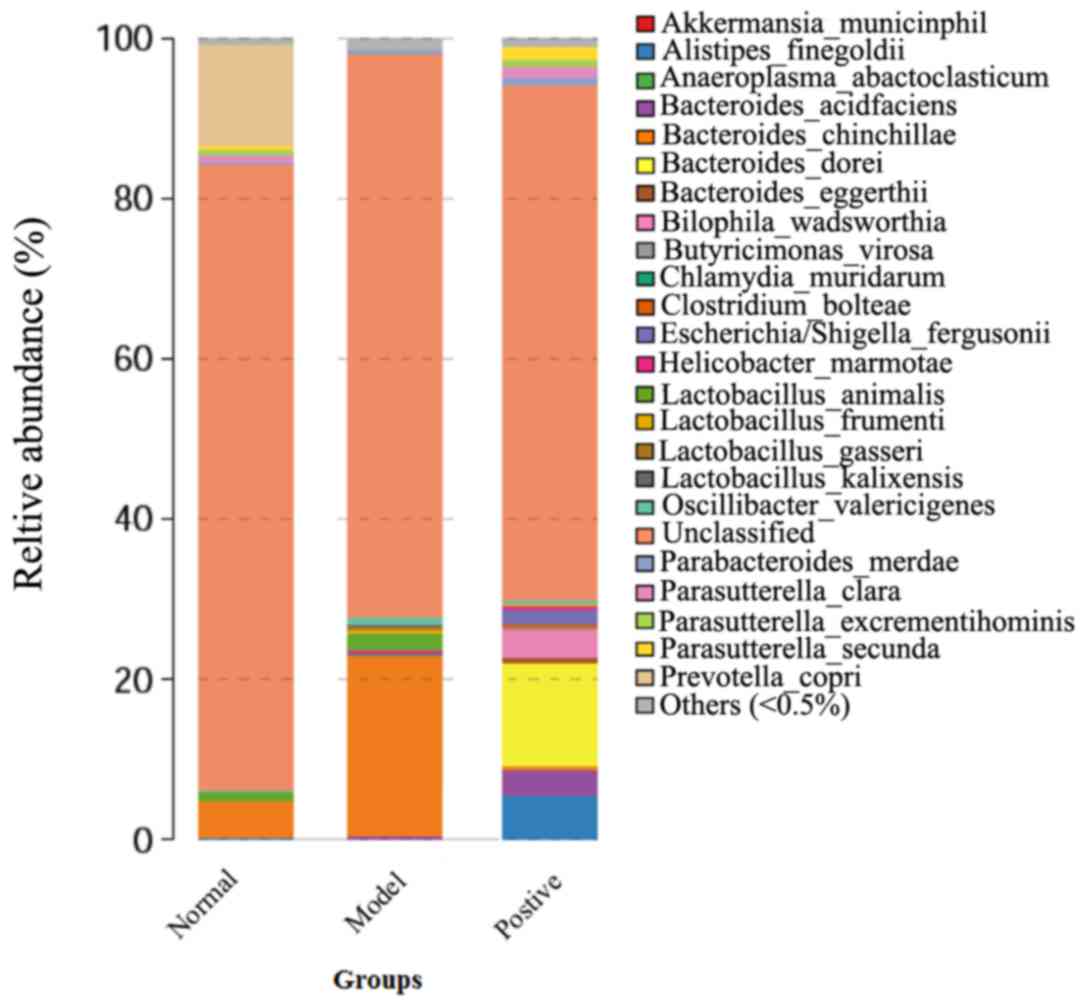
Results of the 16S rDNA assays of the
rat intestine
Following discarding the low-quality sequencing
reads using an inner program (BGI Tech Solutions, Co., Ltd.) to
generate clean data, a detailed result for each sample was
obtained. The operational taxonomic units (OTUs) analysis revealed
the OUT numbers of the model rats were lower than that of normal
and positive-treated rats, indicating the species richness of the
model rats was lower than that of the normal and positive-treated
rats (Table IV). Furthermore, the
number of bacteria constituting normal intestinal flora (including
Bacteroidetes and Prevotella) decreased compared with the
normal and positive groups (Fig.
5). By contrast, the number of Butyricimonas increased in the
model group compared with the normal and positive rats. These
results demonstrated that intestinal flora disorder was observed in
the model rats. All of the above results indicated that a rat model
of allergic asthma complicated with intestinal flora disorder was
successfully established, and was subsequently used for the further
investigation of the underlying molecular mechanisms.
 | Table IV.Results of the OTU analysis of
samples. |
Table IV.
Results of the OTU analysis of
samples.
| Group | Tag number | OTU number | OTU number
(removing singletons) |
|---|
| Normal | 14.751 | 828 | 369 |
| Control | 18.346 | 593 | 346 |
| Positive | 16.844 | 718 | 385 |
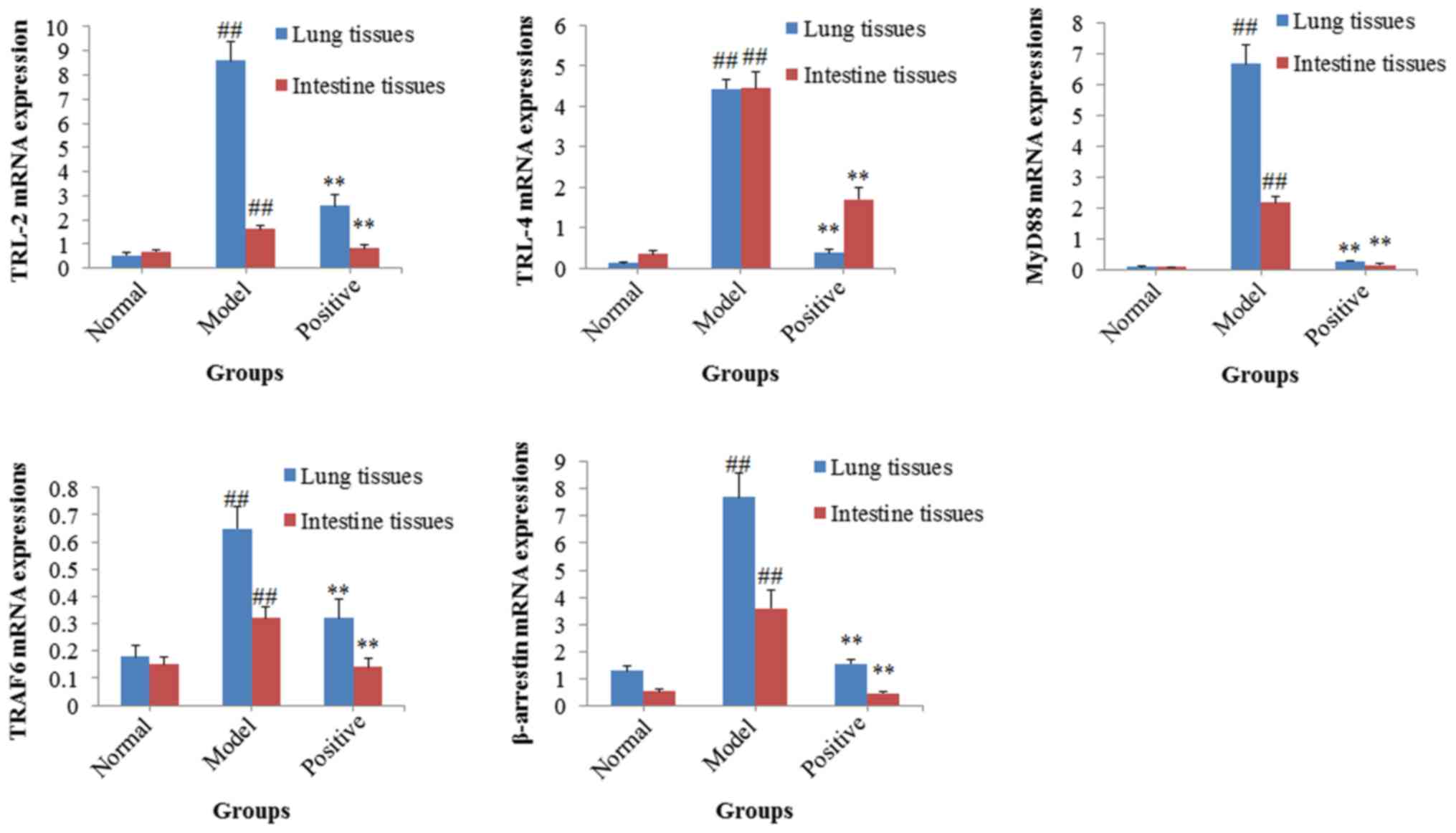
Results of the RT-qPCR assays
Following successful establishment of the rat model
of allergic asthma complicated with intestinal flora disorder, the
potential molecular mechanisms of mutual interactions between the
lung and the large intestine were investigated using RT-qPCR
assays. The mRNA expression levels of TRL-2, TRL-4, MyD88, TRAF6
and β-arrestin were significantly upregulated in both the lung and
intestinal tissues of the model rats, compared with the normal rats
(all P<0.01; Fig. 6). By
contrast, treatment with BEG and ANP significantly decreased the
upregulated mRNA expression levels of TRL-2, TRL-4, MyD88, TRAF6
and β-arrestin (all P<0.01; Fig.
6) in both the lung and intestinal tissues, compared with the
model group.
Discussion
In the present study, using a rat model of allergic
asthma complicated with intestinal flora disorder, the roles of the
TLR/NF-κB signaling pathway in the mutual interactions between the
lung and the large intestine were studied. The results of the
present investigation indicated that allergic asthma was associated
with the intestinal flora disorder and the TLR/NF-κB signaling
pathway may serve a role in this association.
Establishing a suitable and reliable animal model is
the crucial initial step for investigating the mutual interactions
between the lung and the large intestine (18,19).
The present investigation successfully constructed a rat model of
allergic asthma complicated with intestinal flora disorder. The
results indicated that the rats in the model group exhibited
obvious characteristics of allergic asthma and intestinal flora
disorder, which were demonstrated by the pathological alterations,
ELISA results (sIgA, IgE, IFN-γ and IL-4 levels), blood cell count
and bacterial count in feces, as well as the 16S rDNA assay of the
rat intestine. In the model rats, the levels of IL-4 and IgE, and
the number of inflammatory cells and Candida albicans
increased, and obvious inflammatory cell infiltration was observed
in the lung tissues compared with the normal rats.
The sIgA is a marker for the intestinal flora
disorder and damage of intestinal mucosa, and a previous report
revealed that the number of intestinal Bifidobacteria is
closely associated with the level of sIgA (20). In the model group the sIgA
expression levels decreased in both of the intestinal mucosa and
BALF, which supports the ‘exterior-interior relationship between
the lung and the large intestine’ theory. TLRs, pattern recognition
receptors expressed in the cytomembrane, are closely associated
with the body immunocompetence (21). TLRs can recognize specific
conserved molecular components of microorganisms and transfer the
signals into the cell, leading to the activation of NF-κB (21,22).
Furthermore, activation of NF-κB can induce the excessive release
of pro-inflammatory cytokines including IL-1, IL-6, TNF-α and IL-12
(23). The characteristic
pathological alterations associated with asthma include airway
inflammatory reactions and airway remodeling (12). TLRs serve roles in the development
of airway inflammatory reactions and further activate the MyD88-
IRAK-TRAF6-IKK-NF-κB signaling pathway, resulting in excessive
release of pro-inflammatory cytokines and inflammation (24–26).
In addition, TLRs promote the maturation and differentiation of
immune cells but also the transdifferentiation of CD4+ T
cells into T regulatory cells (Tregs). Therefore, the TLR/NF-κB
signaling pathway could further regulate the balance of type 1 T
helper/type 2 T helper cells via Tregs (27). Recently, it has been reported that
the TLR/NF-κB signaling serves roles in the intestinal defense
against pathogens and maintenance of immune system homeostasis and
intestinal flora balance (28). A
previous study indicated that TLR-2 and TLR-4 are closely
associated with the recognition of peptidoglycan and
lipopolysaccharide (29),
respectively. Additionally, TLR-2 and TLR-4 serve roles in the
development of allergic asthma and intestinal flora-associated
diseases (30,31). In addition, β-arrestin was
upregulated in patients with allergic asthma and served a role in
the development of chronic intestinal inflammation (32–34).
Therefore, β-arrestin may serve a role in the development of
allergic asthma and intestinal diseases. In the present study, mRNA
expression levels of TRL-2, TRL-4, MyD88, TRAF6 and β-arrestin
significantly increased in both the lung and intestinal tissues of
the model rats compared with the normal rats. By contrast,
treatment with BEG and ANP could decrease the up-regulated mRNA
expression levels of these genes in both the lung and intestinal
tissues of the model rats. The results if the present study
indicated that the TLR/NF-κB signaling is a potential link between
asthma and intestinal disorders in the rat models and may also be a
molecular mechanism of the ‘exterior-interior relationship between
the lung and the large intestine’. Furthermore the present study
demonstrated, the upregulated TLR/NF-κB signaling is an important
molecular mechanism for the development of lung diseases
complicated with intestinal disorders.
‘Exterior-interior relationship between the lung and
the large intestine’ is a classical basic theory in Traditional
Chinese Medicine and the present study did not prove the direct
mutual interaction between the lung and the large intestine.
However, future studies should aim to investigate the direct mutual
interactions between the two organs in animal models. The present
study indicated that the TLR/NF-κB signaling may serve a role in
the mutual interactions between the lung and the large intestine,
however, alternative signal transduction mechanisms have not been
investigated. Furthermore, the present study analyzed the
alterations of intestinal flora by sequencing 16S rDNA, however,
flora metagenomics analysis of both the lung and intestine would be
an improved strategy for investigating the molecular mechanisms of
the two organs the association between them.
In conclusion, the present experimental results
revealed that the TLR/NF-κB signaling may serve roles in the mutual
interactions between the lung and the large intestine. The results
also support the Traditional Chinese Medicine theory of
‘exterior-interior relationship between the lung and the large
intestine’. Furthermore, the results of the present study suggested
that the TLR/NF-κB signaling is a potential target for the clinical
treatment of lung diseases complicated intestinal disorders.
Acknowledgements
Not applicable.
Funding
The authors are grateful for the financial support
from the National Natural Science Foundation of China (grant no.
81303085) and the Foundation of Science and Technology Department
of Sichuan Province (grant no. 2013JY0067).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YG and CZ designed the experiment; WF, JZ, XL
completed the experiments and analyzed the experimental data; WF
and CZ wrote the paper.
Ethics approval and consent to
participate
All animal experimental protocols were approved by
the Ethics Committee for Laboratory Animal Experimentation of
Chengdu University of Traditional Chinese Medicine.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yin LM, Zhang GQ, Yan XK, Wang Y, Xu YD
and Yang YQ: An in vivo and in vitro evaluation of the mutual
interactions between the lung and the large intestine. Evid Based
Complement Alternat Med. 2013:6956412013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ding H: Huang Di Nei Jing Ling Shu.
Sichuan: Si Chuan Science and Technology Publishing house; pp.
27–28. 2008
|
|
3
|
Yan XK, Wang Y, Zhang GQ, Yang YQ and Cui
LP: The research progress of exterior-interior relationship between
the lung and the large intestine. Shaanxi J TCM. 24:378–380.
2003.
|
|
4
|
Yang F, Wang J and Wang Q: Allergic
diseases and intestinal flora imbalance for allergic constitution
research. J Beijing Univ Tradit Chin Med. 38:509–514. 2015.
|
|
5
|
Jia JJ, Chen X and Jie JP: Modern research
of the exterior-interior relationship between the lung and the
large intestine. Acta Chin Med Pharmacol. 34:23–25. 2011.
|
|
6
|
Lee MY, Shin IS, Jeon WY, Lim HS, Kim JH
and Ha H: Pinellia ternata Breitenbach attenuates ovalbumin-induced
allergic airway inflammation and mucus secretion in a murine model
of asthma. Immunopharmacol Immunotoxicol. 35:410–418. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yacoub MR, Colombo G, Marcucci F, Caminati
M, Sensi L, Di Cara G, Frati F and Incorvaia C: Effects of
sublingual immunotherapy on allergic inflammation: An update.
Inflamm Aller Drug Targets. 11:285–291. 2012. View Article : Google Scholar
|
|
8
|
Nakajima Y, Goldblum RM and Midoro-Horiuti
T: Fetal exposure to bisphenol A as a risk factor for the
development of childhood asthma: An annimal model study. Environ
Health. 11:82012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsabouri S, Mavroudi A, Feketea G and
Guibas GV: Subcutaneous and sublingual immunotherapy in allergic
asthma in children. Front Pediatr. 5:82017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Strachan DP: Hay fever, hygiene, and
household size. BMJ. 299:1259–1260. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim SH, Kim and Lee YC: Effect of Coeni
fructus on ovalbumin-induced airway inflammation and airway
hyper-responsiveness in a mouse of allergic asthma. J Inflamm
(Lond). 9:92012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Roviezzo F, Rossi A, Calazzo E, Orlando P,
Riemma MA, Iacono VM, Guarino A, Ialenti A, Cicala C, Peritore A,
et al: Palmitoylethanolamide supplementation during sensitization
prevents airway allergic symptoms in the mouse. Front Pharmacol.
8:8572017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu WH: Repetitive measurements of enhanced
pause (Penh). Resp Physiol Neurobiol. 206:41–44. 2015. View Article : Google Scholar
|
|
14
|
Peng W, Qiu XQ, Shu ZH, Liu QC, Hu MB, Han
T, Rahman K, Qin LP and Zheng CJ: Hepatoprotective activity of
total iridoid glycosides isolated from Paederia scandens
(Lour.) Merr. var. Tomentosa. J Ethnopharmacol. 174:317–321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu B, Yuan J, Yin SM, Li Z, Xie Y, Chen
Y, Shi Y, Zhang H, Li Y, Lam TW and Luo R: COPE: An accurate
k-mer-based pair-end reads connection tool to facilitate genome.
Bioinformatics. 28:2870–2874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schloss PD, Westcott SL, Ryabin T, Hall
JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH,
Robinson CJ, et al: Introducing mothur: Open-source,
platform-independent, community-supported software for describing
and comparing microbial communities. Appl Environ Microbiol.
75:7537–7541. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu L: Animal models of human diseases.
Dongwuxue Yanjiu. 32:1–3. 2011.(In Chinese). PubMed/NCBI
|
|
19
|
Dai JH, Tan Y and Zhou F: The current
situation of researches on animal model of asthma. Clin J Lab
Animal Sci. 11:167–171. 2001.
|
|
20
|
Sjögren YM, Tomicic S, Lundberg A,
Böttcher MF, Björkstén B, Sverremark-Ekström E and Jenmalm MC:
Influence of early gut microbiota on the maturation of childhood
mucosal and systemic immune responses. Clin Exp Allergy.
39:1842–1851. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duffy L and O'Reilly SC: Toll-like
receptors in the pathogenesis of autoimmune diseases: Recent and
emerging translational developments. Immunotargets Ther. 5:69–80.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Achek A, Yesudhas D and Choi S: Toll-like
receptors: Promising therapeutic targets for inflammatory diseases.
Arch Pharm Res. 39:1032–1049. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee WS, Shin JS, Jang DS and Lee KT:
Cnidilide, an alkylphthalide isolated from the roots of Cnidium
officinale, suppresses LPS-induced NO, PGE2, IL-1β, IL-6 and
TNF-α production by AP-1 and NF-κB inactivation in RAW 264.7
macrophages. Int Immunopharmacol. 40:146–155. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Im EJ, Kim SJ, Hong SB, Park JK and Rhee
MH: Anti-inflammatory activity of Bee Venom in BV2 microglial
cells: Mediation of MyD88-dependent NF-κB signaling pathway. Evid
Based Complement Alternat Med. 2016:37047642016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rana M, Maurya P, Reddy SS, Singh V, Ahmad
H, Dwivedi AK, Dikshit M and Barthwal MK: A standardized chemically
modified curcuma longa extract modulates IRAK-MAPK signaling in
inflammation and potentiates cytotoxicity. Front Pharmacol.
7:2332016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
He A, Ji R, Shao J, He C, Jin M and Xu Y:
TLR4-MyD88-TRAF6-TAK1 complex-mediated NF-κB activation contribute
to the anti-inflammatory effect of V8 in LPS-induced human cervical
cancer SiHa cells. Inflammation. 39:172–181. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Flaherty S and Reynolds JM: TLR function
in murine CD4 (+) T lymphocytes and their role in inflammation.
Methods Mol Biol. 1390:215–227. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sabharwal H, Cichon C, Ölschläger TA,
Sonnenborn U and Schmidt MA: Interleukin-8, CXCL1, and MicroRNA
miR-146a responses to probiotic Escherichia coli nissle 1917
and enteropathogenic E. coli in human intestinal epithelial
T84 and Monocytic THP-1 cells after apical or basolateral
infection. Infect Immun. 84:2482–2492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma YH, Krikun G, Abrahams VM, Mor G and
Guller S: Cell type-specific expression and function of toll-like
receptor 2 and 4 in human placenta: Implications in fetal
infection. Placenta. 28:1024–1031. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding Y, Liao W, He X, Xiang W and Lu Q:
CSTMP exerts anti-Inflammatory effects on LPS-induced human renal
proximal tubular epithelial cells by inhibiting TLR4-mediated NF-κB
pathways. Inflammation. 39:849–859. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Conti F, Boucherit N, Baldassarre V,
Trouplin V, Toman R, Mottola G, Mege JL and Ghigo E: Coxiella
burnetii lipopolysaccharide blocks p38α-MAPK activation through the
disruption of TLR-2 and TLR-4 association. Front Cell Infect
Microbiol. 4:1822015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
DeWire SM, Ahn S, Lefkowitz RJ and Shenoy
SK: Beta-arrestins and cell signaling. Ann Rev Physiol. 69:483–510.
2007. View Article : Google Scholar
|
|
33
|
Ma L and Pei G: Beta-arrestin signaling
and regulation of transcription. J Cell Sci. 120:213–218. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Walker JK, Fong AM, Lawson BL, Savov JD,
Patel DD, Schwartz DA and Lefkowitz RJ: Beta-arrestin-2 regulates
the development of allergic asthma. J Clin Invest. 112:566–574.
2003. View Article : Google Scholar : PubMed/NCBI
|