Introduction
Poly[adenosine diphosphate (ADP)-ribose] (PAR)
polymerase (PARP)-1 is the member of the PARP family that is
considered to be the ‘prototypical’ PARP enzyme (1). PARP1 can be activated by DNA strand
breaks and a set of posttranslational modifications [previously
reviewed in (2)]. Active PARP1
cleaves aldehyde dehydrogenase into nicotinamide and ADP-ribose
(ADPR), and forms ADPR polymers, also known as PAR, on different
acceptor proteins (3,4). PAR chains can modify the function of
the acceptors, enabling PAR-mediated regulation of protein
function. PARP1 is responsible for >80% of all cellular PARP
activities (5,6).
PARP1 is widely recognized as a pro-inflammatory
protein in T helper1-mediated pathologies [previously reviewed in
(7–9)]. The pro-inflammatory properties of
PARP1 have numerous molecular roots. Firstly, PARP1 is a vital
positive co-factor of several pro-inflammatory transcription
factors [previously reviewed in (7)], of which the first to be identified
was nuclear factor-κB (NF-κB) (10). In addition, PARP-mediated
epigenetic changes also contribute to the pro-inflammatory
transcriptional properties of PARP1 (11). The induction of these transcription
factors facilitates the production of pro-inflammatory chemokines,
cytokines and lipid mediators (7,11).
These mediators in turn facilitate the chemotaxis of immune cells
and also have a pivotal role in their activation (12,13).
Adhesion factors (such as intercellular adhesion molecule 1 and
vascular cell adhesion molecule 1) that help immune cells enter the
site of inflammation are also expressed in a PARP1-dependent manner
(14). Finally, there are other
factors, including inducible nitric oxide synthase,
cyclooxygenase-2 and certain matrix metalloproteinases that are
activated in a PARP1-dependent manner as well (15,16).
Taken together, immune cell activation, infiltration, cell
migration and oxidative/nitrosative stress are PARP1-dependent.
Notably, the administration of PARP inhibitors to humans also has
an anti-inflammatory effect (17).
Recent advances in sequencing technology have
largely increased current knowledge on the composition of the
microflora (the collective microfloral genome often referred to as
the microbiome) in various regions of the human body (18–22).
Sequencing-based determination of the microbiome not only revealed
novel bacterial species and enabled the study of the microflora,
but also revealed that bodily cavities (such as lower airways),
previously thought to be sterile, do contain bacteria in low
numbers (18–22). Furthermore, these studies have shed
light on the interactions between the host and the microbiome.
Microbes produce metabolites that enter the systemic circulation,
and can affect the cells of the host (23–29)
and interact directly with the components of the innate immune
system (30–32). In turn, the host influences the
microbial communities through the immune system, feeding behavior
and personal hygiene (18).
Changes in the composition of the microbiome have been associated
with particular diseases, including metabolic diseases, autism and
cancer, where the reduction in the diversity of the microbiome
frequently coincides with the onset of the disease (33).
The molecular determinants of the interaction
between the host's immune system and the microbiome are largely
unknown. Previous studies have revealed the role of the innate
immune system, more precisely the Toll-like receptor (TLR) family
(30,31,34,35).
As PARP1 modulates the TLR-mediated signaling (32,36–40),
the present study was performed to investigate the changes in the
microbiome upon the deletion of PARP1.
Materials and methods
Animals
PARP1 knockout mice with a C57BL/6J background were
used (41), and were generated in
Het-to-Het breeding. A total of 6 mice were housed in each cage
(standard block shape 365×207×140 mm, surface 530 cm2;
1284 L Eurostandard Type II. L from Techniplast) with Lignocel
Select Fine (J. Rettenmaierund Söhne, Germany) as bedding. Mice
were housed under a 12 h light/dark cycle at 22±1°C. Mice had ad
libitum access to food and water (sterilized tap water). The
animal facility was overseen by a veterinarian. Male mice (20
animals, 8–12 weeks old, 22–26 g body weight) were randomly
selected from a larger pool of mice bred at the Animal Facility of
the University of Debrecen (Debrecen, Hungary). Randomization
between groups was not possible since group assignment was based on
genotypes (n=20, 10 per experimental group). Animals were fasted 16
h prior to sampling to exclude the effect of potentially different
eating periods and ingested food quantities. Following this, fresh
fecal samples were collected and stored immediately in liquid
nitrogen. Subsequently, animals were sacrificed by cervical
dislocation. Subsequently the initial 15 mm segment of the duodenum
and ~1/3 of the cecum was removed. Both intestinal samples in
addition to a freshly collected fecal pellet were rapidly frozen in
liquid nitrogen immediately following removal. For long term
storage samples were kept at −80°C (41). All animal experiments were approved
by the local and national ethical board of the University of
Debrecen (reg. 1/2015/DEMÁB).
DNA isolation and sequencing
Total DNA was isolated form each sample using the
DNeasy PowerSoil kit according to the manufacturer's protocol (cat.
no. 12888-100; Qiagen GmbH, Hilden, Germany). Subsequently
identical amounts of DNA from each sample within the groups were
pooled; the use of identical quantities of DNA ensures that each
sample contributes equally to the abundance. From the pooled
samples 16S ribosomal RNA genes were amplified and sequenced.
Samples were assessed for quality and potential contaminants on a
1.5% agarose gel. DNA isolation and sequencing were performed by
UD-GenoMed as a commercial service (UD-GenoMed, Debrecen,
Hungary).
Analysis of the microbiome
Sequence fragments were uploaded to the metagenomics
RAST server, MG-RAST (v4.0, metagenomics.anl.gov/) where paired end joining and
microbiome reconstruction was performed (42). Subsequent analysis was performed
with a specialized standalone software Taxamat (v1.04), which is
freely available at www.taxamat.com. Using Taxamat, data representing food
contaminants and host DNA (Viridiplantae and Metazoa)
was removed. To produce the sequencing depth of each sample to a
comparable level, data were downsampled so that samples with higher
abundance matched the samples with the lowest abundance values.
Source files for the sequencing raw and the curated data can be
found at www.ncbi.nlm.nih.gov/bioproject/411773 (NCBI
Bioproject PRJNA411773).
Statistical analysis
Diversity profiles were created using
Palaeontological Statistics (PAST) (43). Diversity indices were calculated
using PAST and Taxamat (www.taxamat.com). When comparing diversity profiles,
the curve data points were downsampled to eight evenly distributed
values over the whole range and statistical significance was
determined using two tailed Student's t-test for paired samples.
Family and order distributions across samples were compared using
the ‘prop.test’ function (www.rdocumentation.org/packages/mosaic/versions/1.1.1/topics/prop.test)
in RStudio (version 0.99.484; www.rstudio.com) (44,45).
Results
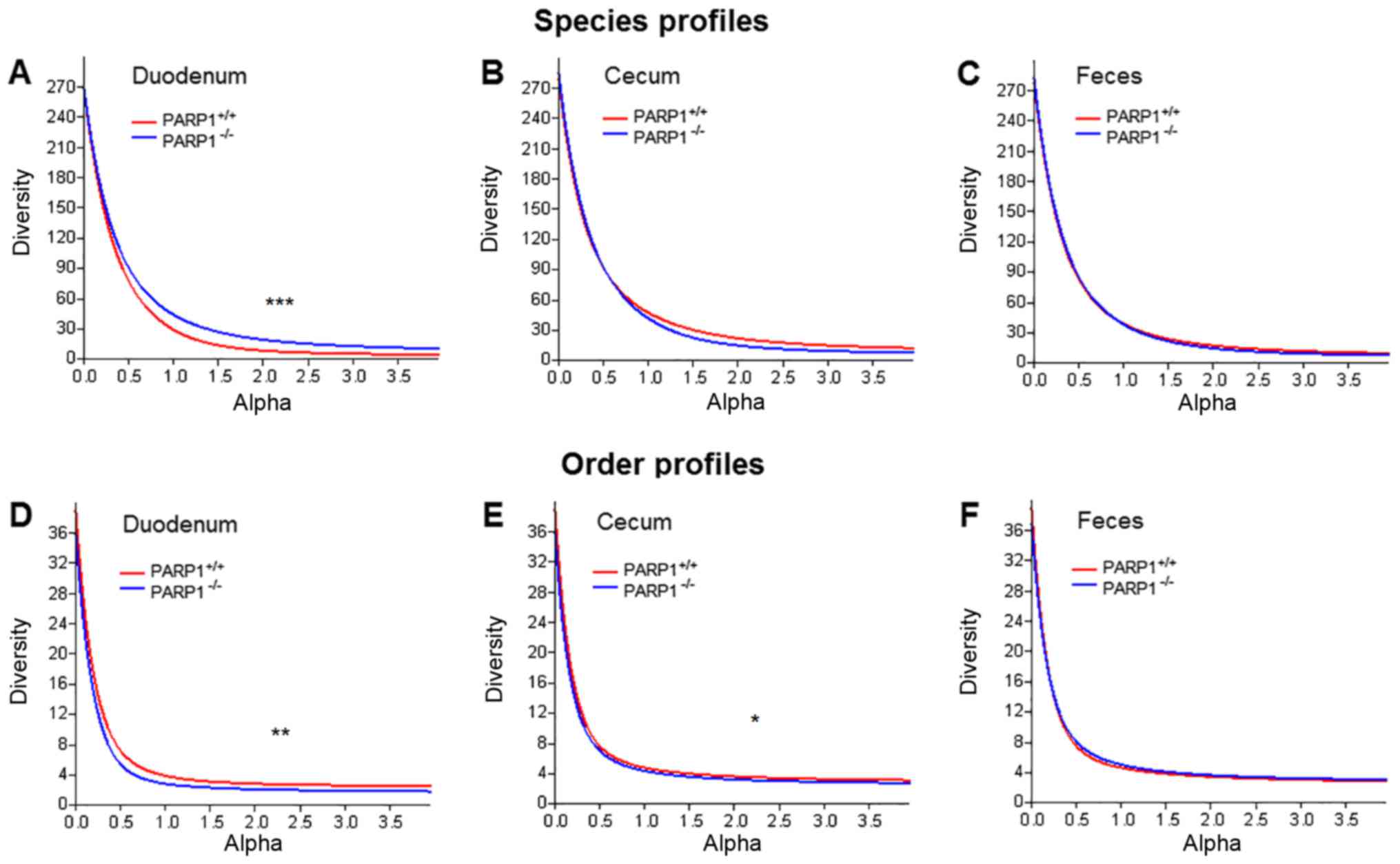
As a first step, changes to microbial diversity were
assessed by comparing the gut samples of the PARP1+/+
and PARP1−/− mice. When comparing diversity indices, the
main pitfall is the arbitrary choice of the used index. To avoid
this issue diversity profiles were plotted in addition to comparing
individual indices (Fig. 1). These
curves use the parameter (α) dependent exponent of the Renyi index
(46). This function returns the
number of taxa at α=0, a number proportional to the Shannon index
at α=1 and a Simpson-like index at α=2. When plotting these
profiles, it was demonstrated that the duodenal samples of
PARP1+/+ and PARP1−/− animals were slightly
different in terms of their diversity profiles (Fig. 1A and D; P<0.001 and P<0.01,
respectively). Notably, on the species level the samples from
PARP1−/− demonstrated higher diversity values while on
order level this trend was the opposite. When comparing diversity
profiles representing the lower gastrointestinal tract (cecum) and
feces, the only significant differences were in the cecal samples
on order level (P<0.05; Fig.
1E). However, even in that case, although statistically
significant, profile curves were very similar. The same trend was
also demonstrated by the traditional Shannon and Simpson indices
(Table I).
 | Table I.Simpson and Shannon indices obtained
in the present study. |
Table I.
Simpson and Shannon indices obtained
in the present study.
|
| Species | Order |
|---|
|
|
|
|
|---|
| Indices | Duodenum | Cecum | Feces | Duodenum | Cecum | Feces |
|---|
| PARP1 | +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | −/− | +/+ | −/− |
| Simpson | 0.883 | 0.948 | 0.955 | 0.933 | 0.942 | 0.931 | 0.651 | 0.528 | 0.726 | 0.693 | 0.710 | 0.727 |
| Shannon | 3.378 | 3.789 | 3.858 | 3.733 | 3.656 | 3.643 | 1.360 | 1.046 | 1.570 | 1.477 | 1.533 | 1.617 |
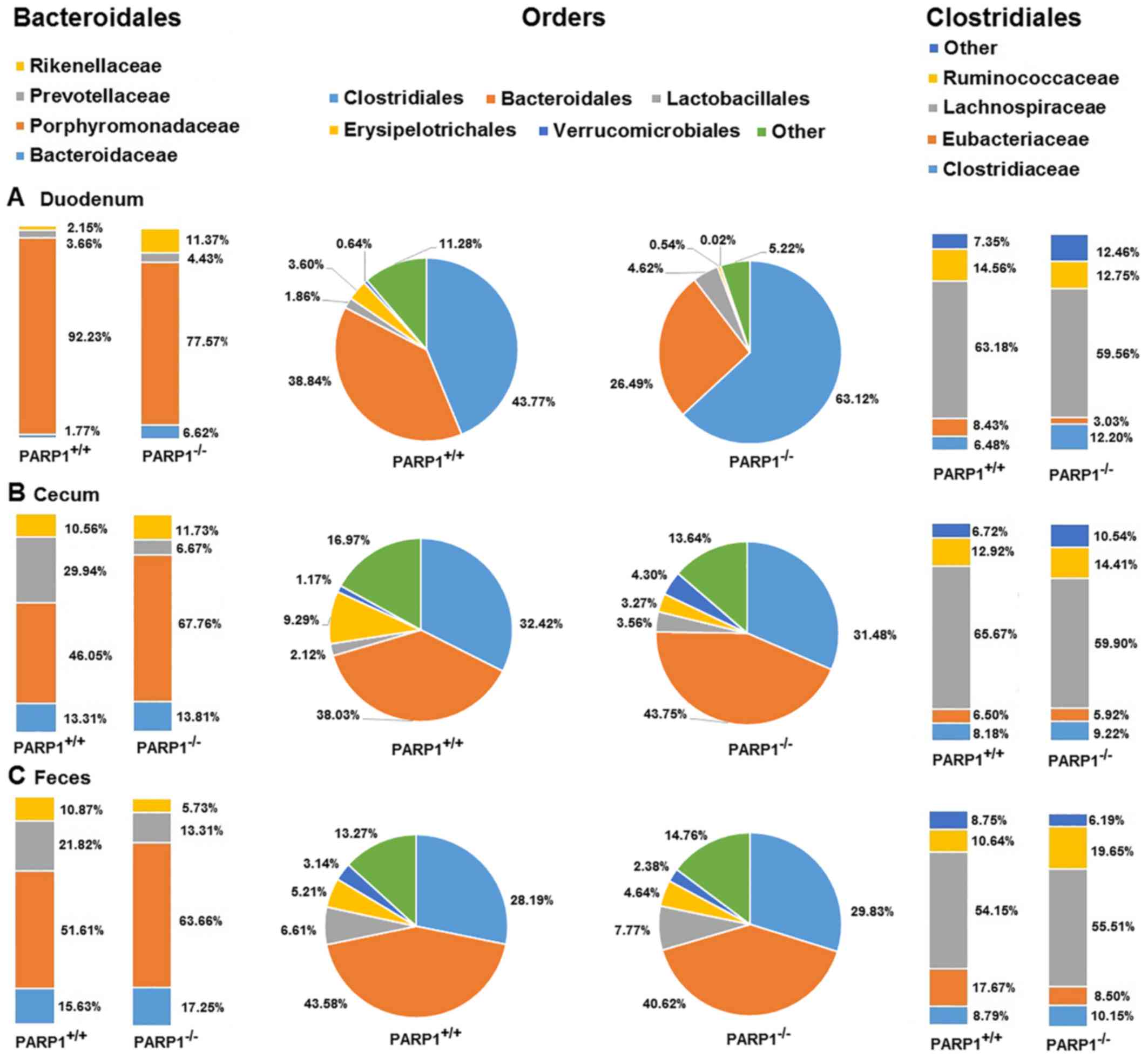
The most abundant orders in all samples were then
investigated. Clostridiales and Bacteroidales represented between
~70 and 90% of all taxa across all samples. The less abundant
orders were Lactobacillales, Erysipelotrichales and
Verrucomicrobiales accounting for a further 5–15% while the rest
(~5–15%) were spread out almost evenly between a further ~30 orders
(Fig. 2). On the order level all
samples appeared to be similar, the only trend worth noting was the
decreased ratio of Clostridiales in cecal and fecal samples when
compared with duodenal ones (Fig.
2, middle panel). The statistical significance levels are
presented in Table II. This was
true for samples from wild type and from PARP1−/−
animals, albeit the duodenal samples from the latter demonstrated a
higher ratio of Clostridiales (~63% in PARP1−/− animals
vs. ~44% in PARP1+/+ animals; Fig. 2A). As these results indicated, the
majority of the other orders proportionally advanced to fill in the
gap left by Clostridiales throughout the gastrointestinal
system.
 | Table II.Taxon proportion significance levels
between compared samples. |
Table II.
Taxon proportion significance levels
between compared samples.
|
| Significance
levels |
|---|
|
|
|
|---|
| Compared
samples | Order | Bacteroidales | Clostridiales |
|---|
| PARP+/+
duodenum vs. PARP+/+ cecum | P<0.05 | P<0.001 | ns |
| PARP+/+
duodenum vs. PARP+/+ feces | P<0.001 | P<0.001 | ns |
| PARP+/+
cecum vs. PARP+/+ feces | ns | P<0.001 | ns |
| PARP−/−
duodenum vs. PARP−/− cecum | P<0.001 | P<0.01 | ns |
| PARP−/−
duodenum vs. PARP−/− feces | P<0.001 | P<0.01 | ns |
| PARP−/−
cecum vs. PARP−/− feces | ns | P<0.05 | ns |
| PARP+/+
duodenum vs. PARP−/− duodenum | P<0.001 | P<0.001 | ns |
| PARP+/+
cecum vs. PARP−/− cecum | ns | P<0.001 | ns |
| PARP+/+
feces vs. PARP−/− feces | ns | P<0.001 | P<0.05 |
Further investigation on the family level revealed
that there was little to no difference in the main composition in
Clostridiales (Fig. 2, right-hand
panel). The main families were Lachnospiraceae, Clostridiaceae,
Eubacteriaceae and Ruminococcaceae. More than half of
the Clostridiales were comprised of members of the
Lachnospiraceae family (54–65%), while the next two,
Clostridiaceae and Ruminococcaceae, were responsible
for a further ~20%. When comparing the family composition of the
different samples, there was little to no changes in these ratios
throughout the gastrointestinal system, nor were any differences
detected between samples from wild type and PARP1−/−
animals, except for a slight elevation of the
Ruminococcaceae ratio in the fecal samples of
PARP1−/− animals (Fig.
2, right-hand panel; Table
II; P<0.05).
When investigating the family composition of the
order Bacteroidales the most abundant species were
Porphyromonadaceae representing ~50–90% of all taxa
(Fig. 2, left-hand panel). The
other families included Rikenellaceae, Bacteroidaceae and
Prevotellaceae. Other taxa not including those already
mentioned reached only a combined ratio of a maximal 0.20% across
all samples. PARP1−/− originated samples demonstrated
little change throughout the gastrointestinal system. The most
noteworthy was the ~14% decrease in the family
Porphyromonadaceae's ratio, which was almost exclusively
made up for by the increase in Prevotellaceae and
Bacteroidaceae between the duodenal and fecal samples. This
14% decrease in the ratio of Porphyromonadaceae was much
less prominent in PARP−/− animals than in their
PARP1+/+ counterpart, where a 46 and 40% decrease was
detected in cecal and fecal samples respectively, when compared
with duodenal samples. In fact, the little change of
Porphyromonadaceae in PARP1−/− animals kept this
family the most abundant one in the Bacteroidales order of the
knockout strain across all samples, while in wild type animals only
the duodenal samples were dominated (92%) by
Porphyromonadaceae (Fig.
2A, left-hand panel). Cecal and fecal samples in the
PARP1+/+ group, consisted only 46 and 51% of
Porphyromonadaceae respectively (Fig. 2B and C, left-hand panel;
statistical significance levels are provided in Table II).
Samples isolated from wild type animals demonstrated
notable changes too. Duodenal samples harbored ~92%
Porphyromonadaceae in the order Bacteroidales, leaving
barely any room for the other families in this order, the second
most abundant being Prevotellaceae (3.6%) followed by
Rikenellaceae (2.1%). Notably, Porphyromonadaceae
contributed to only ~50% of the Bacteroidales family, while the
other most prominent families indicated a ~10-fold increase when
compared with duodenal samples (Bacteroidaceae: 13–17%;
Prevotellaceae: 21–29%; Fig.
2A-C, statistical significance levels are provided in Table II).
Discussion
A recent study demonstrated PARP1-mediated changes
in the fecal microbiome in regard to mucosal injury (47). Following that thread, the
composition of the microbiome on the lower part of the
gastrointestinal tract and feces was assessed in the present
study.
The most prominent result of the present study was
that in the duodenum, in the absence of PARP1, the order of
diversity decreased. These results were similar to those of
Larmonier et al (47) who
also reported a decrease in diversity.
Reference strains of mouse gut bacteria are
practically unavailable and very few studies have attempted to
provide a broad overview. One of these attempts was made in 2016 by
Lagkouvardos et al (48)
who aimed to establish the Mouse Intestinal Bacterial Collection.
Their results demonstrated that certain species are specific to the
mouse intestine. The present results are based on direct sequencing
only, while Lagkouvardos et al (48) utilized culturing in parallel.
Despite the differences in methodology, the present results on
order and family level are very similar with the ones mentioned in
Lagkouvardos et al (48),
thus validating them.
It is of note that the present experimental system
did not challenge the microbiome; in other words, the absence of
PARP1 alone led to visible changes in the microbiome in the absence
of a disease. Furthermore, direct sequencing of 16S ribosomal DNA
was used, which may add a bias to the chemistry prior to the in
silico evaluation as compared with shotgun sequencing; however,
in the upper parts of the gastrointestinal tract the number of the
bacterial DNA is low as compared with the host DNA making shotgun
sequencing cumbersome (20).
What could cause these changes in the microbiome?
Innate immunity is already implicated in the regulation of gut
bacteria through TLRs (22–24).
PARP1 is a positive co-factor of several key inflammatory
transcription factors (such as NF-κB and activator protein-1)
(6) and through that PARP1 may
modulate TLR function (36,37,40,49).
Although, there is no direct evidence, the present study proposed
that the interdependence of PARP1 and TLRs is a likely explanation
for changes in the microbiome in the PARP1−/− mice.
PARP1 is responsible for the majority of the cellular PARP activity
(5,50,51),
therefore, its absence often resembles to PARP inhibitor treatment.
However, there is no evidence for the capability of PARP inhibitors
to influence the microbiome.
At present it is difficult to assess the
physiological relevance of these findings. A body of evidence has
indicated that PARP1 serves a key role in inflammatory pathologies
[such as arthritis (52,53) or type I diabetes (54,55)]
or metabolic diseases [such as type II diabetes (56–58)], where the microbiome has a pivotal
pathogenic role (26,57,58).
Similarly to these, changes in the duodenal flora serve a dominant
role in the pathogenesis of type II diabetes (59). These possibilities require further
assessment in order to verify causal association. Damage to the gut
flora, similarly to certain antibiotics, may contribute to the
diarrhea observed as a side effect of PARP inhibitor treatment in
humans (60). This link between
diarrhea and changes in the microbiome also suggests that the
application of PARP inhibitors may predispose to or aggravate
antibiotic-induced diarrhea in PARP inhibitor-treated patients.
Taken together, understanding the link between PARPs and the
microbiome has importance for the clinical application of these
inhibitors.
Acknowledgements
The authors would like to thank Dr. Zsolt Karányi
(University of Debrecen, Debrecen, Hungary) for his guidance in
statistical analysis.
Funding
The present study was funded by grants from NKFIH
(grant nos. K123975 and GINOP-2.3.2-15-2016-00006) and the Momentum
Fellowship and PROJEKT2017-44 of the Hungarian Academy of
Sciences.
Availability of data and materials
The primary data for microbiome sequencing is
available in the NCBI repository www.ncbi.nlm.nih.gov/bioproject/411773 (NCBI
BioProject PRJNA411773).
Authors' contributions
AV and TK conducted the collection of fecal samples.
AV conducted sample collection and performed data analysis. AV, GK,
BLB and PB wrote the manuscript. PB conceptulized the study and
drafted the manuscript. BLB and GK provided their medical expertise
in understanding and discussing the observed changes. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All animal experiments were approved by the local
and national ethical board of the University of Debrecen (reg.
1/2015/DEMÁB; Debrecen, Hungary).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Amé JC, Spenlehauer C and de Murcia G: The
PARP superfamily. Bioessays. 26:882–893. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cantó C, Sauve A and Bai P: Crosstalk
between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol
Aspects Med. 34:1168–201. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartolomei G, Leutert M, Manzo M, Baubec T
and Hottiger MO: Analysis of chromatin ADP-ribosylation at the
genome-wide level and at specific loci by ADPr-ChAP. Mol Cell.
61:474–485. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gibson BA and Kraus WL: Identification of
protein substrates of specific PARP enzymes using analog-sensitive
PARP mutants and a ‘Clickable’ NAD+ analog. Methods Mol Biol.
1608:111–135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schreiber V, Amé JC, Dollé P, Schultz I,
Rinaldi B, Fraulob V, Ménissier-de Murcia J and de Murcia G:
Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient
base excision DNA repair in association with PARP-1 and XRCC1. J
Biol Chem. 277:23028–23036. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bai P and Virág L: Role of
poly(ADP-ribose) polymerases in the regulation of inflammatory
processes. FEBS Lett. 586:3771–3777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosado MM, Bennici E, Novelli F and Pioli
C: Beyond DNA repair, the immunological role of PARP-1 and its
siblings. Immunology. 139:428–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mangerich A and Bürkle A: Pleiotropic
cellular functions of PARP1 in longevity and aging: Genome
maintenance meets inflammation. Oxid Med Cell Longev.
2012:3216532012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oliver FJ, Ménissier-de Murcia J, Nacci C,
Decker P, Andriantsitohaina R, Muller S, de la Rubia G, Stoclet JC
and de Murcia G: Resistance to endotoxic shock as a consequence of
defective NF-kappaB activation in poly(ADP-ribose) polymerase-1
deficient mice. EMBO J. 18:4446–4454. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Buzzo CL, Medina T, Branco LM, Lage SL,
Ferreira LC, Amarante-Mendes GP, Hottiger MO, De Carvalho DD and
Bortoluci KR: Epigenetic regulation of nitric oxide synthase 2,
inducible (Nos2) by NLRC4 inflammasomes involves PARP1 cleavage.
Sci Rep. 7:416862017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kiss B, Szántó M, Szklenár M, Brunyánszki
A, Marosvölgyi T, Sárosi E, Remenyik É, Gergely P, Virág L, Decsi
T, et al: Poly(ADP) ribose polymerase-1 ablation alters eicosanoid
and docosanoid signaling and metabolism in a murine model of
contact hypersensitivity. Mol Med Rep. 11:2861–2867. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cecchinato V and Uguccioni M: Insight on
the regulation of chemokine activities. J Leukoc Biol. Apr
18–2018.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hughes CE and Nibbs RJB: A guide to
chemokines and their receptors. FEBS J. Apr 10–2018.(Epub ahead of
print). View Article : Google Scholar :
|
|
14
|
Zingarelli B, Szabó C and Salzman AL:
Blockade of Poly(ADP-ribose) synthetase inhibits neutrophil
recruitment, oxidant generation, and mucosal injury in murine
colitis. Gastroenterology. 116:335–345. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin Y, Tang X, Zhu Y, Shu T and Han X:
Identification of PARP-1 as one of the transcription factors
binding to the repressor element in the promoter region of COX-2.
Arch Biochem Biophys. 505:123–129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brunyánszki A, Hegedus C, Szántó M,
Erdélyi K, Kovács K, Schreiber V, Gergely S, Kiss B, Szabó E, Virág
L and Bai P: Genetic ablation of PARP-1 protects against
oxazolone-induced contact hypersensitivity by modulating oxidative
stress. J Invest Dermatol. 130:2629–2637. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Morrow DA, Brickman CM, Murphy SA, Baran
K, Krakover R, Dauerman H, Kumar S, Slomowitz N, Grip L, McCabe CH
and Salzman AL: A randomized, placebo-controlled trial to evaluate
the tolerability, safety, pharmacokinetics, and pharmacodynamics of
a potent inhibitor of poly(ADP-ribose) polymerase (INO-1001) in
patients with ST-elevation myocardial infarction undergoing primary
percutaneous coronary intervention: Results of the TIMI 37 trial. J
Thromb Thrombolysis. 27:359–64. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mikó E, Vida A and Bai P: Translational
aspects of the microbiome-to be exploited. Cell Biol Toxicol.
32:153–156. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Del Vecchio F, Mastroiaco V, Di Marco A,
Compagnoni C, Capece D, Zazzeroni F, Capalbo C, Alesse E and
Tessitore A: Next-generation sequencing: Recent applications to the
analysis of colorectal cancer. J Transl Med. 15:2462017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Quince C, Walker AW, Simpson JT, Loman NJ
and Segata N: Shotgun metagenomics, from sampling to analysis. Nat
Biotechnol. 35:833–844. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shaffer M, Armstrong AJS, Phelan VV,
Reisdorph N and Lozupone CA: Microbiome and metabolome data
integration provides insight into health and disease. Transl Res.
189:51–64. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reuter JA, Spacek DV and Snyder MP:
High-throughput sequencing technologies. Mol Cell. 58:586–597.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tlaskalová-Hogenová H, Stěpánková R,
Kozáková H, Hudcovic T, Vannucci L, Tučková L, Rossmann P, Hrnčíř
T, Kverka M, Zákostelská Z, et al: The role of gut microbiota
(commensal bacteria) and the mucosal barrier in the pathogenesis of
inflammatory and autoimmune diseases and cancer: Contribution of
germ-free and gnotobiotic animal models of human diseases. Cell Mol
Immunol. 8:110–120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kahn SE, Cooper ME and Del Prato S:
Pathophysiology and treatment of type 2 diabetes: Perspectives on
the past, present, and future. Lancet. 383:1068–1083. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Naseer MI, Bibi F, Alqahtani MH, Chaudhary
AG, Azhar EI, Kamal MA and Yasir M: Role of gut microbiota in
obesity, type 2 diabetes and Alzheimer's disease. CNS Neurol Disord
Drug Targets. 13:305–311. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Puertollano E, Kolida S and Yaqoob P:
Biological significance of short-chain fatty acid metabolism by the
intestinal microbiome. Curr Opin Clin Nutr Metab Care. 17:139–144.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Duca FA, Sakar Y, Lepage P, Devime F,
Langelier B, Doré J and Covasa M: Replication of obesity and
associated signaling pathways through transfer of microbiota from
obese prone rat. Diabetes. 63:1624–1636. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xie G, Wang X, Huang F, Zhao A, Chen W,
Yan J, Zhang Y, Lei S, Ge K, Zheng X, et al: Dysregulated hepatic
bile acids collaboratively promote liver carcinogenesis. Int J
Cancer. 139:1764–1775. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yoshimoto S, Loo TM, Atarashi K, Kanda H,
Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et
al: Obesity-induced gut microbial metabolite promotes liver cancer
through senescence secretome. Nature. 499:97–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leifer CA, McConkey C, Li S, Chassaing B,
Gewirtz AT and Ley RE: Linking genetic variation in human Toll-like
receptor 5 genes to the gut microbiome's potential to cause
inflammation. Immunol Lett. 162:3–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Velloso LA, Folli F and Saad MJ: TLR4 at
the crossroads of nutrients, gut microbiota, and metabolic
inflammation. Endocr Rev. 36:245–271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Caballero S and Pamer EG:
Microbiota-mediated inflammation and antimicrobial defense in the
intestine. Annu Rev Immunol. 33:227–256. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goedert JJ, Jones G, Hua X, Xu X, Yu G,
Flores R, Falk RT, Gail MH, Shi J, Ravel J and Feigelson HS:
Investigation of the association between the fecal microbiota and
breast cancer in postmenopausal women: A population-based
case-control pilot study. J Natl Cancer Inst. 107:djv1472015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mu C, Yang Y and Zhu W: Crosstalk between
the immune receptors and gut microbiota. Curr Protein Pept Sci.
16:622–631. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Frosali S, Pagliari D, Gambassi G,
Landolfi R, Pandolfi F and Cianci R: How the intricate interaction
among toll-like receptors, microbiota, and intestinal immunity can
influence gastrointestinal pathology. J Immunol Res.
2015:4898212015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Farez MF, Quintana FJ, Gandhi R, Izquierdo
G, Lucas M and Weiner HL: Toll-like receptor 2 and poly(ADP-ribose)
polymerase 1 promote central nervous system neuroinflammation in
progressive EAE. Nat Immunol. 10:958–964. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zerfaoui M, Errami Y, Naura AS, Suzuki Y,
Kim H, Ju J, Liu T, Hans CP, Kim JG, Abd Elmageed ZY, et al:
Poly(ADP-ribose) polymerase-1 is a determining factor in
Crm1-mediated nuclear export and retention of p65 NF-kappa B upon
TLR4 stimulation. J Immunol. 185:1894–1902. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kauppinen A, Suuronen T, Ojala J,
Kaarniranta K and Salminen A: Antagonistic crosstalk between NF-κB
and SIRT1 in the regulation of inflammation and metabolic
disorders. Cell Signal. 25:1939–1948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Krukenberg KA, Kim S, Tan ES, Maliga Z and
Mitchison TJ: Extracellular poly(ADP-ribose) is a pro-inflammatory
signal for macrophages. Chem Biol. 22:446–452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qin WD, Mi SH, Li C, Wang GX, Zhang JN,
Wang H, Zhang F, Ma Y, Wu DW and Zhang M: Low shear stress induced
HMGB1 translocation and release via PECAM-1/PARP-1 pathway to
induce inflammation response. PLoS One. 10:e01205862015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Menissier-de Murcia J, Niedergang C,
Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M,
Dierich A, LeMeur M, et al: Requirement of poly(ADP-ribose)
polymerase in recovery from DNA damage in mice and in cells.
ProcNatl Acad Sci USA. 94:7303–7307. 1997. View Article : Google Scholar
|
|
42
|
Meyer F, Paarmann D, D'Souza M, Olson R,
Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, et
al: The metagenomics RAST server-a public resource for the
automatic phylogenetic and functional analysis of metagenomes. BMC
Bioinformatics. 9:3862008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hammer O, Harper DAT and Ryan PD: PAST:
Paleontological statistics software package for education and data
analysis. Palaeontologia Electronica. 4:92001.
|
|
44
|
Newcombe RG: Two-sided confidence
intervals for the single proportion: Comparison of seven methods.
Stat Med. 17:857–872. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Newcombe RG: Interval estimation for the
difference between independent proportions: Comparison of eleven
methods. Stat Med. 17:873–890. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tothmeresz B: Comparison of different
methods for diversity ordering. J Veget Sci. 6:283–290. 1995.
View Article : Google Scholar
|
|
47
|
Larmonier CB, Shehab KW, Laubitz D, Jamwal
DR, Ghishan FK and Kiela PR: Transcriptional reprogramming and
resistance to colonic mucosal injury in Poly(ADP-ribose) Polymerase
1 (PARP1)-deficient mice. J Biol Chem. 291:8918–8930. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lagkouvardos I, Pukall R, Abt B, Foesel
BU, Meier-Kolthoff JP, Kumar N, Bresciani A, Martínez I, Just S,
Ziegler C, et al: The mouse intestinal bacterial collection (miBC)
provides host-specific insight into cultured diversity and
functional potential of the gut microbiota. Nat Microbiol.
1:161312016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Davis K, Banerjee S, Friggeri A, Bell C,
Abraham E and Zerfaoui M: Poly(ADP-ribosyl)ation of high mobility
group box 1 (HMGB1) protein enhances inhibition of efferocytosis.
Mol Med. 18:359–369. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Szanto M, Rutkai I, Hegedus C, Czikora Á,
Rózsahegyi M, Kiss B, Virág L, Gergely P, Tóth A and Bai P:
Poly(ADP-ribose) polymerase-2 depletion reduces doxorubicin-induced
damage through SIRT1 induction. Cardiovasc Res. 92:430–438. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bai P, Canto C, Brunyánszki A, Huber A,
Szántó M, Cen Y, Yamamoto H, Houten SM, Kiss B, Oudart H, et al:
PARP-2 regulates sirt1 expression and whole-body energy
expenditure. Cell Metab. 13:450–460. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pascual M, López-Nevot MA, Cáliz R, Ferrer
MA, Balsa A, Pascual-Salcedo D and Martín J: A poly(ADP-ribose)
polymerase haplotype spanning the promoter region confers
susceptibility to rheumatoid arthritis. Arthritis Rheum.
48:638–641. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Masutani M, Nakagama H and Sugimura T:
Poly(ADP-ribosyl)ation in relation to cancer and autoimmune
disease. Cell Mol Life Sci. 62:769–783. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Burkart V, Wang ZQ, Radons J, Heller B,
Herceg Z, Stingl L, Wagner EF and Kolb H: Mice lacking the
poly(ADP-ribose) polymerase gene are resistant to pancreatic
beta-cell destruction and diabetes development induced by
streptozocin. Nat Med. 5:314–319. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pacher P, Beckman JS and Liaudet L: Nitric
oxide and peroxynitrite in health and disease. Physiol Rev.
87:315–424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bai P, Cantó C, Oudart H, Brunyánszki A,
Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al:
PARP-1 Inhibition Increases Mitochondrial Metabolism through SIRT1
Activation. Cell Metab. 13:461–468. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Koren O, Goodrich JK, Cullender TC, Spor
A, Laitinen K, Bäckhed HK, Gonzalez A, Werner JJ, Angenent LT,
Knight R, et al: Host remodeling of the gut microbiome and
metabolic changes during pregnancy. Cell. 150:470–480. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Le Chatelier E, Nielsen T, Qin J, Prifti
E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy
S, et al: Richness of human gut microbiome correlates with
metabolic markers. Nature. 500:541–546. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kamvissi-Lorenz V, Raffaelli M, Bornstein
S and Mingrone G: Role of the Gut on glucose homeostasis: Lesson
learned from metabolic surgery. Curr Atheroscler Rep. 19:92017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fong PC, Boss DS, Yap TA, Tutt A, Wu P,
Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et
al: Inhibition of Poly(ADP-Ribose) polymerase in tumors from BRCA
mutation carriers. N Engl J Med. 361:123–134. 2009. View Article : Google Scholar : PubMed/NCBI
|