Introduction
Autophagy is a process of degrading and recycling
discarded proteins and organelles that is dependent on lysosomes.
The process of autophagy involves formation of autophagosomes,
fusion of autophagosomes and lysosomes into autolysosomes, and
substrate degradation. This dynamic process, called autophagic
flux, is crucial for maintaining intracellular homeostasis and
preventing aging (1,2).
Podocytes are key components of the glomerular
filtration barrier. Podocyte injury is a common pathogenesis for
many glomerular diseases. Since podocytes are a type of terminally
differentiated cells, their ability to regenerate after injury is
weak. Therefore, self-repair mechanisms are particularly important
for podocytes. Podocytes have high constitutive autophagy and
podocyte-specific autophagy knockdown aggravates podocyte damage
and progression of glomerular diseases (3). However, the mechanism of podocyte
autophagy is not completely understood.
Amino acids (AA) are important upstream signals for
autophagy regulation. Amino acid starvation (AAS) is a critical
mechanism for autophagy induction, but the functions of AA in
podocyte autophagy are unclear. Mammalian target of rapamycin
(mTOR) is a serine/threonine phosphorylase that has key functions
in intracellular regulation of cell proliferation and energy. mTOR
is also important in inhibiting autophagy. Short-term AAS can block
mTOR activity and relieve its inhibition of autophagy-related
molecules in downstream pathways, initiating autophagy (4,5). AA
signaling also regulates autophagy at the transcriptional level,
such as through transcription factor EB (TFEB). TFEB promotes
transcription of multiple genes in the autophagy process, including
formation of autophagosomes, autolysosome formation and substrate
degradation. As observed in HeLa cells stably expressing high
levels of TFEB (6) and in HEK-293T
cells (7), AAS promotes nuclear
translocation of TFEB. We hypothesized that AA may also regulate
podocyte autophagy via mTOR and TFEB pathways. Therefore, this
study aimed to investigate the functions and mechanisms of AA
signaling in podocyte autophagy.
Materials and methods
Cell culture and treatment
The conditionally immortalized mouse podocyte cell
line (Mouse Podocyte Clone-5, MPC-5) was a kind gift from Dr.
Jochen Reiser (Rush University Medical Center, Chicago, IL, USA).
Differentiated podocytes are unable to replicate, and primary
podocytes would result in rapid growth arrest when culturing. The
MPC-5 has overcome these difficulties, and conditionally retains a
differentiation potential similar to that of podocytes in
vivo (8), which is widely used
as a cell model in research focusing on human podocyte injury.
Cells were cultured as described previously (9). The recovered cells were cultured at
33°C in RPMI-1640 medium supplemented with 10% fetal bovine serum
(FBS; both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
and recombinant interferon (IFN)-γ (ProSpec, Tany Technogene, Ness
Ziona, Israel) for 2–3 days for proliferation. Podocytes were then
transferred to 100-cm2 culture dishes coated with type-I
collagen (BD Bioscience, Bedford, MA, USA) in IFN-γ free RPMI-1640
medium containing 5% FBS and cultured at 37°C for 10–13 days for
differentiation. To establish an AA-starved podocyte model,
differentiated podocytes were treated with AA-deprivation RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 5%
FBS for 6, 12 or 24 h. Podocytes were treated with complete
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 5% FBS as the control group.
Small interfering RNA (siRNA)
transfection
SiRNA targeting TFEB and control siRNA were designed
and synthesized by RiboBio Co. Ltd (Guangzhou, China). TFEB siRNA
or control scrambled siRNA was transfected into podocytes with
Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's protocol. Podocytes were
transfected for 48 h at 37°C. The sequence of siRNA used in this
study was: siTFEB 5′CCATGGCCATGCTACATATdTdT-3′.
Western blot analysis
Podocytes were washed three times with cold
phosphate-buffered saline (PBS) and lysed with lysis buffer
(Beyotime Institute of Biotechnology, Shanghai, China) premixed
with proteinase inhibitor (Guangzhou Genebase Bioscience,
Guangzhou, China). Protein was measured using bicinchoninic acid
protein quantification kits (Thermo Fisher Scientific, Inc.) and
15–30 micrograms protein was processed on 7.5–10% sodium dodecyl
sulfate-polyacrylamide gels and transferred to polyvinylidene
fluoride membranes (Millipore, Billerica, MA, USA). After blocking
with 5% non-fat dry milk for 1 h at room temperature, membranes
were incubated overnight at 4°C with primary antibodies: Rabbit
anti-light chain (LC)3A/B (1:1,000; cat. no. 4108), rabbit
anti-beclin1 (1:1,000; cat. no. 3495), rabbit anti-p62 (1:1,000;
cat. no. 5114), rabbit anti-phospho-p70s6k (1:1,000; cat. no. 9205;
all Cell Signaling Technology, Danvers, MA, USA); rabbit anti-GAPDH
(1:3,000; cat. no. AP0063; Bioworld Technology, Nanjing, China);
rabbit anti-p70s6k (1:1,000; cat. no. ab32359; Abcam, Cambridge,
MA, USA). Anti-rabbit IgG (1:3,000; cat. no. 31460; Thermo Fisher
Scientific, Inc.) was incubated with membranes at room temperature
for 1 h. Blots were treated with enhanced chemiluminescence reagent
(Advansta, Menlo Park, CA, USA). Images were captured by automatic
imager (General Electric, Fairfield, CT, USA). Densitometric
analyses of protein bands used ImageJ software (National Institutes
of Health, Bethesda, MD, USA).
Transmission electron microscopy
Podocytes were fixed in 2.5% glutaraldehyde. Samples
were dehydrated with alcohol and post-fixed with 1% osmium
tetroxide. Samples were embedded into resin (Epon 812) and cut into
ultrathin slices and double stained with uranyl acetate and
examined by transmission electron microscope (FEI, TECNAI 12,
Lausanne, Netherlands). Numbers of autophagosomes or autolysosomes
per cell were counted blind (10,11).
Green fluorescent protein
(GFP)-monomeric red fluorescent protein monomeric red fluorescent
protein (mRFP)-LC3 adenovirus transduction
Cultured podocytes were seeded onto cover slides in
six-well plates. To detect autophagic flux, podocytes were
transduced with tandem GFP-mRFP-LC3 adenovirus which was purchased
from Hanbio Technology (Shanghai, China) as described previously
(9). Podocytes were transduced
with the GFP-mRFP-LC3 adenovirus at a concentration of
1×107 PFU/well and incubated in RPMI-1640 medium
containing 2% FBS. The transduction medium was replaced with fresh
culture medium after 6 h of transduction. Treatment (with
AA-deprivation RPMI-1640 medium supplemented with 5% FBS or
complete RPMI-1640 medium supplemented with 5% FBS for 6 h) was
performed at 24 h post-transduction.
Immunofluorescent staining
Cultured podocytes transduced with GFP-mRFP-LC3
adenovirus were washed with PBS and fixed with 4% paraformaldehyde
and permeabilized using 0.1% Triton X-100 for 10 min. After washing
with PBS, podocytes were stained with DAPI (Sigma, St. Louis, MO,
USA) for 10 min at room temperature. Photomicrographs were taken by
confocal laser scanning microscopy (KS 400; Zeiss, Postfach,
Germany). GFP signal is sensitive to acidic proteolytic conditions
of the lysosome lumen, whereas mRFP is more stable (12). Colocalization of GFP and mRFP
indicates autophagosomes (yellow). MRFP signal alone indicates
autolysosomes (red).
To observe subcellular localization of TFEB, 4%
paraformaldehyde was used to fix cells at room temperature for 15
min. Samples were permeabilized using 0.1% Triton X-100 for 10 min
and blocked with 5% bovine serum albumin at room temperature for 30
min followed by incubating overnight at 4°C with rabbit anti-TFEB
antibody (1:200; cat. no. A303-673A; Bethyl, Montgomery, TX, USA,).
Samples were washed 3 times with PBS and incubated with goat
anti-rabbit AlexaFluor 555 antibody (1:200; cat. no. A32732; Thermo
Fisher Scientific, Inc.) for 1 h at room temperature. After washing
with PBS, podocytes were stained with DAPI for 10 min at room
temperature and analyzed by confocal laser scanning microscopy.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA samples were extracted using a TRIzol RNA
isolation system (Invitrogen). Purity and concentration of mRNA
samples were measured using a NanoDrop 2000 spectrophotometer
(Thermo Fisher Scientific, Inc., Wilmington, DE, USA). Samples were
reverse transcribed (RT) into cDNAs using PrimeScript™ RT regent
kits (Takara Biotechnology, Shiga, Japan). cDNAs were used for
RT-qPCR analysis using a Power SYBR Green PCR Master Mix (Takara
Biotechnology). qPCR was performed with 2 µl aliquots of cDNA (10
ng/µl) using specific primer pairs. Primers were as follows: LC3A
forward, 5′-GACCGCTGTAAGGAGGTGC-3′ and reverse,
5′-CTTGACCAACTCGCTCATGTTA-3′; LC3B forward,
5′-TTATAGAGCGATACAAGGGGGAG-3′ and reverse,
5′-CGCCGTCTGATTATCTTGATGAG-3′; GAPDH forward,
5′-AGGTCGGTGTGAACGGATTTG-3′ and reverse,
5′-TGTAGACCATGTAGTTGAGGTCA-3′. Samples were amplified by 95°C for 2
min, 40 cycles of 95°C for 30 sec, 95°C for 5 sec, 60°C for 5 sec,
and a 72°C for 10 min. Data were calculated using the
2−ΔΔCq method (13).
Statistical analysis
Values are expressed as mean ± standard error of the
mean. Data were analyzed by the statistical package SPSS for
Windows version 21.0 (IBM Corp., Armonk, NY, USA). Multiple
comparisons were performed with one-way analysis of variance
followed by the least-significant difference post hoc test. Data
from two groups were compared by Student's t-test. P<0.05 was
considered to indicate a statistically significant difference. All
experimental procedures were repeated at least three times.
Results
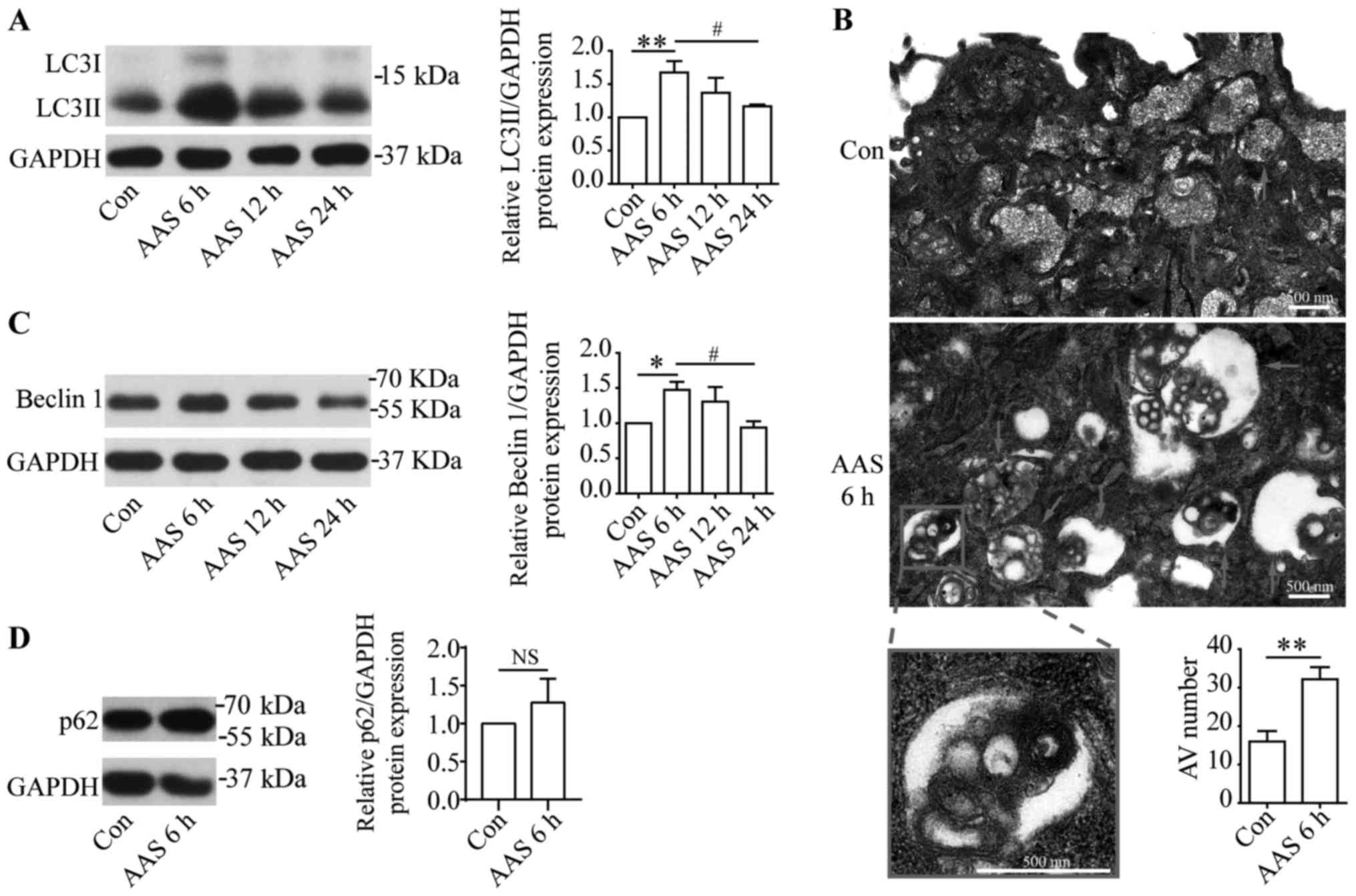
AAS promotes podocyte autophagy
To determine if AAS regulated podocyte autophagy, we
used AA deprivation medium and normal medium to culture podocytes
for indicated times. Western blots showed that after 6-h AAS,
LC3II, an autophagosome marker, increased in podocytes
significantly to peak level (Fig.
1A). After 12-h AAS treatment, LC3II levels were still higher
than the control group, but decreased compared with 6-h AAS
treatment (Fig. 1A). After 24-h
AAS, LC3II levels returned to baseline (Fig. 1A). The effect of AA signal on
autophagy was further studied by transmission electron microscopy,
which showed that 6-h AAS promoted formation of autophagic vacuoles
in podocytes (Fig. 1B). Western
blots found that the changes for beclin1, a marker of autophagy
initiation, at different AAS times coincided with LC3II (Fig. 1C). These results suggested that AAS
increased autophagic activity. p62 serves as a substrate of
autophagy. In certain settings, stimulation of autophagy correlates
with decreased level of p62 (14).
We investigated the p62 expression by western blots and the results
showed that p62 did not altered significantly under the stimulation
of AAS for 6 h (Fig. 1D). This is
inconsistent with the increased autophagic activity under AAS
stimulation (Fig. 1A-C),
indicating that in addition to autophagy, there are other factors
that can affect the level of p62 in this AAS-stimulated model.
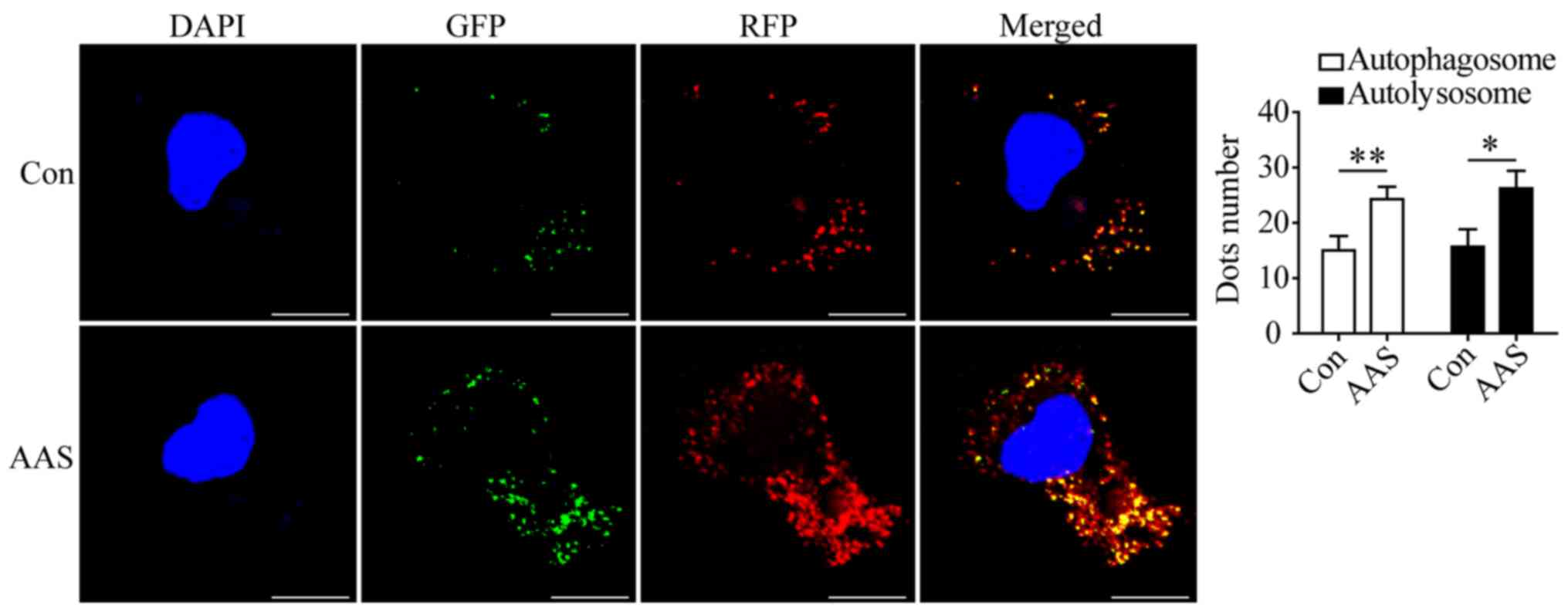
AAS promotes autophagic flux
We evaluated the effect of AAS on autophagic flux in
podocytes. The AAS-induced enhancement of beclin1 expression
(Fig. 1C) suggested the
AAS-induced increase of autophagosomes (Fig. 1A and B) was related to the
enhancement of autophagy initiation. Moreover, in podocytes
transduced with GFP-mRFP-LC3 adenovirus, AAS increased the number
of autophagosomes (Fig. 2) and
autolysosomes (Fig. 2). These
results indicated that AAS contributed to formation and turnover of
autophagosomes.
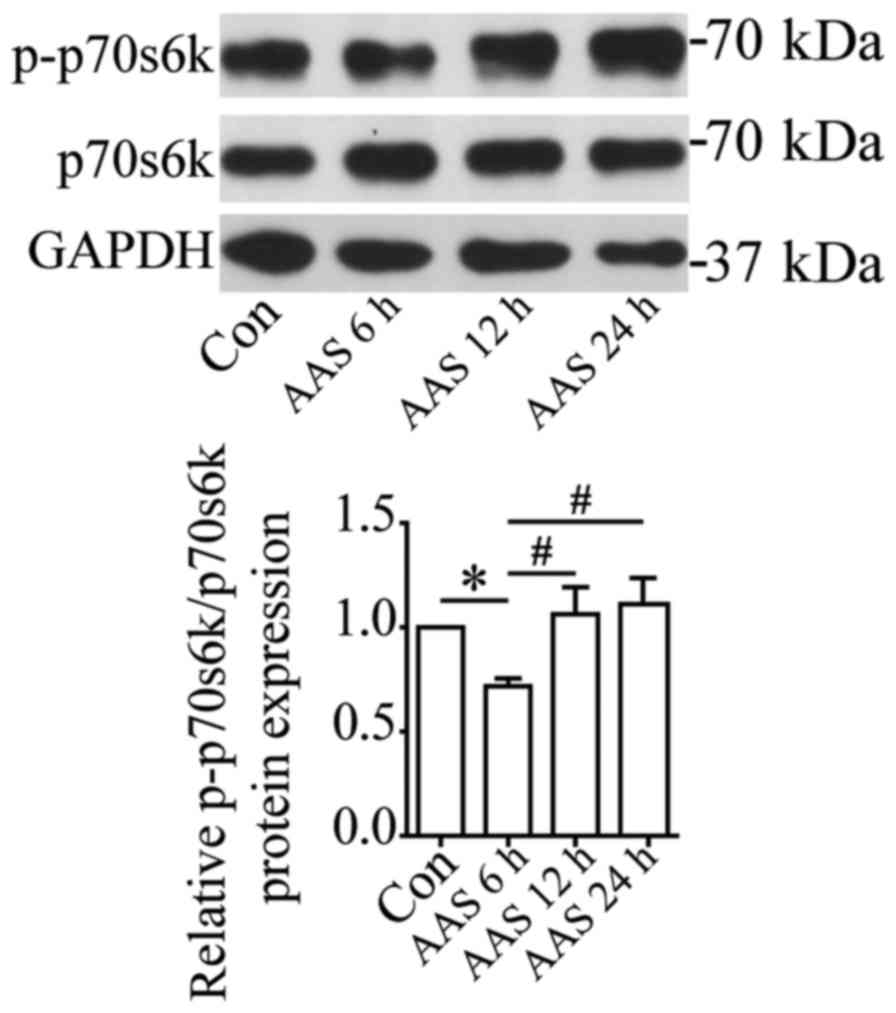
mTOR was inactivated under AAS
We investigated the molecular mechanisms underlying
AAS regulation of autophagy in podocytes. mTOR is a well-known
negative regulator of autophagy. p70S6k (p70 ribosomal protein S6
kinase) is the phosphorylation substrate of mTOR, and the level of
phosphorylated p70S6k reflects mTOR activity (15). Western blots showed that 6-h AAS
inhibited mTOR activity (Fig. 3).
The mTOR activity gradually recovered after 12-h AAS (Fig. 3), contrary to the changes of
autophagy after AAS. This result suggested that AAS promoted
autophagy by inhibiting mTOR activity.
TFEB mediated AAS-induced
autophagy
The above-mentioned results demonstrated AAS promote
autophagic flux at multiple steps. We have reported previously that
TFEB promoted podocyte autophagy through enhancing autophagosome
formation and degradation (16).
Thus, we studied the role of TFEB in AAS-induced autophagy in
cultured podocytes. AAS promoted nuclear translocation of TFEB
(Fig. 4A) and raised levels of
mRNA (Fig. 4B) and protein
(Fig. 1A) for LC3B, a target gene
of TFEB (6,16), indicating increased TFEB activity
by AAS. Next, we utilized TFEB siRNA to observe whether TFEB
mediated AAS-induced autophagy. As we reported previously, TFEB
siRNA significantly inhibited the expression of TFEB at mRNA, total
protein and nuclear protein level in cultured podocytes (16). The present study found that TFEB
siRNA blocked AAS-induced LC3II upregulation (Fig. 4C and D), indicating TFEB mediated
the AAS-induced autophagy in podocytes.
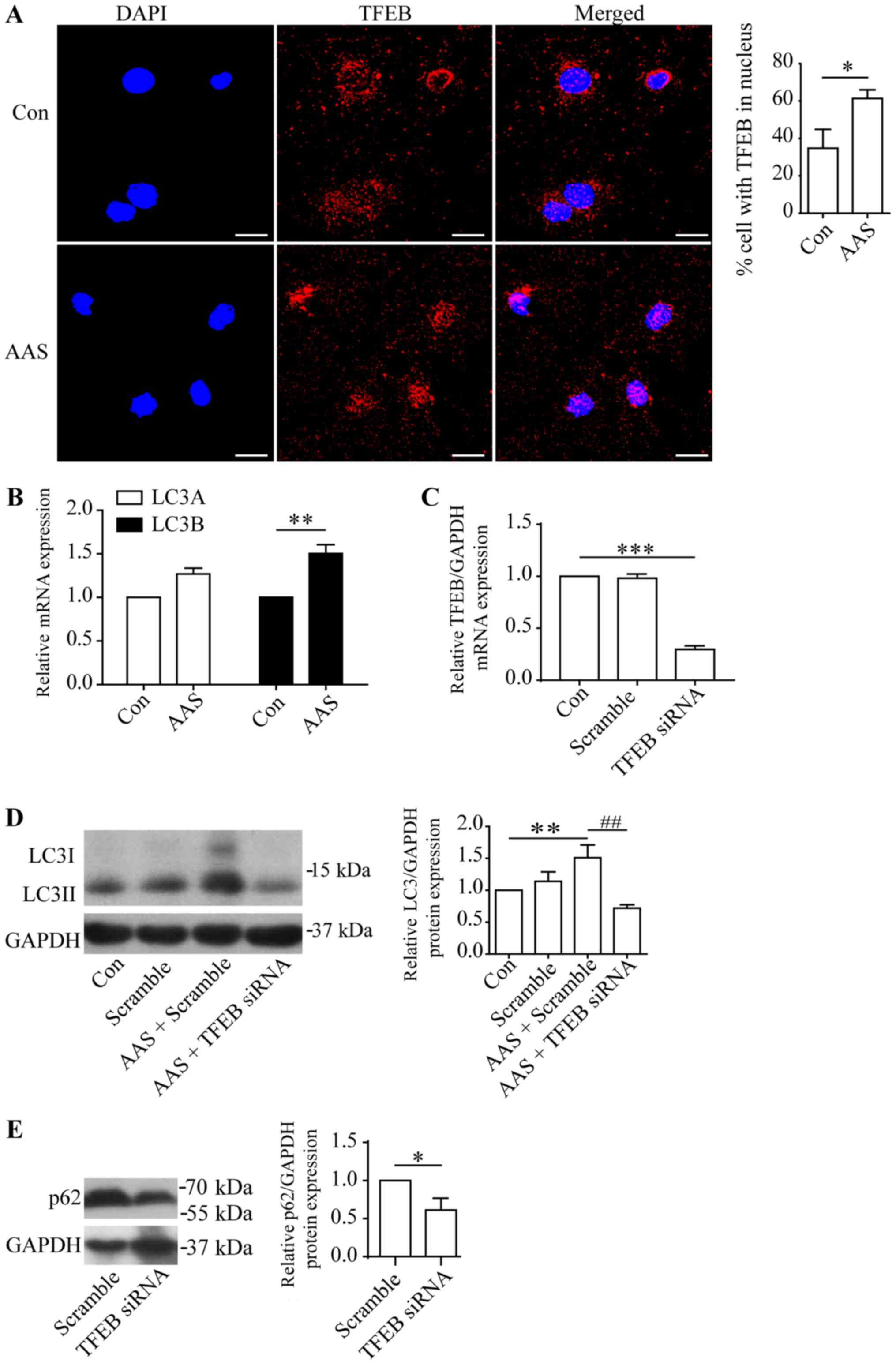 | Figure 4.TFEB-mediated amino acid starvation
induces autophagy in podocytes. (A) Immunofluorescence staining for
TFEB (red staining) and DAPI (blue staining) in cultured podocytes:
Untreated (Control) or amino acid starved for 6 h. AAS enhanced
TFEB nuclear translocation. Scale bars, 20 µm. (B) RT-qPCR analyses
of LC3A and LC3B in cultured podocytes: Untreated (Control) or
amino acid starved for 6 h. (C) RT-qPCR of TFEB mRNA expression.
(D) Western blotting of LC3II in podocytes transfected with
scrambled or TFEB siRNA: Untreated (Control) or amino acid starved
for 6 h. TFEB siRNA blocked AAS-induced autophagy. *P<0.05 and
**P<0.01 vs. control; ##P<0.01 vs. AAS +
scrambled. (E) Western blotting of p62 in podocytes transfected
with scrambled or TFEB siRNA. TFEB siRNA decreased the p62
expression. Values are presented as the mean ± standard error of
the mean (n=3). *P<0.05 vs. Scramble. TFEB, transcription factor
EB; DAPI, 4′,6-diamidino-2-phenylindole; Con, control; AAS, amino
acid starvation; LC3, microtubule-associated protein 1 light chain
3; siRNA, small interfering RNA; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; Scramble,
scrambled siRNA. |
Moreover, p62 serves as a target gene of TFEB
(6,16). We found that TFEB siRNA decreased
the p62 protein expression in cultured podocytes (Fig. 4E). As AAS increased the nuclear
transportation and activity of TFEB, these results suggested a
potential role of TFEB in regulating p62 levels in AAS-stimulated
podocytes.
Discussion
The present study investigated the effect of AA on
podocyte autophagy. We found that AAS promoted autophagy in
podocytes. AAS affected autophagy through at least two pathways,
inhibiting mTOR activity and promoting TFEB nuclear translocation
and activity.
AA regulation of autophagy was time-dependent: 6 h
of AAS promoted podocyte autophagy to reach peak level. With AAS
over 6 h, autophagy decreased gradually and resumed basic levels at
24 h. The changes of mTOR activity were time-dependent under AAS,
but opposite of autophagy. The changes of mTOR under AAS were
similar to a study that found that short-term nutrient deprivation
inhibits the mTOR signaling pathway in normal rat kidney cells, but
mTOR was reactivated with prolonged starvation (17). This result may be due to
starvation-induced autophagy leading to decomposing and recycling
autophagy substrates, which reactivates mTOR (17).
In addition to post-transcriptional regulation of
autophagy through mTOR, AA signaling also regulates autophagy
transcriptionally. Our study found that AAS promoted nuclear
translocation of TFEB and increased TFEB activity in podocytes.
Moreover, TFEB siRNA inhibited the increased autophagy induced by
AAS in podocytes. These results demonstrated that AAS also
contributed to podocyte autophagy via activating TFEB.
Autophagy is a dynamic and multi-step process
orchestrated by series of proteins. Thus, multi-dimensional
assessments of autophagy are recommended to interpret it. The
present study utilized western blots to detect LC3 and beclin1,
GFP-mRFP-LC3 fluorescent probe and TEM to examine autophagy, which
all suggested an increasing autophagic activity by AAS. Autophagy
upregulation often correlates with enhanced autolysosome-mediated
degradation of substrates such as p62 (14). We also investigated the level of
p62 under AAS stimulation. However, p62 did not altered
significantly under AAS stimulation for 6 h, which is inconsistent
with the AAS-induced upregulation of autophagic activity. Moreover,
p62 serves as a target gene of TFEB (6) and we validated the role of TFEB in
regulating p62 protein levels in podocytes. We also observed that
AAS increased TFEB activity. These results suggested that in this
AAS-stimulated model, p62 level was affected not only by the
increased activity of autophagy (led to p62 downregulation) but
also by increased activation of TFEB (led to p62 upregulation).
Therefore, p62 may not serve as an effective marker for autophagy
degradation in this model.
Autophagy in podocytes has been studied often, but
the level of autophagy in podocytes is too weak to be measured
precisely by commonly used detection methods. Nutritional
starvation is a common method to increase autophagic activity
(11). Previous studies reported
serum starvation (18), Hanks
solution induced starvation (11)
or EBSS solution induced starvation (19) promoted podocyte autophagy. These
were autophagy induction models induced by a variety of nutrient
deficiencies. Our study showed that, among these nutrient
components, AA were a key component in inducing autophagy. In
addition, AAS induced high autophagic activity in cultured podocyte
model, providing a new tool for studying podocyte autophagy.
Studies found that podocyte autophagy damage occurs
in many glomerular diseases, contributing to pathogenesis and
progression of renal disease such as diabetic nephropathy (20), focal segmental glomerulosclerosis
(11) and membranous nephropathy
(21). Restoring podocyte
autophagy levels is an urgent problem to be solved. Previous
studies found that serum concentrations of various AAs in patients
with chronic kidney disease and diabetic nephropathy are abnormal
(22,23). Very-low-protein diets (AA
restriction) alleviated renal injury and restored autophagy levels
in diabetic rats (24). Our
results suggested that manipulation of local AA concentrations in
podocytes may be a potential method for regulating podocyte
autophagy.
In conclusion, this study found that short-term AAS
promoted podocyte autophagy by inhibiting mTOR and promoting TFEB
activity.
Acknowledgements
The authors would like to thank Professor Jochen
Reiser (Rush University Medical Center) for providing the podocyte
cell line.
Funding
The present study was supported by the Nationa
Natural Science Foundation of China (grant nos. 81770733 and
81270784).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and WS conceived the present study. YC, XZ and WS
analyzed the data. YC, XZ and XL wrote the manuscript. YC, XZ, JL,
LZ, RL, HZ, RL, SL, WS and XL were all involved in performing the
experiments and had final approval of the version to be
published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mizushima N and Komatsu M: Autophagy:
Renovation of cells and tissues. Cell. 147:728–741. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hartleben B, Gödel M, Meyer-Schwesinger C,
Liu S, Ulrich T, Köbler S, Wiech T, Grahammer F, Arnold SJ,
Lindenmeyer MT, et al: Autophagy influences glomerular disease
susceptibility and maintains podocyte homeostasis in aging mice. J
Clin Invest. 120:1084–1096. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sancak Y, Peterson TR, Shaul YD, Lindquist
RA, Thoreen CC, Bar-Peled L and Sabatini DM: The Rag GTPases bind
raptor and mediate amino acid signaling to mTORC1. Science.
320:1496–1501. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin G, Lee SW, Zhang X, Cai Z, Gao Y, Chou
PC, Rezaeian AH, Han F, Wang CY, Yao JC, et al: Skp2-mediated RagA
ubiquitination elicits a negative feedback to prevent
amino-acid-dependent mTORC1 hyperactivation by recruiting GATOR1.
Mol Cell. 58:989–1000. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Settembre C, Di Malta C, Polito VA, Garcia
Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D,
Colella P, et al: TFEB links autophagy to lysosomal biogenesis.
Science. 332:1429–1433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Settembre C, Zoncu R, Medina DL, Vetrini
F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et
al: A lysosome-to-nucleus signalling mechanism senses and regulates
the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mundel P, Reiser J, Zúñiga Mejía Borja A,
Pavenstädt H, Davidson GR, Kriz W and Zeller R: Rearrangements of
the cytoskeleton and cell contacts induce process formation during
differentiation of conditionally immortalized mouse podocyte cell
lines. Exp Cell Res. 236:248–258. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan X, Chen Y, Liang X, Yu C, Lai Y, Zhang
L, Zhao X, Zhang H, Lin T, Li R and Shi W:
Lipopolysaccharide-induced podocyte injury is mediated by
suppression of autophagy. Mol Med Rep. 14:811–818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei Q and Dong Z: HDAC4 blocks autophagy
to trigger podocyte injury: Non-epigenetic action in diabetic
nephropathy. Kidney Int. 86:666–668. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeng C, Fan Y, Wu J, Shi S, Chen Z, Zhong
Y, Zhang C, Zen K and Liu Z: Podocyte autophagic activity plays a
protective role in renal injury and delays the progression of
podocytopathies. J Pathol. 234:203–213. 2014.PubMed/NCBI
|
|
12
|
Inoki K, Mori H, Wang J, Suzuki T, Hong S,
Yoshida S, Blattner SM, Ikenoue T, Rüegg MA, Hall MN, et al: mTORC1
activation in podocytes is a critical step in the development of
diabetic nephropathy in mice. J Clin Invest. 121:2181–2196. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Holz MK, Ballif BA, Gygi SP and Blenis J:
mTOR and S6K1 mediate assembly of the translation preinitiation
complex through dynamic protein interchange and ordered
phosphorylation events. Cell. 123:569–580. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao X, Chen Y, Tan X, Zhang L, Zhang H,
Li Z, Liu S, Li R, Lin T, Liao R, et al: Advanced glycation
end-products suppress autophagic flux in podocytes by activating
mammalian target of rapamycin and inhibiting nuclear translocation
of transcription factor EB. J Pathol. 245:235–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu L, McPhee CK, Zheng L, Mardones GA,
Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al: Termination of
autophagy and reformation of lysosomes regulated by mTOR. Nature.
465:942–946. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fang L, Li X, Luo Y, He W, Dai C and Yang
J: Autophagy inhibition induces podocyte apoptosis by activating
the pro-apoptotic pathway of endoplasmic reticulum stress. Exp Cell
Res. 322:290–301. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mao N, Tan RZ, Wang SQ, Wei C, Shi XL, Fan
JM and Wang L: Ginsenoside Rg1 inhibits angiotensin II-induced
podocyte autophagy via AMPK/mTOR/PI3K pathway. Cell Biol Int.
40:917–925. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tagawa A, Yasuda M, Kume S, Yamahara K,
Nakazawa J, Chin-Kanasaki M, Araki H, Araki S, Koya D, Asanuma K,
et al: Impaired podocyte autophagy exacerbates proteinuria in
diabetic nephropathy. Diabetes. 65:755–767. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu WJ, Li ZH, Chen XC, Zhao XL, Zhong Z,
Yang C, Wu HL, An N, Li WY and Liu HF: Blockage of the
lysosome-dependent autophagic pathway contributes to complement
membrane attack complex-induced podocyte injury in idiopathic
membranous nephropathy. Sci Rep. 7:86432017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Laidlaw SA, Berg RL, Kopple JD, Naito H,
Walker WG and Walser M: Patterns of fasting plasma amino acid
levels in chronic renal insufficiency: Results from the feasibility
phase of the Modification of Diet in Renal Disease Study. Am J
Kidney Dis. 23:504–513. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pena MJ, Lambers Heerspink HJ, Hellemons
ME, Friedrich T, Dallmann G, Lajer M, Bakker SJ, Gansevoort RT,
Rossing P, de Zeeuw D and Roscioni SS: Urine and plasma metabolites
predict the development of diabetic nephropathy in individuals with
Type 2 diabetes mellitus. Diabet Med. 31:1138–1147. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kitada M, Ogura Y, Suzuki T, Sen S, Lee
SM, Kanasaki K, Kume S and Koya D: A very-low-protein diet
ameliorates advanced diabetic nephropathy through autophagy
induction by suppression of the mTORC1 pathway in Wistar fatty
rats, an animal model of type 2 diabetes and obesity. Diabetologia.
59:1307–1317. 2016. View Article : Google Scholar : PubMed/NCBI
|