Introduction
Lung cancer is the leading cause of
cancer-associated mortality worldwide (1). Identifying novel risk factors is
essential in order to prevent the disease. Abnormal overexpression
of the metabolic enzyme glycine dehydrogenase (GLDC) has been
associated with lung cancer and other types of tumors (2–4).
GLDC is critical for the formation of cancer initiating cells in
non-small cell lung cancer (NSCLC) (2). In animal models, its overexpression
can induce malignant transformation of lung normal cells and
promote the formation of tumors (2). However, there is currently no
perspective study on the associations between GLDC and lung
cancer.
DNA methylation is a covalent chemical modification,
resulting in the addition of a methyl (CH3) group at the
carbon 5 position of the cytosine ring, which is a hallmark of
human diseases such as lung cancer (5–7).
Methylation of DNA at position 5 of the cytosine ring is catalyzed
by DNA methyltransferases (DNMTs) and is the predominant epigenetic
modification in mammals (8). The
mammalian DNMT family includes 4 active members: DNMT1, DNMT3A,
DNMT3B and DNMT3L (9,10). DNMT1 is the most abundant DNMT and
is involved in the maintenance of methylation (8,11,12).
DNMT3 functions as a de novo methyltransferase and consists
of 2 associated proteins encoded by the distinct genes, DNMT3A and
DNMT3B (11). The expression
levels of these DNMTs are reportedly elevated in cancers of the
colon, prostate, breast, liver and in leukemia (13–16).
Aberrant methylation of the tumor suppressive gene (TSG) is an
early event in the development of lung cancer (17,18).
MicroRNAs (miRNA/miRs) are a group of small non-coding RNAs (~22
nucleotides) that regulate gene expression (19–21).
The expressions of miR-29a, −29b and −29c were downregulated in
NSCLCs (7). Expression of the
miR-29 family is inversely associated with DNMTs expression in lung
cancer tissues and the miR-29 family directly targets DNMTs; the
miR-29 family can revert aberrant methylation in lung cancer by
targeting DNMTs (7). In addition,
the enforced expression of the miR-29 family in lung cancer cell
lines restored normal patterns of DNA and promoted the
re-expression of TSGs silenced by methylation (7). However, the regulatory mechanism
associated with miR-29 family expression has not been fully
elucidated. The aim of the study was to assess the association
between serum GLDC and lung cancer risk and study the mechanism
underlying the effects of GLDC in lung cancer.
Materials and methods
Study cohort and serum samples
A nested case-control study was conducted in the
well-characterized Chinese Cohort (22). The project included 300 invasive
lung cancer cases, each of which were matched with 2 controls
(n=600). The participants were recruited in Shandong Cancer
Hospital and Shanghai cancer institute between 1998 and 2013, when
they received physical examination and the physical characteristics
are presented in Table I. Each
participant donated more than one blood sample at the recruitment.
For each case-subject match set, 2 control subjects closest to the
case (based on matching criteria, age at time of sampling) with an
available blood sample were chosen among the appropriate risk sets
consisting of all cohort members alive and free of cancer at the
time of diagnosis of the index case. Serum aliquots of 500 µl were
stored at −180°C for measurements of GLDC. The matching criterion
was age at the time blood was drawn. The Shandong (China) Ethical
Board of the Shandong Academy of Occupational Health and
Occupational Medicine (Shandong, China) approved the present study
and written informed consent was obtained from each individual
recruited.
 | Table I.Baseline characteristics of the study
cohort. |
Table I.
Baseline characteristics of the study
cohort.
|
| Mean |
|
|---|
|
|
|
|
|---|
| Characteristics | Cases (n=300) | Controls (n=600) | aP-value |
|---|
| Age, median
(years) | 60.1 | 59.2 |
|
| Sex (%) |
|
|
|
| Men | 82.7 | 80.2 |
|
|
Women | 17.3 | 19.8 | 0.65 |
| Height (cm,
median) | 170.1 | 169.9 |
0.87 |
| Weight (kg,
median) | 70.1 | 74.5 |
0.23 |
| BMI
(kg/m2, median) | 24.5 | 25.2 |
0.62 |
| Smoking
history |
|
|
|
| No. of
cigarette/day (median) | 22.9 | 17.2 | <0.01 |
| Years of smoking
(median) | 32.9 | 27.3 | <0.01 |
| Asbestos exposure
(%) | 2.4 | 2.1 |
0.02 |
| Daily dietary
intake |
|
|
|
| Total
energy (kcal) | 2,787 | 2,760 |
0.88 |
|
Carbohydrates (g, median) | 285.4 | 297.3 |
0.09 |
| Protein
(g, median) | 101.3 | 102.8 |
0.74 |
|
Saturated fat (g, median) | 54.7 | 53.2 |
0.34 |
| Education (%;
>elementary school) | 19.1 | 21.1 |
0.04 |
| Physical activity
(%; >3 h/week) | 10.7 | 12.1 |
0.67 |
Cell culture
Normal human bronchial epithelial (NHBE) cells were
obtained from the America Type Culture Collection (ATCC; Manassas,
VA, USA) were grown in RPMI-1640 medium (Sigma, Shanghai, China)
containing 10% fetal bovine serum (FBS; Shanghai ExCell Biology,
China) and 100 mg/ml penicillin and streptomycin (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified
atmosphere with 5% CO2 (23).
Pre-miR-29a/-29b/29c/Control miR, GLDC
expressing plasmids/empty vectors and transfection experiments
Pre-miR-29a/-29b/29c/Control miR were purchased from
Ambion, (Thermo Fisher Scientific, Inc.). GLDC expressing plasmids
and empty vectors (mock) were purchased from Tiangen (Beijing,
China). For transfection experiments, the cells were cultured in
serum-free medium without antibiotics at 60% confluence for 24 h,
and then transfected with transfection reagent (Lipofectamine 2000;
Thermo Fisher Scientific, Inc.) according to manufacturer's
instructions. After incubation for 6 h, the medium was removed and
replaced with normal culture medium for 48 h.
Enzyme-linked immunosorbent assay
(ELISA)
ELISA analysis was employed to detect the levels of
serum GLDC protein in study cohort and was performed as described
previously (24). The intra-batch
and inter-batch coefficients of variation for GLDC protein were
4.54 and 8.73%, respectively. Multivariate unconditional logistic
regression was performed to calculate the odds ratios (OR) and
corresponding 95% confidence intervals (95% CI) for lung cancer
occurrence, calculating ORs over the quartile levels and on a
continuous log2 scale of circulating GLDC. The final
multivariate models shown included 3 factors (years of smoking,
asbestos exposure and sex) that affect the exposure and disease
relation ≥10%. Ptrends was calculated by using the
median values of GLDC quartiles.
Western blot analysis
Western blot analysis was performed as described
previously (25). For membrane
incubation the following primary antibodies were used: Rabbit
anti-GLDC (cat. no. ab232989; 1:500), anti-DNMT1 (cat. no. ab87654;
1:500) anti-DNMT3A (cat. no. EPR18455; 1:500), anti-DNMT3B (cat.
no. ab2851; 1:500) and anti-β-actin (cat. no. ab5694; 1:500; all
Abcam, Cambridge, MA, USA) antibodies for overnight incubation at
4°C. Membranes were also incubated with IRDye™-800 conjugated
anti-secondary antibodies (cat. no: ab6721; 1:10,000; Abcam) for 30
min at room temperature. For analysis, β-actin was a loading
control. The specific proteins were visualized using the Odyssey™
Infrared Imaging System (Gene Company, Ltd., Hong Kong, China).
Colony formation assay
The colony formation assay was performed as
described previously (26).
MTT assay
The effect on cell proliferation was assessed using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and was performed
as described previously (27).
Briefly, 1×104 cells were seeded onto 96-well plates and
were incubated for 24 h in a 37°C, 5% CO2 cell culture
incubator. MTT reagent (50 µl; 5 mg/ml) was added to each well and
cells were incubated for a further 4 h. Then the formazan
precipitate was dissolved in 150 µl dimethylsulfoxide and the
absorbance rate was measured in a microplate reader at a wave
length of 570 nm, with the reference wavelength set at 630 nm.
Absorbance was directly proportional to the number of surviving
cells. The viability of the control group.
Microarray analysis of miRNA
Microarray Analysis of miRNA was performed as
described previously (28).
Northern blotting analysis
Northern blot analysis of miRNAs was performed as
described previously (29,30). The probe sequences were as follows:
miR-29a, 5′-TAACCGATTTCAGATGGTGCTA-3′; miR-29b,
5′-AACACTGATTTCAAATGGTGCTA-3′; miR-29c,
5′-TAACCGATTTCAAATGGTGCTA-3′; U6 small nuclear RNA,
5′-CCATGCTAATCTTCTCTGTATCGTTCCAA-3′.
Statistical analysis
The results of the colony formation and MTT assays
were statistically analyzed, using a Student's t-test. Data were
expressed as the mean ± standard error. Spearman correlation was
performed for all other results in order to analyze the association
between serum GLDC protein concentration and other variables (age,
BMI and years of smoking). Multivariate logistic regression
analysis was applied to assess the association between serum GLDC
and lung cancer risk. All P-values presented are 2-sided and
P<0.05 was considered to indicate a statistically significant
difference. Statistical analyses were conducted using SAS software,
version 9.3 (SAS Institute, Inc., Cary, NC, USA).
Results
Higher serum GLDC is associated with
increased risk in lung cancer
Baseline characteristics comparing 300 lung cancer
cases and 600 matched controls are outlined in Table I; the matched variable was age. The
median age was 60.1 years for lung cancer cases and 59.2 years for
matched controls. The majority of the lung cancer cases and matched
controls were men (82.7% for cases and 80.2% for controls). The
cases had a lower average level of education, as well as a higher
proportion of smokers (number of cigarettes per day or years of
smoking) and greater occupational exposure to asbestos associated
with lung cancer risk. However, no differences in body mass index
(BMI) and physical activity were observed between the 2 groups. In
addition, those with lung cancer had a higher energy intake rate as
well as a lower consumption of fruit/carotenoids. In those with
lung cancer, ~33% of cases were adenocarcinomas and 22% of the
cases were squamous cell carcinomas (Table II). All other histological types
of lung cancer accounted for <20% (Table II).
 | Table II.Distribution of lung cancer by
histological type. |
Table II.
Distribution of lung cancer by
histological type.
| Histological
type | Number of cases
(n=282) | Percentage of total
(%) |
|---|
| Squamous cell
carcinoma | 62 | 22 |
| Adenocarcinoma | 94 | 33 |
| Small cell
carcinoma | 40 | 14 |
|
Large/undifferentiated cell carcinoma | 50 | 18 |
| Other carcinoma or
carcinoma unspecified | 21 | 7 |
| Unspecified
morphology | 15 | 5 |
Serum GLDC was positively associated with the
overall risk of lung cancer (Table
III; OR=1.48; 95% CI, 1.01–2.04). The risk was elevated in the
highest quartile (OR=1.59; 95% CI, 1.15–2.54) when compared with
the lowest quartile (OR=0.99; 95% CI, 0.76–1.23). Tables IV and V present the associations between age,
BMI, years of smoking and GLDC in the lung cancer cases and the
matched controls. Serum GLDC levels were positively correlated with
years of smoking (Spearman's ρ=0.81; Table IV); however, the association was
attenuated in the sera of matched controls (Spearman's ρ=0.48;
Table V).
 | Table III.Adjusted odds ratios for lung cancer
by quartile levels and on a continuous log2 scale of
circulating prolactin (n=300). |
Table III.
Adjusted odds ratios for lung cancer
by quartile levels and on a continuous log2 scale of
circulating prolactin (n=300).
|
| Quartile
number |
|
|
|---|
|
|
|
|
|
|---|
| Variable | 1 | 2 | 3 | 4 | Log2
odds ratio (95% CI) |
Ptrend |
|---|
| Ca/Co | 48/145 | 53/148 | 80/159 | 119/148 | 1.48
(1.01–2.04) | 0.01 |
| OR | 1 | 0.99
(0.76,1.23) | 1.15
(078,1.43) | 1.59
(1.15–2.54) |
|
|
 | Table IV.Spearman's correlation coefficients
between age, body mass index, years of smoking and serum glycine
dehydrogenase in patients with lung cancer (n=300). |
Table IV.
Spearman's correlation coefficients
between age, body mass index, years of smoking and serum glycine
dehydrogenase in patients with lung cancer (n=300).
| Variable | Age | BMI | Years of
smoking |
|---|
| Glycine
dehydrogenase | −0.28 | 0.08 | 0.81 |
 | Table V.Spearman's correlation coefficients
between age, body mass, years of smoking and serum glycine
dehydrogenase in matched controls (n=600). |
Table V.
Spearman's correlation coefficients
between age, body mass, years of smoking and serum glycine
dehydrogenase in matched controls (n=600).
| Variable | Age | BMI | Years of
smoking |
|---|
| Glycine
dehydrogenase | −0.16 | 0.21 | 0.48 |
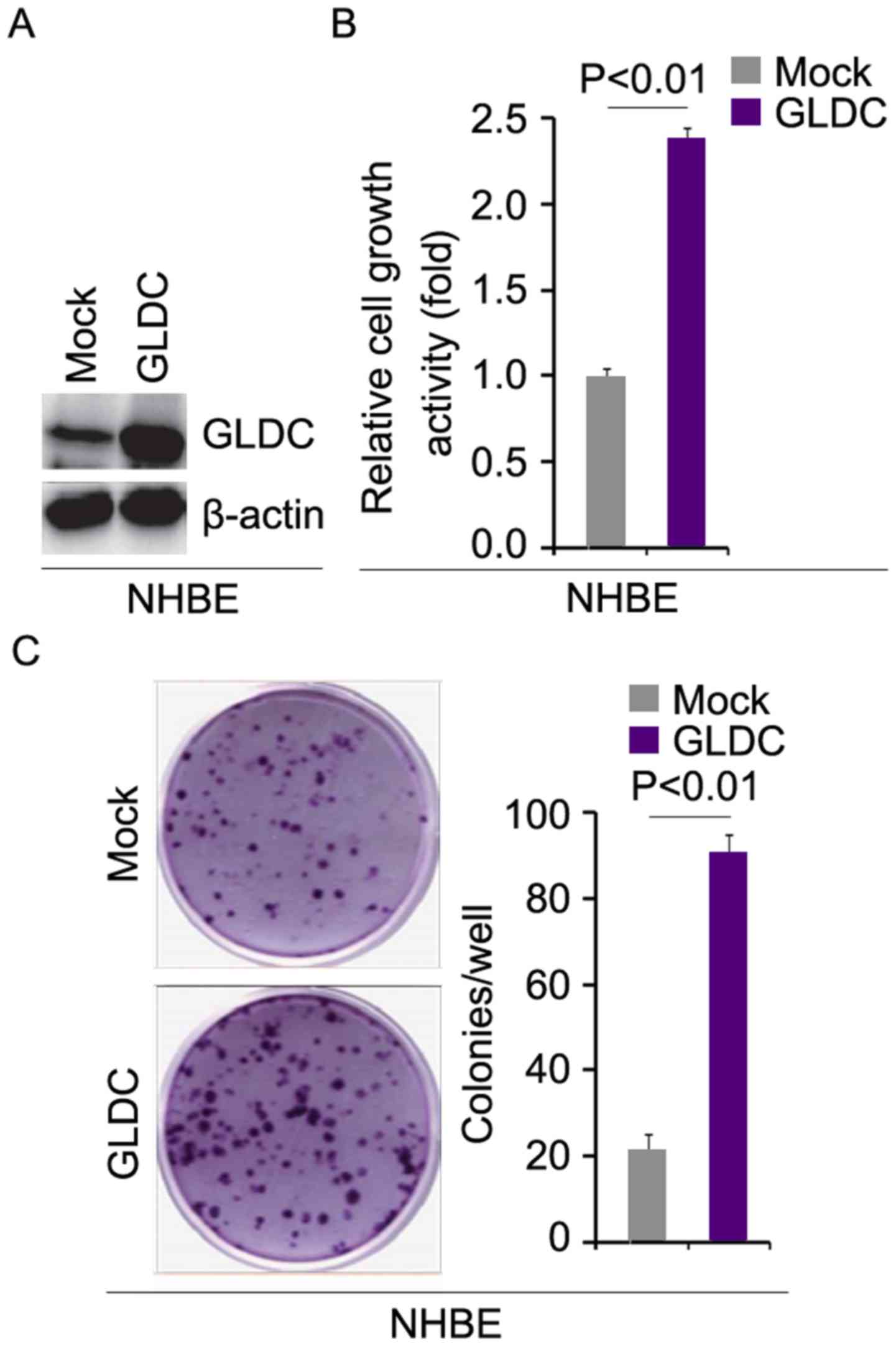
GLDC promotes tumorigenesis in normal
human bronchial epithelial (NHBE) cells
To investigate whether GLDC promotes proliferation
in NHBE cells, western blotting was performed to determine whether
GLDC expressing plasmids can upregulate GLDC protein expression in
NHBE cells. The results of western blotting showed that GLDC
protein was upregulated by GLDC expressing plasmids in cells
(Fig. 1A). To identify the role of
GLDC in regulating proliferation, an MTT assay was performed.
Overexpressing GLDC significantly promoted proliferation in NHBE
cells (Fig. 1B). A colony
formation assay was employed to detect whether GLDC protein
affected the colony formation rate of the cells. The results
demonstrated that GLDC promoted colony formation in NHBE cells
(Fig. 1C).
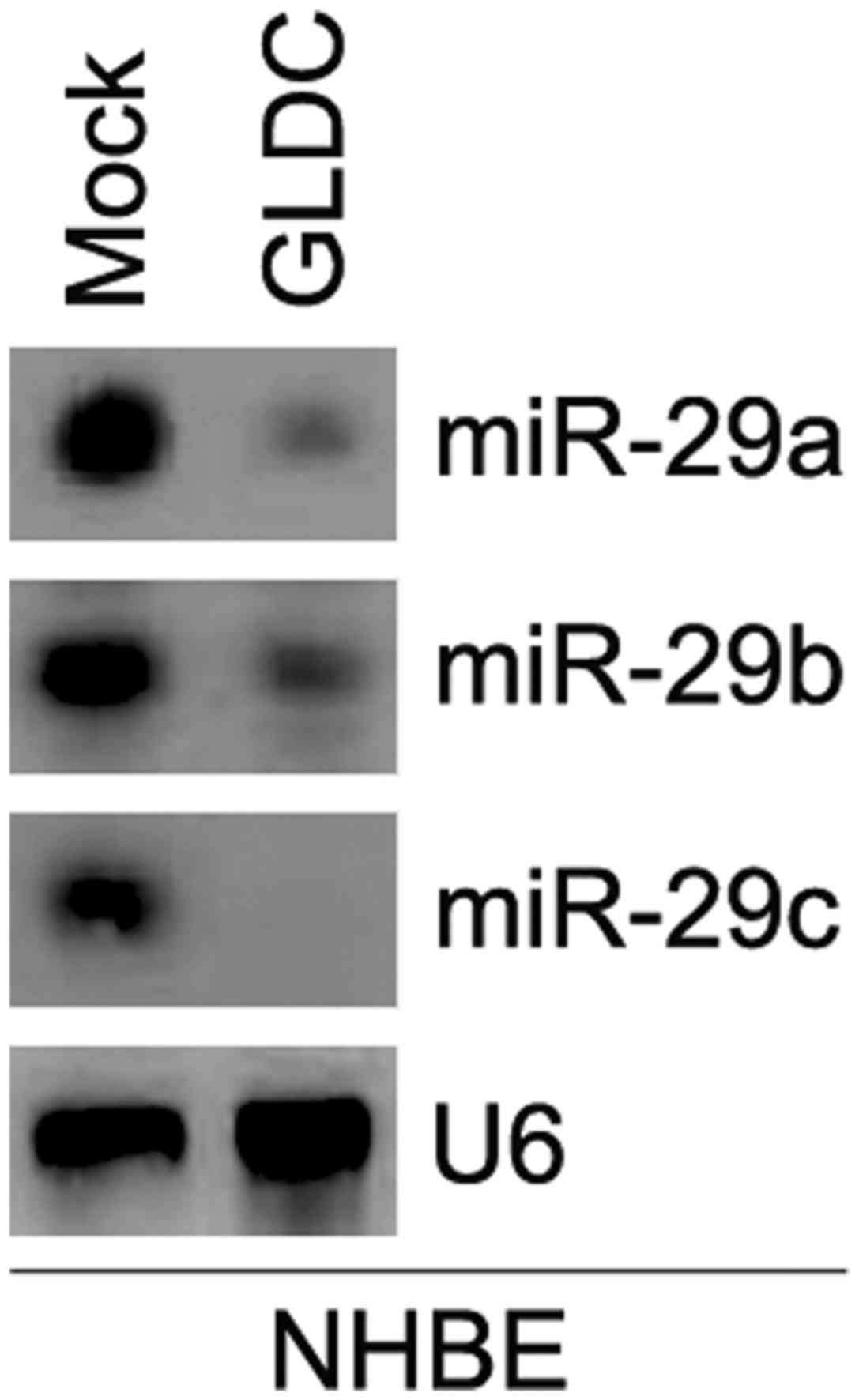
GLDC inhibits miR-29a/b/c expression
in NHBE cells
To examine the role of GLDC in the regulation of
miRNA expression, microarray analysis was performed in NHBE cells.
A total of 19 evidently altered miRNAs were identified; 9 miRNAs
were downregulated and 10 miRNAs were upregulated with >5 fold
changes in cells transfected with GLDC expressing plasmids compared
with control cells (Table VI).
Northern blot analysis was conducted to confirm whether
overexpressing GLDC affected miR-29 a/b/c expression in NHBE cells.
The results revealed that overexpressing GLDC markedly
downregulated their expression in cells (Fig. 2).
 | Table VI.microRNA expression and glycine
dehydrogenase regulation in human bronchial epithelial cells. |
Table VI.
microRNA expression and glycine
dehydrogenase regulation in human bronchial epithelial cells.
| miRNA | Fold Change (GLDC
vs. control)a | P-value |
|---|
| hsa-miR-29a | −105.34 |
3.54×10−4 |
| hsa-miR-29c | −75.07 |
3.01×10−4 |
| hsa-miR-29b | −63.04 |
1.63×10−4 |
| hsa-miR-1226 | −32.85 |
1.67×10−4 |
|
hsa-miR-146b-5p | −22.7 |
4.57×10−4 |
| has-miR-504 | −12.55 |
3.85×10−5 |
| hsa-miR-98 | −12.21 |
7.63×10−5 |
| hsa-miR-30b | −7.08 |
2.45×10−5 |
| has-miR-224 | −6.05 |
4.40×10−4 |
| has-miR-297 | 5.68 |
5.66×10−6 |
|
hsa-miR-1915-5p | 5.84 |
1.53×10−6 |
| hsa-miR-183 | 15.25 |
4.76×10−6 |
| hsa-miR-1268 | 16.88 |
3.68×10−4 |
| hsa-miR-31 | 26.05 |
3.70×10−5 |
| hsa-miR-21 | 27.86 |
4.10×10−6 |
| hsa-miR-572 | 37.38 |
1.09×10−5 |
| has-miR-3188 | 41.89 |
1.31×10−4 |
|
hsa-miR-1225-5p | 43.14 |
2.00×10−5 |
| hsa-miR-328 | 46.30 |
5.48×10−4 |
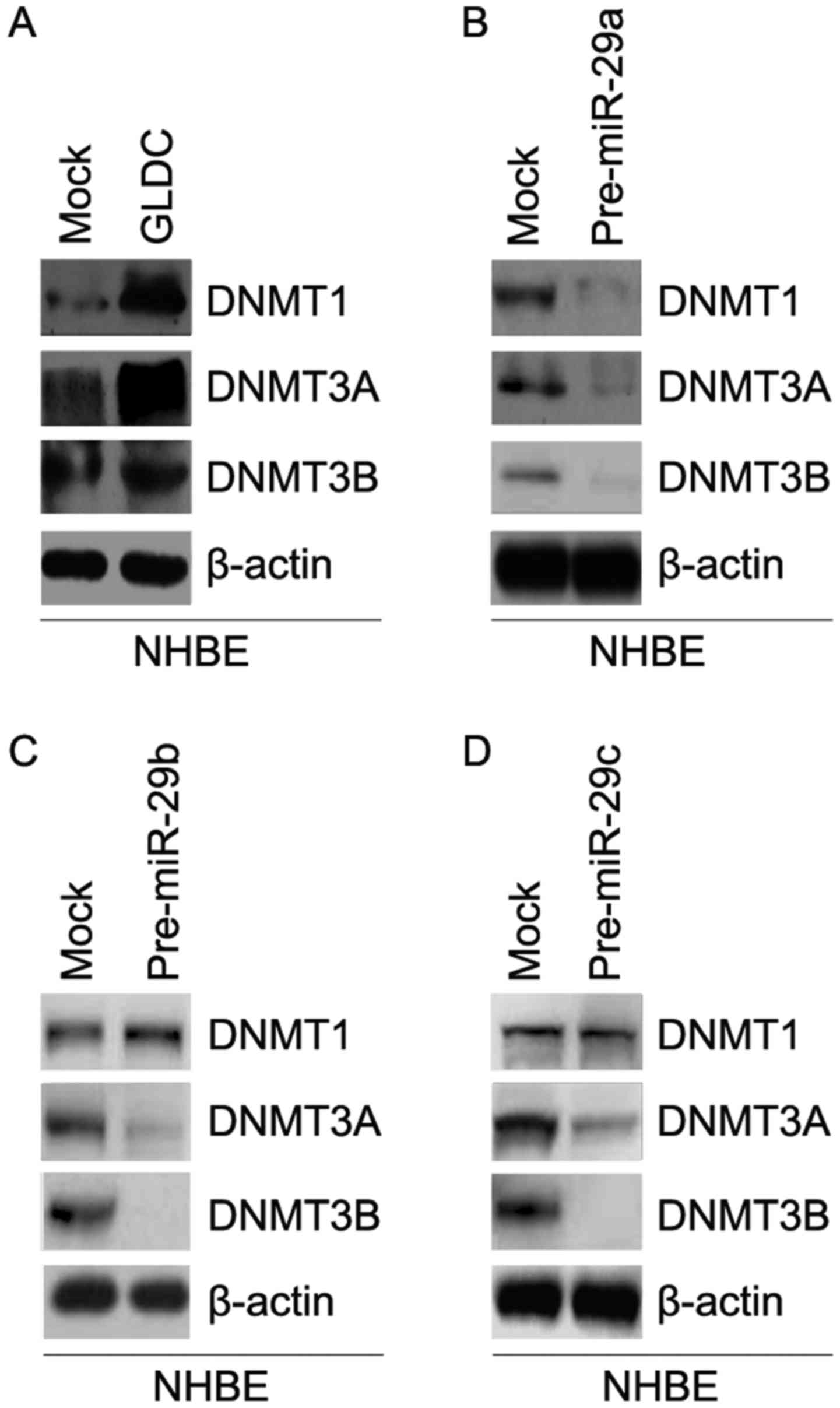
GLDC promotes DNMTs protein expression
and miR-29a/b/c inhibits their expression in NHBE cells
In order to detect whether GLDC can affect DNMT1,
DNMT3A and DNMT3B protein expression, western blot analysis was
performed in NHBE cells transfected with GLDC expressing plasmids
and empty vectors. The results of western blotting showed that
DNMT1, DNMT3A and DNMT3B protein expression were upregulated by
GLDC (Fig. 3A).
In addition, the results of western blotting were
used to detect whether miR-29a/b/c can regulate DNMT1, DNMT3A and
DNMT3B protein expression in NHBE cells. The results showed that
miR-29a inhibited DNMT1, DNMT3A and DNMT3B protein expression in
NHBE cells (Fig. 3B). However,
miR-29b/c only downregulated DNMT3A and DNMT3B protein expression
in NHBE cells (Fig. 3C and D).
GLDC is negatively correlated with
miR-29a/b/c expression in the sera of participants
Having demonstrated that GLDC inhibited miR-29a/b/c
expression in NHBE cells, the present study then used Spearman's
correlation to analyze whether GLDC protein is negatively
correlated with serum miR-29a/b/c expression. The results
demonstrated that GLDC protein was negatively correlated with serum
miR-29a/b/c expression in the serum of participants (Table VII).
 | Table VII.Spearman's correlation coefficients
between serum glycine dehydrogenase and microRNA-29 family in all
participants (n=891a). |
Table VII.
Spearman's correlation coefficients
between serum glycine dehydrogenase and microRNA-29 family in all
participants (n=891a).
| Variable | miR-29a | miR-29b | miR-29c |
|---|
| GLDC | −0.81 | −0.70 | −0.58 |
Discussion
Experimental evidence has revealed that GLDC may be
an oncogene in lung cancer (2);
however, up to now, there has not been a perspective study that
determined whether it can promote the initiation of lung cancer. In
this prospective study, an increased lung cancer risk was
associated with higher serum GLDC concentrations. The results were
in line with previous experimental evidence that demonstrated that
exposure to GLDC can transform normal breast cells and primary NHBE
cells to malignancy-like status (2). In order to reduce lung cancer
mortality, prevention is one of the most effective strategies.
Smoking is an important risk factor for lung cancer (31). Elucidating how carcinogens are
produced by smoking and gaining a better understanding of this
process will improve the scientific basis for the assessment of
mechanisms associated with lung cancer development.
The present study showed that years of smoking were
positively associated with the serum concentration of GLDC, which
can increase lung cancer risk. The results implied that smoking may
promote the initiation of lung cancer by upregulating serum GLDC
concentration. Thus, developing an antagonist for serum GLDC may be
helpful to prevent lung cancer in smokers.
In line with previous perspective and lab results
(2), the present study revealed
that GLDC promoted malignant transformation in NHBE cells.
Increased proliferation and colony formation abilities are
hallmarks of cancer (32). The
results of the present study demonstrated that GLDC can promote
proliferation and colony formation abilities in NHBE cells.
Aberrant methylation of TSG is an early event in the
development of lung cancer (17,18).
DNMTs control changes in methylation and 3 catalytically active
DNMTs (DNMT1, DNMT3A and DNMT3B) have been identified (33). Recently, it has been reported that
miRNA-29a/b/c can revert aberrant methylation in lung cancer by
regulating DNMT3A and DNMT3B (7).
In the present study, GLDC expression in serum, induced by smoking,
was an upstream regulator of the miR-29 family. In addition, GLDC
promoted DNMTs protein expression. Consistent with a previous
report (7), the results of the
present study showed that the miR-29 family may inhibit DNMTs
protein expression in NHBE cells. Thus, the smoking/GLDC/miR-29
family/DNMTs signaling pathway may serve an important role in the
early malignant transformation of normal lung cells.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TSG
|
tumor suppressive gene
|
|
GLDC
|
glycine dehydrogenase
|
|
NSCLC
|
non-small cell lung cancer
|
References
|
1
|
Juergens RA and Brahmer JR: Adjuvant
treatment in non-small cell lung cancer: Where are we now? J Natl
Compr Cancer Netw. 4:595–600. 2006. View Article : Google Scholar
|
|
2
|
Xia J, Wu Z, Yu C, He W, Zheng H, He Y,
Jian W, Chen L, Zhang L and Li W: miR-124 inhibits cell
proliferation in gastric cancer through down-regulation of SPHK1. J
Pathol. 227:470–480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Go MK, Zhang WC, Lim B and Yew WS: Glycine
decarboxylase is an unusual amino acid decarboxylase involved in
tumorigenesis. Biochemistry. 53:947–956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li X, Cui C, Guo Y and Yang G: Glycine
decarboxylase expression increased in p53-mutated B cell lymphoma
mice. Oncol Res Treat. 38:586–589. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jones PA and Baylin SB: The epigenomics of
cancer. Cell. 128:683–692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chuang JC and Jones PA: Epigenetics and
microRNAs. Pediatr Res. 61:R24–R29. 2007. View Article : Google Scholar
|
|
7
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Subramaniam D, Thombre R, Dhar A and Anant
S: DNA methyltransferases: A novel target for prevention and
therapy. Front Oncol. 4:802014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ren J, Singh BN, Huang Q, Li Z, Gao Y,
Mishra P, Hwa YL, Li J, Dowdy SC and Jiang SW: DNA hypermethylation
as a chemotherapy target. Cell Signal. 23:1082–1093. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jin B, Li Y and Robertson KD: DNA
methylation: Superior or subordinate in the epigenetic hierarchy?
Genes Cancer. 2:607–617. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ferguson-Smith AC and Greally JM:
Epigenetics: Perceptive enzymes. Nature. 449:148–149. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miremadi A, Oestergaard MZ, Pharoah PD and
Caldas C: Cancer genetics of epigenetic genes. Hum Mol Genet.
16:R28–R49. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Girault I, Tozlu S, Lidereau R and Bièche
I: Expression analysis of DNA methyltransferases 1, 3A, and 3B in
sporadic breast carcinomas. Clin Cancer Res. 9:4415–4422.
2003.PubMed/NCBI
|
|
14
|
Saito Y, Kanai Y, Nakagawa T, Sakamoto M,
Saito H, Ishii H and Hirohashi S: Increased protein expression of
DNA methyltransferase (DNMT) 1 is significantly correlated with the
malignant potential and poor prognosis of human hepatocellular
carcinomas. Int J Cancer. 105:527–532. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Patra SK, Patra A, Zhao H and Dahiya R:
DNA methyltransferase and demethylase in human prostate cancer. Mol
Carcinog. 33:163–171. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Danenberg PV and Laird PW: CpG island hypermethylation in human
colorectal tumors is not associated with DNA methyltransferase
overexpression. Cancer Res. 59:2302–2306. 1999.PubMed/NCBI
|
|
17
|
Belinsky SA, Nikula KJ, Palmisano WA,
Michels R, Saccomanno G, Gabrielson E, Baylin SB and Herman JG:
Aberrant methylation of p16INK4a is an early event in lung cancer
and a potential biomarker for early diagnosis. Proc Natl Acad Sci
USA. 95:11891–11896. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Soria JC, Lee HY, Lee JI, Wang L, Issa JP,
Kemp BL, Liu DD, Kurie JM, Mao L and Khuri FR: Lack of PTEN
expression in non-small cell lung cancer could be related to
promoter methylation. Clin Cancer Res. 8:1178–1184. 2002.PubMed/NCBI
|
|
19
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
London SJ, Yuan JM, Chung FL, Gao YT,
Coetzee GA, Ross RK and Yu MC: Isothiocyanates, glutathione
S-transferase M1 and T1 polymorphisms, and lung-cancer risk: A
prospective study of men in Shanghai, China. Lancet. 356:724–729.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang A, Wang X, Shan X, Li Y, Wang P,
Jiang P and Feng Q: Curcumin reactivates silenced tumor suppressor
gene RARβ by reducing DNA methylation. Phytother Res. 29:1237–1245.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gehrmann M, Cervello M, Montalto G,
Cappello F, Gulino A, Knape C, Specht HM and Multhoff G: Heat shock
protein 70 serum levels differ significantly in patients with
chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma.
Front Immunol. 5:3072014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang D, Liang T, Gu Y, Zhao Y, Shi Y, Zuo
X, Cao Q, Yang Y and Kan Q: Protein N-arginine methyltransferase 5
promotes the tumor progression and radioresistance of
nasopharyngeal carcinoma. Oncol Rep. 35:1703–1710. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kataoka J, Shiraha H, Horiguchi S,
Sawahara H, Uchida D, Nagahara T, Iwamuro M, Morimoto H, Takeuchi
Y, Kuwaki K, et al: Loss of Runt-related transcription factor 3
induces resistance to 5-fluorouracil and cisplatin in
hepatocellular carcinoma. Oncol Rep. 35:2576–2582. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiang Y, Lu DL, Li JP, Yu CX, Zheng DL,
Huang X, Wang ZY, Hu P, Liao XH and Zhang TC: Myocardin inhibits
estrogen receptor alpha-mediated proliferation of human breast
cancer MCF-7 cells via regulating MicroRNA expression. IUBMB Life.
68:477–487. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gong J, Li J, Wang Y, Liu C, Jia H, Jiang
C, Wang Y, Luo M, Zhao H, Dong L, et al: Characterization of
microRNA-29 family expression and investigation of their
mechanistic roles in gastric cancer. Carcinogenesis. 35:497–506.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu J, Ryan DG, Getsios S,
Oliveira-Fernandes M, Fatima A and Lavker RM: MicroRNA-184
antagonizes microRNA-205 to maintain SHIP2 levels in epithelia.
Proc Natl Acad Sci USA. 105:19300–19305. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peto R, Darby S, Deo H, Silcocks P,
Whitley E and Doll R: Smoking, smoking cessation, and lung cancer
in the UK since 1950: Combination of national statistics with two
case-control studies. BMJ. 321:323–329. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jeltsch A: Beyond Watson and Crick: DNA
methylation and molecular enzymology of DNA methyltransferases.
Chembiochem. 3:274–293. 2002. View Article : Google Scholar : PubMed/NCBI
|