Introduction
Current diagnostic tools for detection of breast
cancer have contributed to reduce its mortality. However, in 2015,
the National Institute of Statistics and Geography (INEGI) in
Mexico reported that breast cancer was the first cause of death
among women between 35 and 44 years old (1). Additionally, ovarian cancer was
within the top ten causes of cancer death in Mexican women
(2).
Despite recent advances in science and new
approaches for the detection of ovarian cancer through liquid
biopsy, such as salivary samples, (3) in Mexico there are still no effective
methods to reduce mortality due to ovarian cancer. Transvaginal
ultrasound is the most commonly used technique but it might not
always detect early stage abnormalities. In addition, there is no
self-examination method as there is for breast cancer (4). In most cases, ovarian cancer is
diagnosed when the tumor is already in an advanced stage.
Both breast and ovarian cancer share several risk
factors, such as advanced age, nulliparity or having a full term
pregnancy after 30–35 years old, family history of breast or
ovarian cancer, mutations in BRCA1 and BRCA2 genes,
early menarche, not having breastfed, late menopause and obesity
(5–10). Among 5–10% of breast cancer and
10–15% of ovarian cancer cases are due to a hereditary mutation
(11). Hereditary Breast and
Ovarian Cancer syndrome (HBOC) is characterized by a high risk of
developing breast or ovarian cancer (12). Most of the mutations found in HBOC
are localized in the tumor suppressor genes BRCA1 and
BRCA2, which participate in DNA repair. Carriers of a
BRCA1 mutation have a risk of 65–85% and 39–44% of
developing breast and ovarian cancer, respectively, whereas having
a BRCA2 mutation confers a risk of 45–80% of developing
breast cancer and 11–27% for ovarian cancer (11). Several mutations have been
identified in specific populations, for example, in Ashkenazi Jews,
the BRCA1 mutations 185delAG and 8382insC, and the
BRCA2 mutation 6174delT are found within 2–3% of the
population (13). These mutations
have been identified as founder mutations, because they were
originated in a common ancestor and then inherited through
generations (14). In 2007, the
Mexican founder mutation BRCA1 ex9-12del, a deletion of
exons 9–12 of around 14.7 kb, was reported for the first time
(15). Approximately, 42% of the
BRCA1 gene is composed by Alu repetitive sequences,
which can lead to errors during homologous recombination (16). Consequently, the BRCA1
ex9-12del mutation arises due to a recombination event between an
AluSp element in intron 8 and an AluSx element in
intron 12, with the subsequent loss of exons 9–12 (15).
BRCA1 ex9-12del mutation was first documented
in a sample of women with Hispanic ancestry and family history of
breast and ovarian cancer. The deletion was found in four
non-related families, all of them with Mexican ancestry (15). The same research group performed
another study in Hispanics, in which individuals with personal or
family cancer history were shown to have BRCA mutations; 21
of 189 were specifically large rearrangement (LR) mutations, in
which 13 were carriers of the BRCA1 ex9-12del mutation
(17). In a different study
involving Mexican women with breast or ovarian cancer, the
BRCA1 ex9-12del mutation was present in 35% of ovarian
cancer cases associated with BRCA and in 29% of breast
cancer cases associated with BRCA (18). In a later study where only women
with breast cancer participated, the deletion was found in 24% of
cases associated with BRCA mutations, in patients between 35
and 56 years old (19). A more
recent report showed that the BRCA1 ex9-12del mutation
represented 42% of the cases with BRCA mutations among young
Mexican women diagnosed with triple negative breast cancer
(20). Regarding the origin of the
BRCA1 ex9-12del mutation, it has been suggested that it
arose 1480 years ago (95% CI, 920–2260 years) in Puebla, Mexico
(17). Studies performed in high
breast cancer risk individuals from Spain, Colombia, Germany and
Pakistan, have shown the absence of the mutation in those
populations (21,22). Moreover, reports from Myriad
Genetic Laboratories (Salt Lake City, UT) indicated that patients
with HBOC of Latin American/Caribbean ancestry commonly carry large
rearrangements (21.4% of all BRCA mutations). In these
reports, the Mexican founder mutation BRCA1 ex9-12del
accounted for 37% of those LR (23).
Taken together, these data show the high frequency
of the BRCA1 ex9-12del mutation among young Mexican women
with HBOC. Therefore, the detection of this deletion in relatives
of the mutation carriers might encourage strategies for early
detection and prevention of breast and ovarian cancer, such as
recurrent clinical and imaging examinations, lifestyle
modifications or prophylactic surgeries and/or chemoprevention use.
Moreover, analyzing the presence of this deletion in Mexican
patients suspicious of carrying a BRCA mutation will
definitely be a cheaper option, before searching for mutations and
LR in complete BRCA genes. Sequencing of both BRCA
genes costs ~1,000–4,000 USD (24). Myriad Genetic Laboratories
currently uses quantitative multiplex endpoint polymerase chain
reaction (PCR) to detect a set of common LR, including the
BRCA1 ex9-12del mutation, at a cost of 700 USD, under the
name BRACAnalysis Large Rearrangement Test (BART) (23,25).
Unfortunately, most Mexican patients do not have access to such
expensive tests. Other techniques can be used to detect LR, for
example Multiplex Ligation-dependent Probe Amplification (MLPA),
Protein Truncation Test (PTT), long range PCR, Fluorescence In
Situ Hybridization and end-point PCR (26). However, these techniques can be
time consuming, difficult to interpret and expensive since
different equipment and reagents are required. Most of the research
papers that have screened patients for the BRCA1 ex9-12del
mutation have used end-point PCR (15,18,20),
MLPA (19) and one included the
Myriad diagnostic test (17).
The aim of the present study was to develop a new
method to detect the Mexican founder mutation BRCA1
ex9-12del by qPCR and TaqMan® probes. The method we
propose is economic, specific, sensitive, fast, easy to interpret,
and it can be used in both research and diagnostic scenarios.
Materials and methods
Study samples
A total of four DNA samples from previously known
carriers of the BRCA1 ex9-12del mutation were obtained,
three from the Hospital Zambrano Hellion TecSalud and one from the
Laboratorio Nacional Biobanco. Control samples were obtained from
six female volunteers without a family history of cancer, who work
at the Universidad de Monterrey. Three ml of peripheral blood were
collected in a tube with EDTA. The study complied with the
guidelines of the Declaration of Helsinki and was approved by the
Research and Ethics Committee from the University of Monterrey
(registration no. 01072017-CIE). Written informed consent was
obtained from all subjects.
DNA extraction
The Wizard Genomic DNA Purification kit (Promega
Corp., Madison, WI, USA) was used to isolate DNA from the six
volunteers' blood samples. DNA extraction was performed according
to manufacturer's instructions. Genomic DNA was quantified by UV
absorbance using Nanodrop (Thermo Fisher Scientific, Inc.,
Wilmington, MA, USA). The DNA quality was evaluated with the
A260/280 and A260/230 ratios. Samples were stored at −20°C.
Characterization of the recombination
region between introns 8 and 12 of BRCA1
The key contribution of our work was the design of a
TaqMan® probe which is complementary to the
recombination region between introns 8 and 12 of BRCA1, for
the detection of the BRCA1 ex9-12del mutation. In order to
achieve this, the first step was to identify the DNA sequence of
the recombination site. Therefore, an end-point PCR assay was
designed (Fig. 1), and then the
PCR product was sequenced. The DNA of a sample from a BRCA1
ex9-12del mutation carrier was amplified using the Go-taq flexi DNA
polymerase kit (Promega Corp.). The PCR was carried out in a total
volume of 15 µl containing 100 ng of DNA, 1X buffer, 1.5 nM
MgCl2, 0.2 mM dNTPs, 0.2 µM primer F, 0.2 µM primer R,
1.25 units of DNA polymerase and nuclease-free water. Initial
denaturation was set at 94°C for 3 min, then 30 cycles of
denaturation at 94°C for 30 sec, annealing 40 sec at 60°C for
primers F and R1 or at 55°C for primers F and R2, extension at 72°C
for 1 min. A final extension was included at 72°C for 5 min. The
reaction was performed in a Thermocycler Eppendorf Mastercycle EP
gradient S Thermal (Eppendorf, Hamburg, Germany). The PCR products
were analyzed by electrophoresis on a 1.5% agarose gel and stained
with GelGreen™ (Biotium, Fremont, CA, USA). Primers were designed
using the BRCA1 DNA sequence with accession no. L78833.1
from the GenBank of the National Center of Biotechnology
Information (NCBI; http://www.ncbi.nlm.nih.gov/nuccore/1698398/).
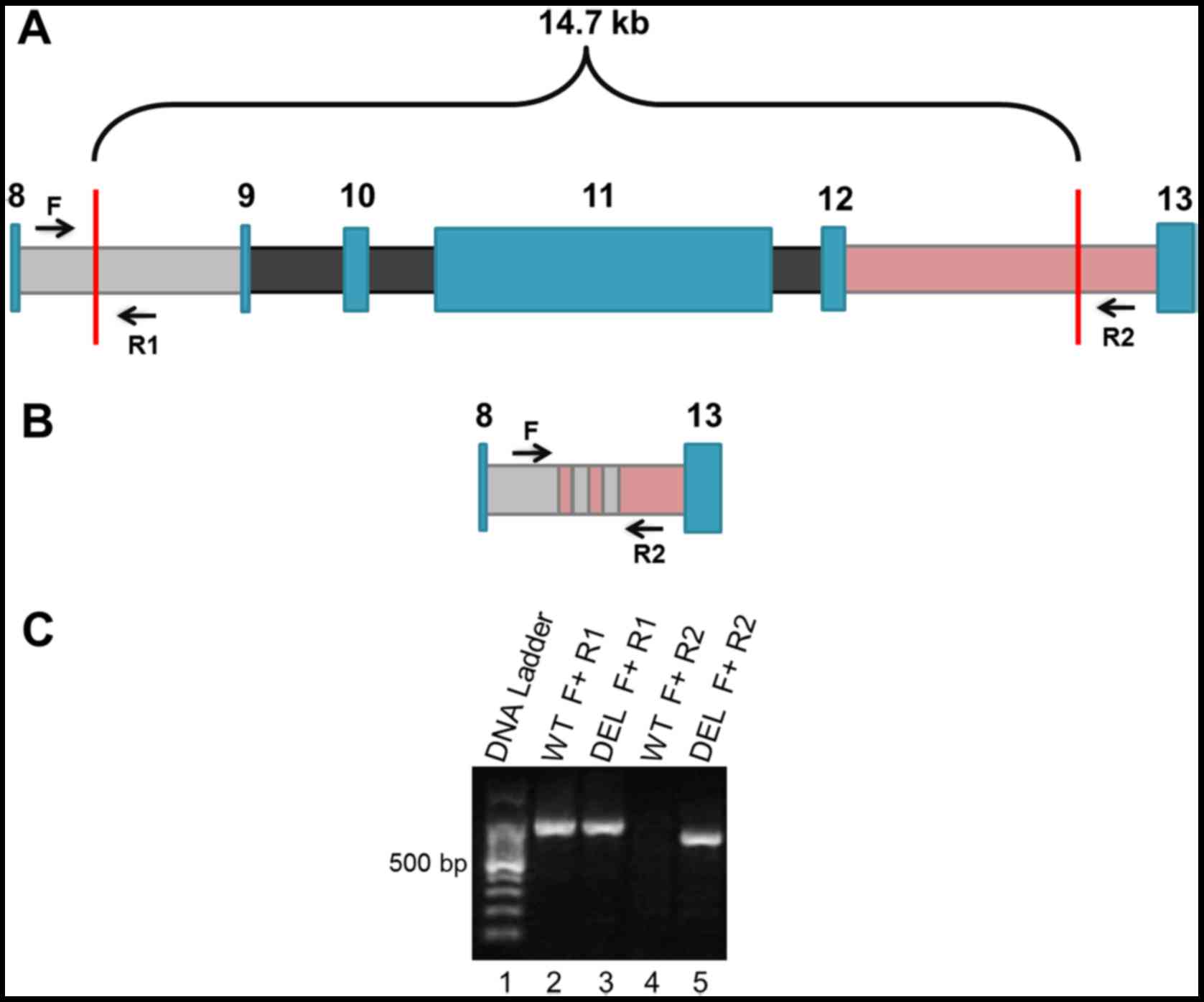 | Figure 1.Location of the BRCA1
ex9-12del mutation and design of the end-point PCR method for its
detection. (A) Wild-type allele of the BRCA1 gene showing
exons (blue color) and introns (8 gray color; 9, 10 and 11 black
color, 12 pink color). In the BRCA1 ex9-12del mutation, the
missing DNA includes exons 9 to 12 (14.7 kb) in one single copy of
the gene (heterozygous), having the breakpoint somewhere in introns
8 and 12 (vertical red lines). Black arrows indicate the design and
position of the primers used for the end-point PCR assay (F, R1 and
R2). (B) Variant allele with the deletion of exons 9–12 and the
recombination of introns 8 and 12 (pink/gray zone). Black arrows
indicate position of primers F and R2. (C) Agarose gel
electrophoresis after the end-point PCR of a sample carrying the
mutation (DEL) and a wild-type sample (WT). Lane 1, DNA ladder.
Lanes 2 and 3 represent the amplification of samples WT and DEL
with primers F and R1, showing an amplified product corresponding
to a region of the wild-type allele (~840 bp). Lanes 4 and 5 are
the PCR products of a WT sample and a DEL sample respectively, with
primers F and R2, amplifying the allele with deletion only (~630
bp). This PCR fragment can be seen only in samples DEL. Primers F
and R2 in a wild-type allele would give an amplified PCR product
>14.47 kb, an impossible task for our end-point PCR. PCR,
polymerase chain reaction. |
Sequencing of the recombination region
between introns 8 and 12 of BRCA1
The PCR product amplified with primers F and R2 of a
sample from a BRCA1 ex9-12del mutation was purified with
QIAquick kit (Qiagen, Hilden, Germany) following manufacturer's
instructions. The sample was sequenced in a Genetic Analyzer 3100
(Thermo Fisher Scientific, Inc.) by an external supplier, the
Instituto Potosino de Investigación Científica y Tecnológica
(IPICYT, San Luis Potosí, Mexico). Sequences were analyzed with
CodonCode Aligner software (CodonCode Corp., Centerville, MA, USA).
The obtained recombination sequence was the same as the one
previously published in 2007 when the BRCA1 ex9-12del
mutation was reported (15).
Design of the qPCR primers and
TaqMan® probes
Using the recombination sequence, primers were
designed to amplify a 164 bp fragment flanking this recombination
site. Therefore, the F primer was designed to bind to a sequence in
intron 8, and the R primer to intron 12. Melting temperature of
both primers was 60°C. Primers were tested in samples with the
BRCA1 ex9-12del mutation, resulting in the amplification of
a single 164 bp DNA fragment. The TaqMan® probe
complementary to the recombination region was designed using the
software Custom qPCR Probes from Integrated DNA Technologies (IDT,
Coralville, IA, USA). The probe was ordered with FAM™
dye and the quenchers ZEN™/IB®FQ.
As a DNA control, a PCR was included to amplify a
region of exon 11 from BRCA1. Design of primers and
TaqMan® probe specific for exon 11 was carried out using
the reference sequence (L78833.1) the same way as for the
BRCA1 ex9-12del mutation, but HEX™ dye was used
instead.
qPCR with designed primers and
TaqMan® probes
Thermocycler StepOnePlus™ Real-Time PCR
System (Thermo Fisher Scientific, Inc.) was used to perform the
qPCR with TaqMan® probes. The PCR was prepared with 1X
TaqMan® Universal PCR Master Mix (Thermo Fisher
Scientific, Inc.), 0.2 µM primer F, 0.2 µM primer R, 250 nM probe,
10 ng DNA, and nuclease-free water to a total volume of 15 µl.
Thermal cycling conditions were as follows: 60°C for 30 sec, 50°C
for 2 min, 95°C for 10 min, 40 cycles of 95°C for 15 sec and 60°C
for 1 min. A final post-read method was set at 60°C for 30 sec. The
passive reference dye included in the TaqMan® Universal
Master Mix was ROX.
Analysis of results
The fluorescent signal of the TaqMan®
qPCR was analyzed using the StepOnePlus™ software v2.3
(Thermo Fisher Scientific, Inc.) under presence/absence and
genotyping methods. In the presence/absence analysis, a positive
result for a deleterious mutation (BRCA1 ex9-12del) was the
amplification signal of the recombination region given by the probe
carrying the FAM™ dye, displayed in an amplification
plot. We considered a positive signal at a Ct between 27 and 27.5.
As a control we used the amplification signal of the exon 11 given
by the probe carrying the HEX™ dye.
TaqMan® Genotyper software was used to
analyze raw data from qPCR in an allelic discrimination (AD) plot.
The regular AD plot shows each sample well as an individual dot.
The AD plot can show different clusters of dots such as homozygous
and heterozygous samples, no-template controls (NTC) and
undetermined samples. The dots of each cluster are grouped closely
and each cluster is located far from the others. In a typical AD
plot, the cluster of homozygous samples for allele 1 labeled with
HEX™ dye is located in the lower right corner. The
cluster of homozygous samples for the allele 2 labeled with
FAM™ dye is located in the upper left corner and the
cluster of heterozygous (allele 1/allele 2) samples is located
approximately midway between both homozygous clusters. NTC samples
are located in the lower left corner. The undetermined samples are
represented by an X and are located in any region of the AD plot
outside of the described above. Nonetheless, the clusters displayed
in the AD plot from our TaqMan® qPCR assay are
different, as explained below. Carriers of at least a single copy
of the wild-type allele (exon 11) are located in the lower right
corner (HEX™). Carriers of at least one allele with the
BRCA1 ex9-12del mutation are clustered in the upper left
corner (FAM™).
Results
DNA samples
A total of ten DNA samples were subjected to
different experiments in order to detect the BRCA1 ex9-12del
mutation. Four of them were previously known as carriers of the
BRCA1 ex9-12del mutation, three obtained from females and
one from a male. Three of these samples of mutation carriers were
family members (mother, daughter and son) and one from an unrelated
female. Samples used as controls were obtained from six unrelated
females not suspected to carry the BRCA1 ex9-12del
mutation.
End-point PCR
Positive and negative samples of the BRCA1
ex9-12del mutation were analyzed with the end-point PCR method. A
unique band of ~630 bp was observed in the agarose gel with primers
F and R2 in the four positive samples. This band was not observed
in the six negative samples (Fig.
1).
qPCR with TaqMan® probes
using the presence/absence method
The thermocycler allows performing a PCR under
different types of methods, such as presence/absence, genotyping
and relative standard curve. The positive and negative samples of
the BRCA1 ex9-12del mutation were analyzed with qPCR and the
TaqMan® probe complementary to the recombination region
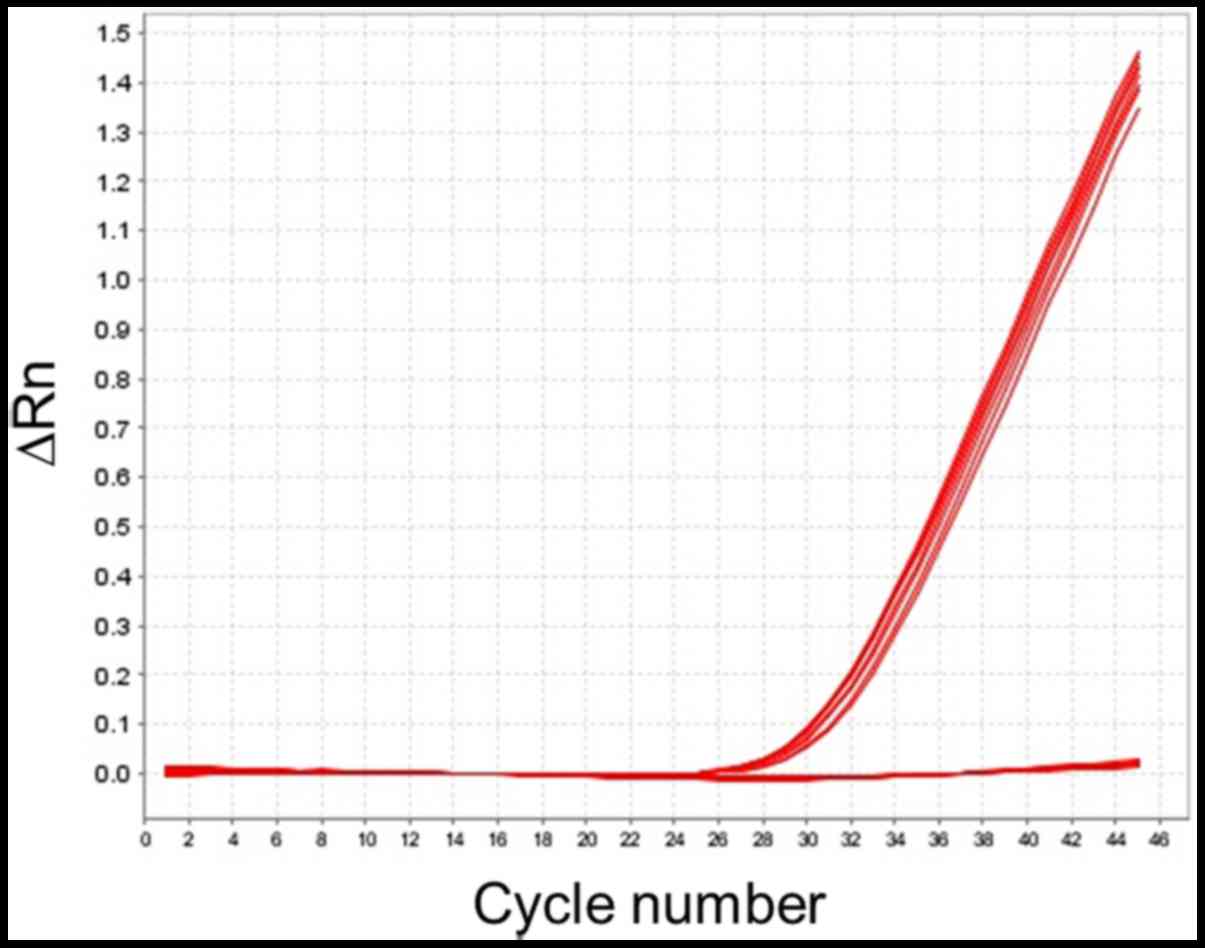
under the presence/absence method. Four DNA samples showed
amplification signal from FAM™ dye, indicating the
presence of the BRCA1 ex9-12del mutation. These positive
samples showed amplification curves at Ct mean 27.46 (Fig. 2). This signal was absent in the six
control samples.
qPCR with TaqMan® probes
using the genotyping method
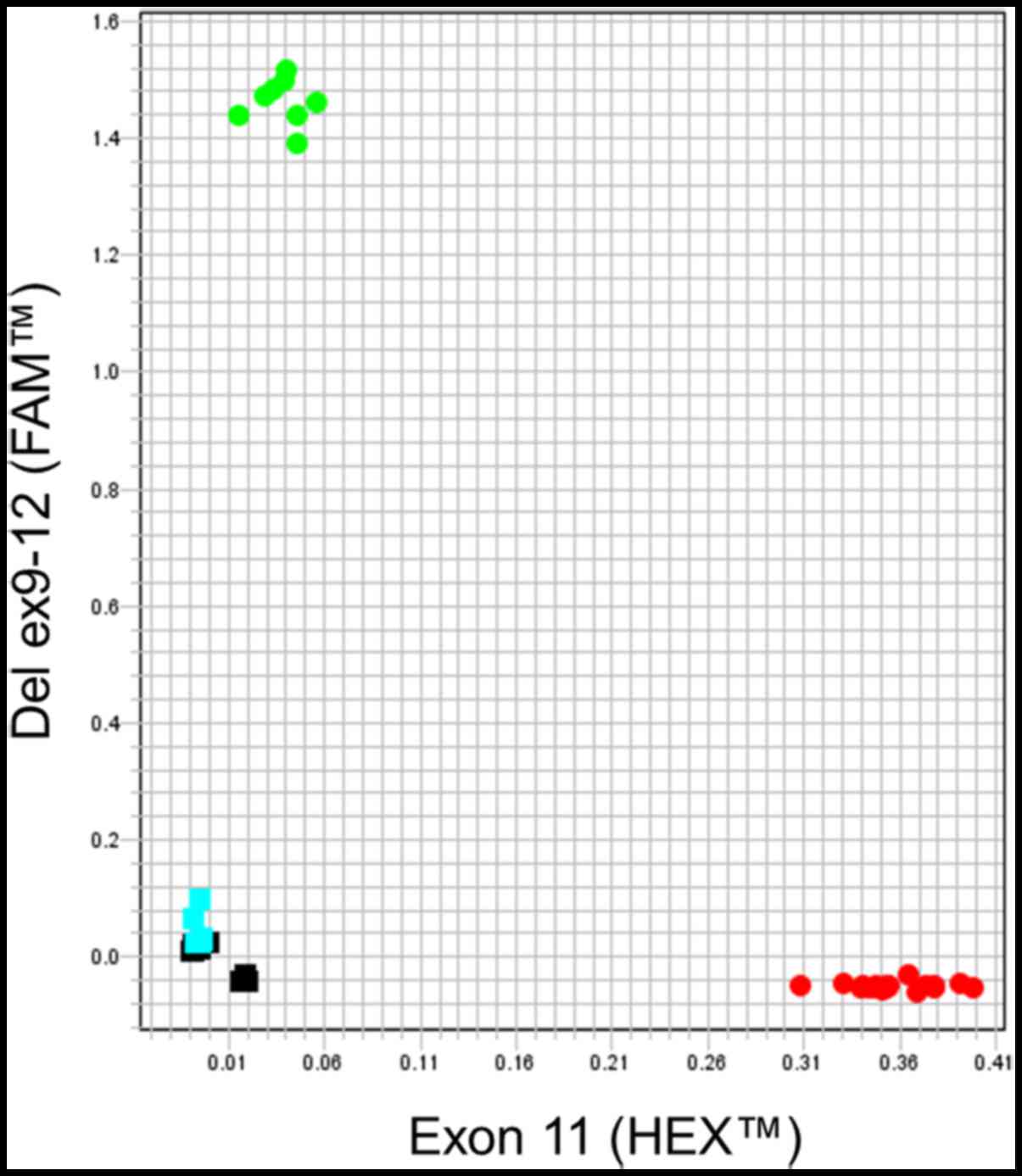
In order to identify heterozygous from homozygous
carriers in a single analysis, the ten DNA samples were tested by
the genotyping method. Six subjects showed fluorescent signal only
from HEX™ dye, meaning they carry only the wild-type
allele (exon 11). In the AD plot, these samples are located in a
single cluster in the lower right corner (Fig. 3). Four subjects showed both
fluorescent signals, from HEX™ and FAM™ dyes,
indicating the presence of exon 11 and the BRCA1 ex9-12del
mutation, respectively. Thereby, these are heterozygous samples
which were located in two clusters, one corresponding to the
HEX™ signal (lower right corner) and one to the
FAM™ signal (duplicates in the upper left corner)
(Fig. 3).
Discussion
Currently, different methods are used for the early
detection of cancer, such as analysis of circulating tumor cells
(CTCs), as well as RNA exosomes (27), proteins (3) and genomic profiles of circulating
cell-free tumor DNA (ctDNA) from liquid biopsy (27,28).
Although the application of these methods can be useful to improve
prognosis, there are still some limitations (28). For example, they are more effective
for early detection of certain types of cancer, such as ovarian
cancer, but less effective to detect breast cancer (29). Therefore, there are no well-defined
biomarkers for all types of cancer (28). However, the effectiveness of breast
cancer detection can be improved by including tests that identify
mutations in the BRCA genes (30).
The inclusion of BRCA screening in the health
care system is highly affected by the cost of BRCA testing
(30). Therefore, it would be
desirable to have alternative low-cost options for the detection of
mutations. The proposed method for the detection of the
BRCA1 ex9-12del mutation is based on qPCR and
TaqMan® probes. This method was capable of detecting
four positive BRCA1 ex9-12del mutation carriers, which were
previously analyzed by our end-point PCR method. This was the
expected result, since the probe was designed to be complementary
to the mutant allele, specifically in the recombination region
between introns 8 and 12, which is a unique sequence in the human
DNA.
Among the main advantages of this method is the low
cost, which is 2–3 USD per sample, compared to a price of ~500 USD
for the determination of a specific mutation. High prices are a
limitation for those who wish to know their genetic status
regarding BRCA1 ex9-12del mutation. Using qPCR allows high
sensitivity since the reaction can be carried out only with 2.5 ng
of DNA (31). In addition, a
TaqMan® probe allows high specificity since it binds
only to the sequence to which is complementary, even if a single
base pair differs from the probe and the DNA region. The fact that
a small amount of DNA is required for the assay gives an extra
strength to this method, since DNA can be extracted from small
biological samples taken in a non-invasive manner, such as urine,
epithelial cells from the mouth or hair roots. Peripheral blood
guarantees a high DNA yield, but its extraction is painful and
uncomfortable for the patient, especially when the quality of the
veins is not good due to prior chemotherapy. It is a fast method
since in 2 h a total of 90 samples can be analyzed in a single
experiment, including the interpretation.
Biallelic mutations in both BRCA genes are
rare (32,33). Therefore, deleterious mutations in
these genes are mainly heterozygous (34). However, biallelic pathogenic
mutations have been reported in individuals with ovarian cancer.
Nevertheless, those patients are compound heterozygous, this is,
they carry mutations in both BRCA1 copies, but in different
regions (for example: c.2457delC/c.5207T>C) (33). Biallelic mutations in BRCA
genes could increase the risk of cancer, as well as leading to more
severe phenotypes (32,35). To date, no individual has been
found homozygous for the BRCA1 ex9-12del mutation. Most
probably the latter would not be compatible with life since great
quantity of genetic material is lost. However, it is important to
detect any region of the wild-type allele at the same time that we
detect the mutant allele. This is also helpful as a control of DNA
presence in the sample. Consequently, we also designed an assay
based on qPCR and a TaqMan® probe to detect a region
from exon 11 of BRCA1.
When performing a TaqMan® qPCR run under
the genotyping method, the amplification of both variant and
wild-type alleles is ideally carried out in the same reaction well;
this is, in a multiplex fashion. Nonetheless, in our study, we
performed the PCR of the variant allele and the PCR of the
wild-type allele in separate reaction wells. So, a sample will be
defined as heterozygous only if it appears in both clusters in the
AD plot (Fig. 3). Consequently, as
future study, we aim to standardize a multiplex qPCR capable of
detecting the variant allele (recombination region) and the
wild-type allele (a region from exons 9 to 12) in one single
reaction, saving time and reagents.
Since BRCA1 ex9-12del mutation is considered
as a founder mutation, and the existing reports have shown that the
recombination region is the same among all carriers of this
mutation; our method is considered highly specific because the
TaqMan® probe binds to this recombination DNA sequence.
However, if a different recombination region is discovered, or if
any additional mutations arise in this region, our method would
have to be modified accordingly and readapted to the newly reported
genetic variants.
Compared to the reported methods used to detect the
BRCA1 ex9-12del mutation, our propose represents a suitable
option for testing given the advantages already mentioned.
The software of the qPCR thermocycler analyzes the
results at the end of the run, displaying the presence or absence
of the mutation and therefore, making the interpretation an easy
step. Moreover, working with qPCR avoids the risk of
post-amplification contamination, since only one process is needed.
On the other hand, end-point PCR requires a first step of
amplification, followed by the run of the samples on an agarose
gel, a process in which the samples can be cross-contaminated
(36). Comparing to the PCR method
reported by Weitzel et al, in 2007 (15), our design requires less DNA input,
2.5 ng (31), whereas the Weitzel
method requires 50–100 ng. Additionally, it does not require
further manipulation of the sample, for example purification of the
amplified product, sequencing-PCR and capillary electrophoresis.
The analysis of results is faster and the identification of the
heterozygous carriers is an easier task (15). Another method used to detect
deletions is MLPA, which has the great advantage of detecting more
than one mutation since it analyzes different regions of the genome
simultaneously. However, for the detection of the BRCA1
ex9-12del mutation, this method has the disadvantage of requiring a
large amount of DNA input (250–450 ng), increased sample handling,
carrying an additional risk of cross-contamination. It also
requires capillary sequencer and the analysis is more complex
(37) compared to our
TaqMan® real-time method. A comparison between current
methods for the detection of the BRCA1 ex9-12del mutation is
summarized in Table I.
 | Table I.Comparison between qPCR, end-point
PCR and MLPA techniques for the detection of the BRCA1
ex9-12del mutation (15,31,36,15). |
Table I.
Comparison between qPCR, end-point
PCR and MLPA techniques for the detection of the BRCA1
ex9-12del mutation (15,31,36,15).
| Variable | qPCR and
TaqMan® probes | End-point PCR | MLPA |
|---|
| Required
equipment | Real-time PCR
thermocycler | Conventional PCR
thermocycler, electrophoresis chambers, electrophoresis power
supply, microwave oven and UV transilluminator to visualize agarose
gels. | Conventional PCR
thermocycler and capillary sequencer. |
| Amount of DNA
required per reaction | 2.5 ng | ≥80 ng | 50-250 ng |
| Processing time
until results (excluding | 2 h | 3.5 h | 24 h |
| DNA extraction)
Sample number to be simultaneously processed (fitting to the
processing time) | 90 | 25 | 96 |
The method we propose will allow the determination
of a high number of samples at the same time, which will be a great
advantage in future studies of association or prevalence.
Additionally, it is a safe, specific and more affordable option for
those wishing to be tested for the mutation.
Acknowledgements
The authors would like to thank Professor Marcelino
Aguirre Garza from Center of Molecular Diagnostics and Personalized
Medicine, Department of Basic Sciences, Division of Health
Sciences, University of Monterrey, San Pedro Garza Garcia, Nuevo
Leon, Mexico, for performing the blood extraction procedure of the
participants.
Funding
The present study was supported by the University of
Monterrey (grant no. UIN-17528).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DAMT designed the sequences of primers and probes,
performed DNA isolation, PCR experiments and wrote the manuscript.
RBRLC conceived the study, directed, planned and analyzed the data
and wrote manuscript. CVG and DAM performed the participants'
selection, clinical evaluation and interpreted the results. EAM and
HABS analyzed the data, interpreted the results and critically
reviewed the manuscript.
Ethics approval and consent to
participate
The protocol was approved by Ethics, Research and
Biosecurity Committees of the University of Monterrey, San Pedro
Garza Garcia, Nuevo Leon, Mexico, registry no. 01072017-CIE.
Written informed consent was obtained from all subjects.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
National Institute of Statistics and
Geography (INEGI): Main causes of mortality by habitual residence,
age groups and gender of the deceasedQuery results: Basic
tabulations. INEGI; Mexico City: 2015, http://www.inegi.org.mx/est/contenidos/proyectos/registros/vitales/mortalidad/tabulados/pc.asp?t=14&c=11817November
15–2017
|
|
2
|
National Institute of Statistics and
Geography (INEGI): General Mortality. INEGI; Mexico City: 2015,
http://www.inegi.org.mx/sistemas/olap/Proyectos/bd/continuas/mortalidad/MortalidadGeneral.aspNovember
15–2017
|
|
3
|
Tajmul M, Parween F, Singh L, Mathur SR,
Sharma JB, Kumar S, Sharma DN and Yadav S: Identification and
validation of salivary proteomic signatures for non-invasive
detection of ovarian cancer. Int J Biol Macromol. 108:503–514.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Doubeni CA, Doubeni AR and Myers AE:
Diagnosis and management of ovarian cancer. Am Fam Physician.
93:937–944. 2016.PubMed/NCBI
|
|
5
|
Aguilar-Cordero MJ, González-Jimenez M,
Álvarez-Ferre J, Padilla-López CA, Mur-Villar N, García-López PA
and Valenza-Peña MC: Breast feeding: An effective method to prevent
breast cancer. Nutr Hosp. 25:954–958. 2010.(In Spanish). PubMed/NCBI
|
|
6
|
Lacey JV Jr, Kreimer AR, Buys SS, Marcus
PM, Chang SC, Leitzmann MF, Hoover RN, Prorok PC, Berg CD and
Hartge P: Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer
Screening Trial Project Team: Breast cancer epidemiology according
to recognized breast cancer risk factors in the Prostate, Lung,
Colorectal and Ovarian (PLCO) Cancer Screening Trial Cohort. BMC
Cancer. 9:842009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ligibel J: Obesity and breast cancer.
Oncology (Williston Park). 25:994–1000. 2011.PubMed/NCBI
|
|
8
|
Luan NN, Wu QJ, Gong TT, Vogtmann E, Wang
YL and Lin B: Breastfeeding and ovarian cancer risk: A
meta-analysis of epidemiologic studies. Am J Clin Nutr.
98:1020–1031. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Russo J, Balogh GA and Russo IH: Full-term
pregnancy induces a specific genomic signature in the human breast.
Cancer Epidemiol Biomarkers Prev. 17:51–66. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cárdenas-Sánchez J, Bargalló-Rocha E,
Erazo-Valle A, Maafs-Molina E and Poitevin-Chacón A: Mexican
Consensus on diagnosis and treatment of breast cancer. 5th
revision. Masson Doyma México S.A; Mexico City: 2013
|
|
12
|
McCarthy AM and Armstrong K: The role of
testing for BRCA1 and BRCA2 mutations in cancer prevention. JAMA
Intern Med. 174:1023–1024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Levy-Lahad E, Catane R, Eisenberg S,
Kaufman B, Hornreich G, Lishinsky E, Shohat M, Weber BL, Beller U,
Lahad A and Halle D: Founder BRCA1 and BRCA2 mutations in Ashkenazi
Jews in Israel: Frequency and differential penetrance in ovarian
cancer and in breast-ovarian cancer families. Am J Hum Genet.
60:1059–1067. 1997.PubMed/NCBI
|
|
14
|
Narod SA and Rodríguez AA: Genetic
predisposition for breast cancer. BRCA1 and BRCA2 genes. Salud
Publica Mex. 53:420–429. 2011.PubMed/NCBI
|
|
15
|
Weitzel JN, Lagos VI, Herzog JS, Judkins
T, Hendrickson B, Ho JS, Ricker CN, Lowstuter KJ, Blazer KR,
Tomlinson G and Scholl T: Evidence for common ancestral origin of a
recurring BRCA1 genomic rearrangement identified in high-risk
Hispanic families. Cancer Epidemiol Biomarkers Prev. 16:1615–1620.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smith TM, Lee MK, Szabo CI, Jerome N,
McEuen M, Taylor M, Hood L and King MC: Complete genomic sequence
and analysis of 117 kb of human DNA containing the gene BRCA1.
Genome Res. 6:1029–1049. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weitzel JN, Clague J, Martir-Negron A,
Ogaz R, Herzog J, Ricker C, Jungbluth C, Cina C, Duncan P, Unzeitig
G, et al: Prevalence and type of BRCA mutations in Hispanics
undergoing genetic cancer risk assessment in the southwestern
United States: A report from the Clinical cancer genetics community
research network. J Clin Oncol. 31:210–216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Villarreal-Garza C, Alvarez-Gomez RM,
Perez-Plasencia C, Herrera LA, Herzog J, Castillo D, Mohar A,
Castro C, Gallardo LN, Gallardo D, et al: Significant clinical
impact of recurrent BRCA1 and BRCA2 mutations in Mexico. Cancer.
121:372–378. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Torres-Mejia G, Royer R, Llacuachaqui M,
Akbari MR, Giuliano AR, Martinez-Matsushita L, Angeles-Llerenas A,
Ortega-Olvera C, Ziv E, Lazcano-Ponce E, et al: Recurrent BRCA1 and
BRCA2 mutations in Mexican women with breast cancer. Cancer
Epidemiol Biomarkers Prev. 24:498–505. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Villarreal-Garza C, Weitzel JN,
Llacuachaqui M, Sifuentes E, Magallanes-Hoyos MC, Gallardo L,
Alvarez-Gomez RM, Herzog J, Castillo D, Royer R, et al: The
prevalence of BRCA1 and BRCA2 mutations among young Mexican women
with triple-negative breast cancer. Breast Cancer Res Treat.
150:389–394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de la Hoya M, Gutierrez-Enriquez S,
Velasco E, Osorio A, de Abajo Sanchez A, Vega A, Salazar R, Esteban
E, Llort G, Gonzalez-Sarmiento R, et al: Genomic rearrangements at
the BRCA1 locus in Spanish families with breast/ovarian cancer.
Clin Chem. 52:1480–1485. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Torres D, Rashid MU, Seidel-Renkert A,
Weitzel JN, Briceno I and Hamann U: Absence of the BRCA1 del (exons
9–12) mutation in breast/ovarian cancer families outside of Mexican
Hispanics. Breast Cancer Res Treat. 117:679–681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Judkins T, Rosenthal E, Arnell C, Burbidge
LA, Geary W, Barrus T, Schoenberger J, Trost J, Wenstrup RJ and Roa
BB: Clinical significance of large rearrangements in BRCA1 and
BRCA2. Cancer. 118:5210–5216. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cartwright-Smith L: Patenting genes: What
does Association for Molecular Pathology v. Myriad Genetics mean
for genetic testing and research? Public Health Rep. 129:289–292.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Myriad, . BRACAnalysis® Large
Rearrangement Test (BART). FAQ. https://webapps.myriad.com/lib/brac/BART-faq.pdfNovember
30–2017
|
|
26
|
Ewald IP, Ribeiro PL, Palmero EI, Cossio
SL, Giugliani R and Ashton-Prolla P: Genomic rearrangements in
BRCA1 and BRCA2: A literature review. Genet Mol Biol. 32:437–446.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Siravegna G, Marsoni S, Siena S and
Bardelli A: Integrating liquid biopsies into the management of
cancer. Nat Rev Clin Oncol. 14:531–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
National Cancer Institute (NCI), .
National Institute of Health (NHI)Liquid Biopsy: Using DNA in blood
to detect, track and treat cancer. NCI Staff; United States: 2017,
https://www.cancer.gov/news-events/cancer-currents-blog/2017/liquid-biopsy-detects-treats-cancerFebruary
12–2018
|
|
29
|
Cohen JD, Li L, Wang Y, Thoburn C, Afsari
B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, et al:
Detection and localization of surgically resectable cancers with a
multi-analyte blood test. Science. 359:926–930. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
D'Andrea E, Marzuillo C, De Vito C, Di
Marco M, Pitini E, Vacchio MR and Villari P: Which BRCA genetic
testing programs are ready for implementation in health care? A
systematic review of economic evaluations. Genet Med. 18:1171–1180.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Martinez-Trevino DA, Moreno-Trevino MG,
Salinas-Santander M, Wohn L, Herrera-Gonzalez S, Aguirre-Garza M,
Rojas OC and Leon-Cachon RB: Rapid Detection of the GSTM3 A/B
polymorphism using real-time PCR with TaqMan® Probes.
Arch Med Res. 47:142–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sawyer SL, Tian L, Kähkönen M,
Schwartzentruber J, Kircher M; Univeristy of Washington Centre for
Mendelian Genomics; FORGE Canada Consortium, ; Majewski J, Dyment
DA, Innes AM, et al: Biallelic mutations in BRCA1 cause a new
Fanconi anemia subtype. Cancer Discov. 5:135–142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Domchek SM, Tang J, Stopfer J, Lilli DR,
Hamel N, Tischkowitz M, Monteiro AN, Messick TE, Powers J, Yonker
A, et al: Biallelic deleterious BRCA1 mutations in a woman with
early-onset ovarian cancer. Cancer Discov. 3:399–405. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rebbeck TR, Friebel TM, Mitra N, Wan F,
Chen S, Andrulis IL, Apostolou P, Arnold N, Arun BK, Barrowdale D,
et al: Inheritance of deleterious mutations at both BRCA1 and BRCA2
in an international sample of 32,295 women. Breast Cancer Res.
18:1122016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Howlett NG, Taniguchi T, Olson S, Cox B,
Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals
G, et al: Biallelic inactivation of BRCA2 in Fanconi anemia.
Science. 297:606–609. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Armour JA, Barton DE, Cockburn DJ and
Taylor GR: The detection of large deletions or duplications in
genomic DNA. Hum Mutat. 20:325–337. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jeuken J, Cornelissen S, Boots-Sprenger S,
Gijsen S and Wesseling P: Multiplex ligation-dependent probe
amplification: A diagnostic tool for simultaneous identification of
different genetic markers in glial tumors. J Mol Diagn. 8:433–443.
2006. View Article : Google Scholar : PubMed/NCBI
|