Introduction
As one of the most prevalent and malignant types of
cancer, lung cancer gives rise to a large number of
cancer-associated mortalities globally each year (1). Among all lung cancer cases, non-small
cell lung cancer (NSCLC) accounts for nearly 80% with an annual
incidence of over 160,000 newly diagnosed cases (2). In previous decades, certain advances
have been achieved in NSCLC therapeutic treatment, including
surgery, chemotherapy and molecular-targeted therapy. However, most
NSCLC patients are diagnosed at an advanced stage and with local
invasion and/or distant metastasis, which lead to largely
unsatisfactory outcomes for NSCLC patients (3). The five-year overall survival rate is
as low as 15% (4). Therefore,
there is an urgent requirement to understand the underlying
mechanism of NSCLC progression.
Long noncoding RNAs (lncRNAs) belong to the family
of noncoding RNAs and are over 200 nucleotides (nt) in length with
no or limited protein-coding ability (5). In the past decade, increasing
evidence has demonstrated that lncRNAs are important regulators in
a diverse number of biological processes, including immunity
(6), differentiation (7,8) and
cancer (9). In human cancer, the
roles of lncRNAs are gradually being uncovered. Previous findings
showed that lncRNAs are involved in the regulation of cancer cell
proliferation, apoptosis and migration (10). For instance, Zhang et al
(11) reported that lncRNA
CPS1-IT1 suppresses cell proliferation, invasion and metastasis in
colorectal cancer. Chen et al (12) demonstrated that lncRNA SNHG20
promotes NSCLC cell proliferation and migration by the epigenetic
silencing of P21 expression. Aberrant expression of several lncRNAs
has been observed in NSCLC, including LINC00339 (13). However, the function and mechanism
of most lncRNAs remain to be investigated in NSCLC.
Recently, LINC00978 has been reported to be involved
in the progression of breast cancer (14) and gastric cancer (15). Nevertheless, the functions of
LINC00978 in NSCLC are largely unknown. In the present study, it
was demonstrated that LINC00978 was significantly upregulated in
NSCLC tissues compared with the adjacent normal tissues.
Furthermore, knockdown of LINC00978 suppressed NSCLC cell
proliferation, migration and invasion while promoting apoptosis. In
terms of the mechanism involved, it was demonstrated that LINC00978
served as a competing endogenous RNA sponge to microRNA
(miR)-6754-5p. In conclusion the results of the present study
demonstrated that LINC00978 exerts an oncogenic role in NSCLC via
inhibiting miR-6754-5p.
Materials and methods
Patient samples
A cohort of 39 NSCLC tissues (29 male and 10 female;
aged 31–63 years; median age, 51 years) and adjacent non-tumor
tissues were collected at Yan'an Hospital Affiliated to Kunming
Medical University from January 2015 to December 2016. The study
was approved by the clinical research ethics committee of Yan'an
Hospital Affiliated to Kunming Medical University. Written informed
consent was obtained from all the patients. Diagnoses of NSCLC were
given by two pathologists. All patients recruited to this study did
not receive any preoperative treatment. NSCLC patients were staged
according to the tumor node metastasis (TNM) staging system of the
American Joint Committee on Cancer.
Cell culture and transfection
The human NSCLC cell lines (H1299, H1650, A549 and
PC9) and human bronchial epithelial cell line (16HBE) were
purchased from the Chinese Academy of Sciences (Shanghai, China)
and cultivated in Dulbecco's modified Eagle's medium (DMEM;
Hyclone; Ge Healthcare Life Sciences, Logan, UT, USA) supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and streptomycin (100 µg/ml), penicillin
(100 U/ml). All the cells were cultured at 37°C in a humidified
incubator with 5% CO2.
According to the manufacturer's protocol,
1×106 A549 or H1299 cells were transfected using
Lipofectamine 2000 Reagent (Thermo Fisher Scientific, Inc.) and 48
h after transfection, subsequent experiments were performed.
Scrambled small interfering (si)RNAs (siNC;
5′-UUCUCCGAACGUGUCACGU-3′) or LINC00978 siRNAs (siLINC00978;
5′-GGUGGCUAUGAGUCAGCAU-3′) were purchased from Invitrogen (Thermo
Fisher Scientific, Inc.) and 100 nM was used for transfection.
miR-6754-5p inhibitors (5′-TCCTCCAAACCAGCCTCCCTGG-3′), mimics
(5′-CCAGGGAGGCUGGUUUGGAGGA-3′) and controls
(5′-GCGUAACUAAUACAUCGGAUUCGU-3′) were purchased from Genecopoeia
Inc., (Rockville, MD, USA) and 100 nM was used for
transfection.
Cell proliferation
For the Cell Counting Kit-8 (CCK-8) assay, A549 or
H1299 cells were seeded in 96-well plates and then cells were
cultured for 24, 48, 72 and 96 h prior to performing the CCK-8
assay (Dojindo Molecular Technologies, Inc., Kumamoto, Japan).
Following incubation with CCK-8 at 37°C, absorbance (optical
density value) at a wavelength of 450 nm was detected and used for
calculating cell viability.
In vitro invasion assays
Cell invasion assay was performed using 24-well
Transwell chambers with polycarbonate membranes containing 8-mm
diameter pores (Corning Inc., Corning, NY, USA). A total of
1×105 A549 or H1299 cells were seeded on the top side of
the membrane precoated with Matrigel in DMEM without serum (BD
Biosciences, Franklin Lakes, NJ, USA). The lower chambers were
filled with DMEM with 10% FBS. Following incubation at 37°C for 24
h, the non-invasive cells on the top side of the membrane were
removed by scraping. Invasive cells on the lower membrane were
fixed with 20% methanol for 30 min at 25°C and stained with 0.1%
crystal violet for 15 min at 25°C. Invasion was quantified by
counting cells in 6 randomized fields of view in each well under a
light microscope (Olympus Corporation, Tokyo, Japan) at the level
of ×100 magnification.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cultured cells (H1299,
H1650, A549, PC9 and 16HBE) or NSCLC tissues utilizing the TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). First strand
complementary DNA was generated with a Reverse Transcription System
kit (Takara Biotechnology Co., Ltd., Dalian, China), according to
the manufacturer's protocol. The expression levels of LINC00978 and
miR-6754-5p were detected using SYBR® Premix Ex Taq™
(Takara Biotechnology Co., Ltd.) on ABI 7500 system (Applied
Biosystems; Thermo Fisher Scientific, Inc.), respectively. The
thermocycling conditions were as follows: Denaturation at 95°C for
10 min, followed by 40 cycles of denaturation at 95°C for 15 sec
and elongation at 60°C for 1 min. U6 small nuclear RNA (snRNA) was
used as the internal controls for LINC00978 and miR-6754-5p,
respectively. The RT-qPCR results were calculated using the
2−ΔΔCq method (16).
The primer sequences were as follows: LINC00978 (forward,
5′-TATGCCCAGGTTCTTCTGAGC-3′; reverse, 5′-GCATGGTGTGGTTCATTCTGG-3′),
U6 (forward, 5′-AACGAGACGACGACAGAC-3′; reverse,
5′-GCAAATTCGTGAAGCGTTCCATA-3′) and miR-6754-5p (forward,
5′-AACGAGACGACGACAGAC-3′; reverse,
5′-CCAGGGAGGCTGGTTTGGAGGA-3′).
RNA immunoprecipitation (RIP)
The RIP experiment was performed using the Magna
RIP™ RNA-Binding Protein Immunoprecipitation kit (EMD Millipore,
Billerica, MA, USA) following the manufacturer's protocol. A total
of 2×106 A549 cells were lysed using complete RIP lysis
buffer and 100 µl of the whole A549 cell extract was incubated with
RIPA buffer (Thermo Fisher Scientific Inc.) containing magnetic
beads conjugated with human anti-Argonaute2 (Ago2) antibody (cat.
no. 07-590; EMD Millipore) for 6–8 h at 4°C. A total of 4 µg normal
mouse IgG (cat. no. PP542-K; EMD Millipore) conjugated to magnetic
beads were used as a negative control. Samples were washed with
washing buffer and incubated with 1 µg/ml proteinase K
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 55°C for 30 min
to isolate the RNA-protein complexes from beads. Then
immunoprecipitated RNA was extracted and subjected to RT-qPCR
analysis.
Bioinformatics analysis
The miRBD online tool (http://mirdb.org/miRDB/custom.html) was used to
predict the potential miR targets of LINC00978.
Apoptosis assay
Apoptosis was determined by staining with Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI)
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Apoptosis was analyzed using a flow
cytometer and BD Accuri C6 Software version 1.0.264.21 (BD
Biosciences). The apoptosis rate was the sum of %
FITC+/PI− and FITC+/PI+
cells.
Luciferase reporter assay
The mutant segments of LINC00978 were constructed.
The wild type (Wt) and mutated type (Mut) of LINC00978 were
inserted into luciferase reporter assay vector pmirGLO (Promega
Corporation, Madison, WI, USA) was co-transfected with miR-NC or
miR-6754-5p mimic into 2×104 A549 cells using
Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The luciferase activity of each sample was
measured using the Dual-Luciferase® Reporter Assay
system (Promega Corporation) and normalized to the Renilla
luciferase activity.
Statistical analysis
Statistical analyses were performed using SPSS 20.0
(IBM, Corp., Armonk, NY, USA) and GraphPad Prism version 6
(GraphPad, Inc., La Jolla, CA, USA. The results were from three
independent experiments and are presented as the mean ± standard
deviation. A student's t-test or one-way analysis of variance
followed by Tukey's post hoc test was used to analyze 2 or multiple
groups, respectively, for statistical significance. Pearson's
correlation coefficient analysis was used to determine the
correlations. P<0.05 was considered to indicate a statistically
significant difference.
Results
LINC00978 is expressed at a high level
in NSCLC tissues
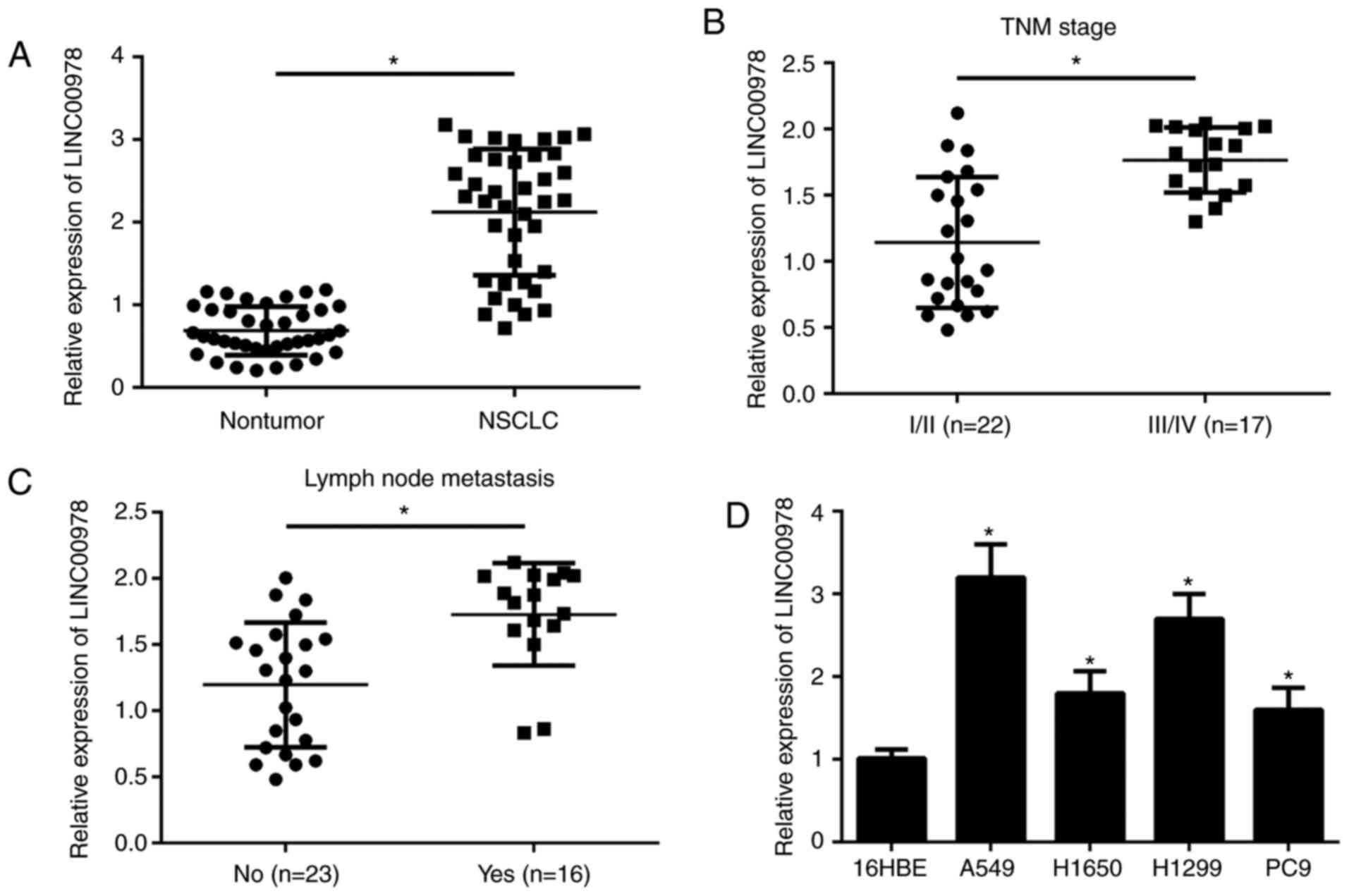
To investigate the function of LINC00978 in NSCLC
progression, RT-qPCR was used to measure its expression patterns.
Results demonstrated that LINC00978 expression was significantly
upregulated in NSCLC tissues compared with the adjacent normal
tissues (P<0.05; Fig. 1A).
Additionally, it was demonstrated that LINC00978 expression was
significantly positively associated with grade III/IV tumor TNM
stage and lymph node metastasis (P<0.05; Fig. 1B and C). Consistently, it was
demonstrated that the expression of LINC00978 was also
significantly upregulated in NSCLC cell lines, including H1299,
H1650, A549 and PC9 cells compared with the 16HBE cell line
(P<0.05; Fig. 1D). The data
from the present study indicated that LINC00978 may exert an
oncogenic role in NSCLC progression.
LINC00978 knockdown suppressed NSCLC
cell proliferation, migration and invasion
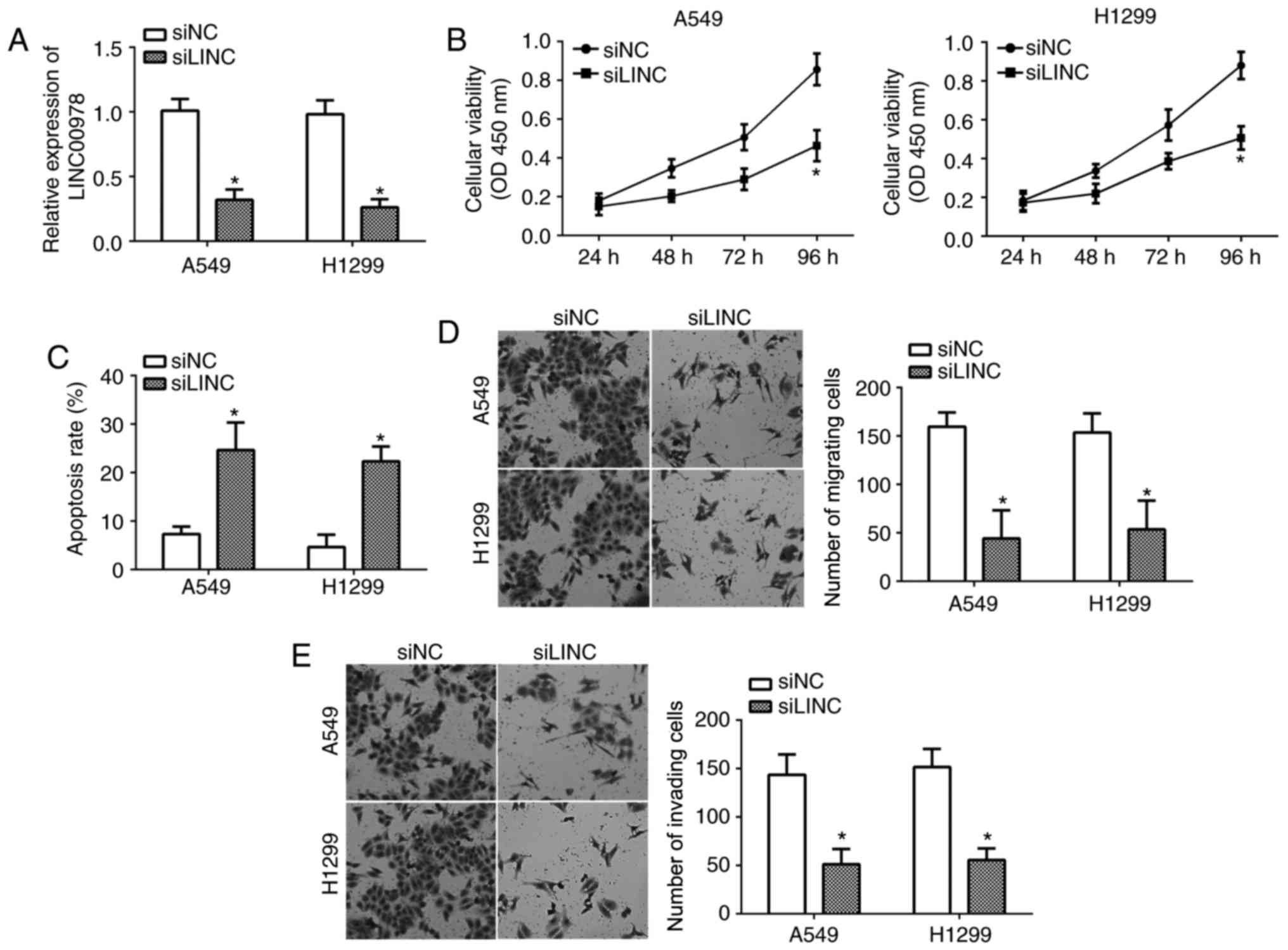
To investigate the influence of LINC00978 on NSCLC,
LINC00978 was knocked down in A549 and H1299 cells by transfection
with specific siRNA against LINC00978. RT-qPCR analysis indicated
that LINC00978 expression was significantly downregulated following
transfection (P<0.05; Fig. 2A).
CCK-8 assays demonstrated that the proliferation of A549 and H1299
cells in the siLINC00978 group was decreased compared with the
control group (Fig. 2B).
Additionally, it was demonstrated that LINC00978 knockdown
significantly promoted cell apoptosis by flow cytometry (P<0.05;
Fig. 2C). Furthermore, the effects
of LINC00978 on NSCLC cell migration and invasion were analyzed by
transwell assays. It was demonstrated that LINC00978 silencing
significantly inhibited the cell numbers of migration and invasion
(P<0.05; Fig. 2D and E). These
results suggested that LINC00978 contributed to the malignancy of
NSCLC.
miR-6754-5p is a direct target of
LINC00978
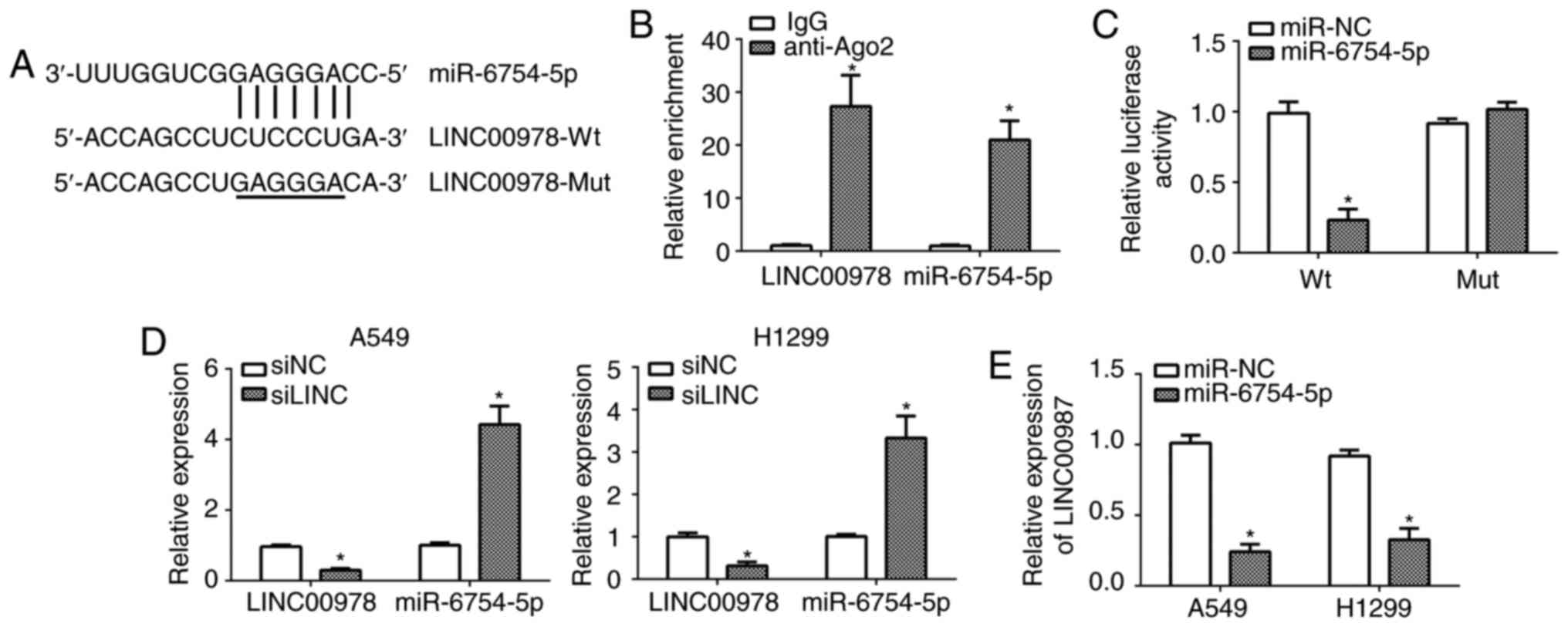
It has been acknowledged that lncRNA could function
as a competing endogenous RNA to regulate gene expression by
binding miR (17). Therefore, it
was hypothesized that LINC00978 possesses a similar function in
NSCLC. Through bioinformatics analysis, it was demonstrated that
LINC00978 may directly interact with miR-6754-5p (Fig. 3A). To determine whether LINC00978
and miR-6754-5p were in the same RNA-induced silencing complex, RIP
assays were performed with A549 cell lysates using anti-Ago2 or
IgG. It was demonstrated that LINC00978 and miR-6754-5p were both
significantly enriched in Ago2-containing miRs relative to control
group (P<0.05; Fig. 3B).
Furthermore, luciferase reporter assays were performed with
reporter plasmids containing the wild-type (WT) or mutated
miR-6754-5p binding sites in LINC00978. As presented, it was
demonstrated that the overexpression of miR-6754-5p significantly
inhibited the luciferase activity in A549 cells transfected with
WT-LINC00978 (P<0.05; Fig. 3C).
In addition, RT-qPCR analysis demonstrated that knockdown of
LINC00978 promoted the expression of miR-6754-5p in A549 and H1299
cells (Fig. 3D). Consistently, the
ectopic expression of miR-6754-5p significantly repressed the
expression of LINC00978 in A549 and H1299 cells (P<0.05;
Fig. 3E). Taken together, these
data indicated that LINC00978 could directly act as a sponge for
miR-6754-5p.
miR-6754-5p is downregulated in NSCLC
tissues
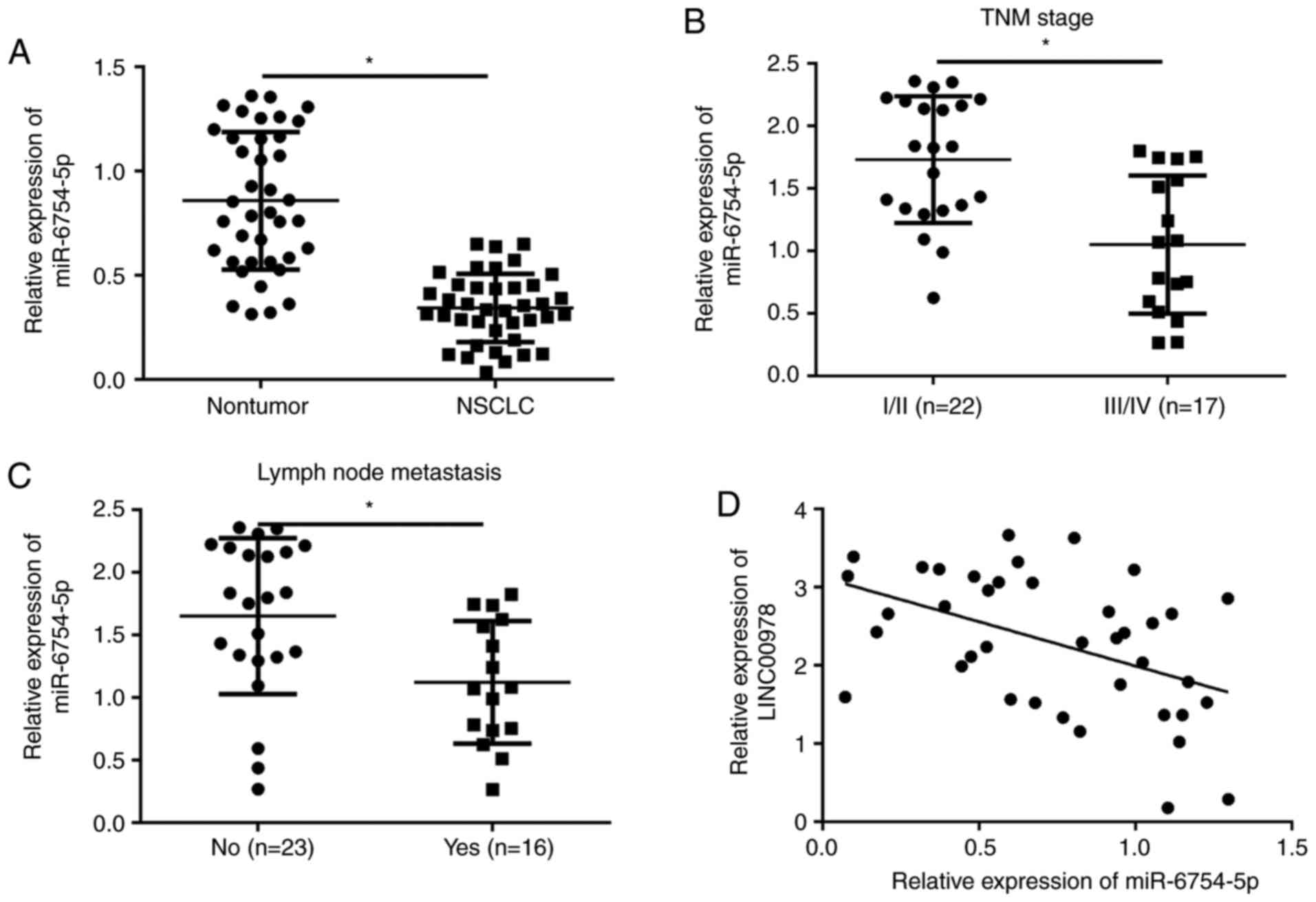
The function of miR-6754-5p remains largely unknown.
To determine the function of miR-6754-5p in NSCLC, its expression
patterns were examined using RT-qPCR. It was demonstrated that
miR-6754-5p was significantly downregulated in NSCLC tissues
compared with the adjacent normal tissues (P<0.05; Fig. 4A). Furthermore, it was demonstrated
that the expression of miR-6754-5p was significantly inversely
associated with TNM stage and tumor metastasis in NSCLC (P<0.05;
Fig. 4B and C). In addition, it
was demonstrated that there was a negative correlation between
miR-6754-5p and LINC00978 expressions in NSCLC tissues (Fig. 4D). The above data suggested that
miR-6754-5p may possess an opposite role to LINC00978 in NSCLC
progression.
Repression of miR-6754-5p reversed the
function of LINC00978 knockdown on NSCLC cells
To further investigate whether LINC00978 regulates
NSCLC progression by inhibiting miR-6754-5p expression, A549 cells
were transduced with siLINC00978 and siLINC00978+miR-6754-5p
inhibitor. RT-qPCR analysis indicated that miR-6754-5p expression
was downregulated in siLINC00978+miR-6754-5p inhibitor group
compared with the siLINC00978 group (Fig. 5A). Then CCK-8 assays, flow
cytometry and transwell assays were performed to determine cell
viability, apoptosis, migration and invasion. The results
demonstrated that LINC00978 knockdown repressed the proliferation,
migration and invasion while promoting cell apoptosis in A549 cells
(Fig. 5B-E). However, inhibition
of miR-6754-5p at the same time reversed the effects of LINC00978
knockdown on A549 cells at least in part (Fig. 5B-E). In conclusion, the results of
the present study indicated that LINC00978 promotes NSCLC cell
proliferation, migration an invasion by inhibiting miR-6754-5p
expression.
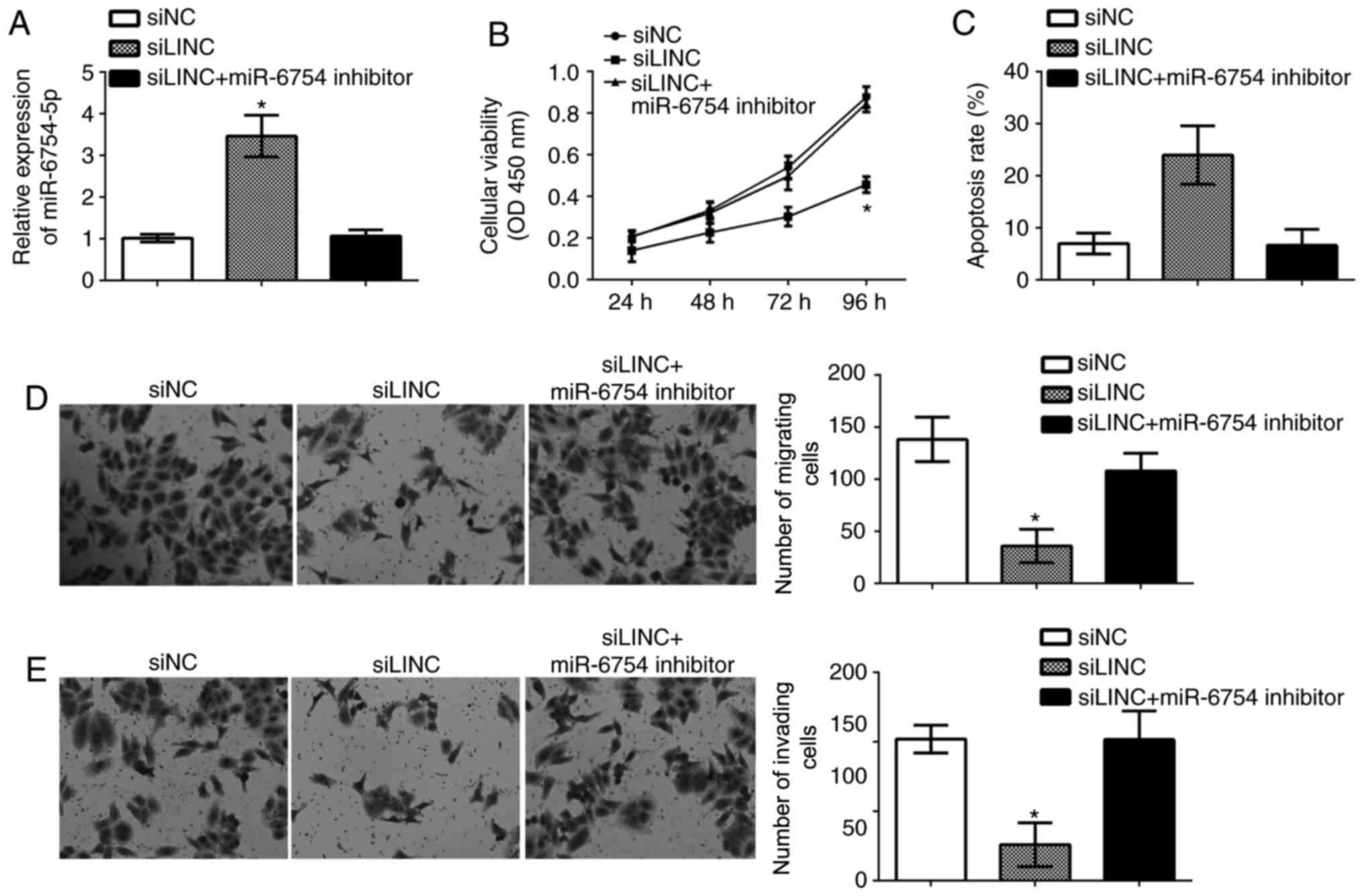 | Figure 5.Repression of miR-6754-5p reversed the
function of LINC00978 knockdown on NSCLC cells. (A) RT-qPCR was
used to determine miR-6754-5p expression in NSCLC cells transfected
with control, siLINC, or miR-17-5p-inhibitor + siLINC. (B) Cell
Counting Kit-8 assay was used to investigate cell viability in
NSCLC cells transfected with a control, siLINC, or
miR-17-5p-inhibitor + siLINC. (C) Cell apoptosis was measured by
fluorescence activated cell sorting in NSCLC cells transfected with
a control, siLINC, or miR-17-5p-inhibitor + siLINC. (D) Cell
migration and (E) invasion were determined by transwell assays in
NSCLC cells transfected with control, siLINC, or
miR-17-5p-inhibitor + siLINC (magnification, ×100). All data are
representative of three independent experiments and expressed as
mean ± standard deviation. *P<0.05 vs. the control group. si,
small interfering; NSCLC, non-small cell lung cancer; miR,
microRNA; LINC, LINC00978. |
Discussion
NSCLC has become a major public health problem and
leads to large amounts of cancer-associated mortalities worldwide
(1). However, the underlying
mechanism of NSCLC development and progression remains elusive.
Several lncRNAs have been reported to participate in NSCLC,
including linc00460 (18) and
ASAP1-IT1 (19). However, the
functions and mechanisms of most lncRNAs in NSCLC have not been
defined. In the present study, it was demonstrated that LINC00978
was expressed at a high level in NSCLC tissues compared with the
adjacent normal tissues. Furthermore, the expression of LINC00978
was positively associated with TNM stage and lymph node metastasis
in NSCLC. The data suggested that LINC00978 may serve an oncogenic
role in NSCLC progression.
Previously, lncRNAs were considered to be
transcriptional noise. However, in the past decade, lncRNA has
become an extremely hot topic in the field of biology. Increasing
evidence indicates that lncRNAs exert essential functions in almost
all kinds of biological processes (20). In human cancer, lncRNAs have been
demonstrated to be involved in tumor development and progression by
regulating survival, proliferation and metastasis (21). In NSCLC, several lncRNAs are
abnormally expressed. For example, Li et al (10) reported that lncRNA MALAT1 is
upregulated and contributes to NSCLC development by modulating
miR-124/STAT3 axis. Jiang et al (22) reported that lncRNA-HOTAIR is
overexpressed and affects tumorigenesis and metastasis of NSCLC by
upregulating miR-613. Jiang et al (23) demonstrated that NEAT1 acts as an
inducer of cancer stem cell-like phenotypes in NSCLC by inhibiting
EGCG-upregulated CTR1. As for LINC00978, certain studies
demonstrated that LINC00978 promotes cancer growth and acts as a
diagnostic or prognostic biomarker in gastric cancer and breast
cancer (14,15). Nevertheless, whether LINC00978
serves a role in NSCLC needs to be elucidated. In the present
study, it was demonstrated that LINC00978 was upregulated in NSCLC
tissues and cell lines. Furthermore, through functional experiments
it was demonstrated that LINC00978 knockdown significantly
inhibited NSCLC cell proliferation, migration and invasion while
promoting apoptosis. The data suggested that overexpression of
LINC00978 may contribute to NSCLC progression.
miRs also belong to the noncoding RNA (ncRNA)
family. However, miRs are a class of short ncRNAs with a length of
~22 nucleotides and that can regulate gene expression by binding to
the complementary sequence in the 3′-untranslated region of target
mRNAs (24). Increasing evidence
demonstrated that miRNAs are expressed in nearly all cell types and
regulate various biological processes (25). By regulating cell proliferation,
migration and invasion, miRs are associated with the occurrence and
progression of human cancer (26).
In NSCLC, a number of miRs have been demonstrated to serve
important roles in regulating malignant behaviors (27). Emerging evidence demonstrates that
the expression of miR could be regulated by lncRNA in cancer cells
(28). In the present study, it
was demonstrated that LINC00978 could act as a sponge for
miR-6754-5p in NSCLC cells. The function of miR-6754-5p was
unknown; however, the results of the present study demonstrated
that miR-6754-5p was significantly downregulated in NSCLC tissues
and its expression was suppressed by LINC00978. Furthermore, CCK-8,
FACS and transwell assays indicated that LINC00978-induced
regulation of NSCLC cell proliferation, migration, invasion and
apoptosis is dependent on the inhibition of miR-6754-5p.
Furthermore, a previous study (15) demonstrated that LINC00978 activates
transforming growth factor (TGF)-β/mothers against decapentaplegic
homolog (SMAD) signaling pathway to promote gastric cancer
progression. Whether LINC00978 has a similar effect on the
TGF-β/SMAD signaling pathway in NSCLC remains unclear. miRs exert
functions via targeting mRNAs for degradation. Through
bioinformatics analysis using TargetScan version 7 (http://www.targetscan.org/vert_71/), a number of
oncogenes including MYC, CCND3, MMP2 and LGR4, were demonstrated to
be potential targets of miR-6754-5p (data not shown). Whether
miR-6754-5p suppresses NSCLC progression via inhibiting these
oncogenes require further investigation.
In conclusion, the results of the present study
demonstrated that LINC00978 promotes the proliferation, migration
and invasion of NSCLC cells by repression of miR-6754-5p
expression. LINC00978 expression was also identified to be
positively associated with clinical severity and tumor metastasis
in NSCLC patients, suggesting LINC00978 may be a promising
prognostic biomarker and therapeutic target for NSCLC
treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XL and YR designed the present study, performed
experiments, analyzed and interpreted the results. TZ conceived the
present study and wrote this manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
For the use of human samples, the protocol for this
study was approved by the Institutional Ethics Committee of Yan'an
Hospital Affiliated to Kunming Medical University and all enrolled
patients signed a written informed consent document.
Patient consent for publication
All patients within this study provide consent for
the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sun C, Li S, Zhang F, Xi Y, Wang L, Bi Y
and Li D: Long non-coding RNA NEAT1 promotes non-small cell lung
cancer progression through regulation of miR-377-3p-E2F3 pathway.
Oncotarget. 7:51784–51814. 2016.PubMed/NCBI
|
|
3
|
Goldstraw P, Ball D, Jett JR, Le Chevalier
T, Lim E, Nicholson AG and Shepherd FA: Non-small-cell lung cancer.
Lancet. 378:1727–1740. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ridge CA, McErlean AM and Ginsberg MS:
Epidemiology of lung cancer. Semin Intervent Radiol. 30:93–98.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang G, An X and Zhao H, Zhang Q and Zhao
H: Long non-coding RNA HNF1A-AS1 promotes cell proliferation and
invasion via regulating miR-17-5p in non-small cell lung cancer.
Biomed Pharmacother. 98:594–599. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu
P, Du Y, Wu J, Qin X, Chen R, et al: Long noncoding RNA lncKdm2b is
required for ILC3 maintenance by initiation of Zfp292 expression.
Nat Immunol. 18:499–508. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X,
Wu B, Xu R, Liu W, Yan P, et al: Divergent lncRNAs regulate gene
expression and lineage differentiation in pluripotent cells. Cell
Stem Cell. 18:637–652. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W,
Zhu P, Wang Y, Wang S, Xia P, et al: LncKdm2b controls self-renewal
of embryonic stem cells via activating expression of transcription
factor Zbtb3. EMBO J. 37:e971742018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L and Fan Z: LncBRM initiates YAP1
signalling activation to drive self-renewal of liver cancer stem
cells. Nat Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li S, Mei Z, Hu HB and Zhang X: The lncRNA
MALAT1 contributes to non-small cell lung cancer development via
modulating miR-124/STAT3 axis. J Cell Physiol. 233:6679–6688. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang W, Yuan W, Song J, Wang S and Gu X:
LncRna CPS1-IT1 suppresses cell proliferation, invasion and
metastasis in colorectal cancer. Cell Physiol Biochem. 44:567–580.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B,
Zhang T, Zhou Y, Chen Q, Wei C, et al: Long non-coding RNA SNHG20
promotes non-small cell lung cancer cell proliferation and
migration by epigenetically silencing of P21 expression. Cell Death
Dis. 8:e30922017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan Y, Haiying G, Zhuo L, Ying L and Xin
H: Long non-coding RNA LINC00339 facilitates the tumorigenesis of
non-small cell lung cancer by sponging miR-145 through targeting
FOXM1. Biomed Pharmacother. 105:707–713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Deng LL, Chi YY, Liu L, Huang NS, Wang L
and Wu J: LINC00978 predicts poor prognosis in breast cancer
patients. Sci Rep. 6:379362016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu M, Huang Z, Zang X, Pan L, Liang W,
Chen J, Qian H, Xu W, Jiang P and Zhang X: Long noncoding RNA
LINC00978 promotes cancer growth and acts as a diagnostic biomarker
in gastric cancer. Cell Prolif. 51:e124252018. View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao L, Kong H, Sun H, Chen Z, Chen B and
Zhou M: LncRNA-PVT1 promotes pancreatic cancer cells proliferation
and migration through acting as a molecular sponge to regulate
miR-448. J Cell Physiol. 233:4044–4055. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y,
Zhou JK, Guo C, Xia Z, Liu L, et al: Long non-coding RNA linc00460
promotes epithelial-mesenchymal transition and cell migration in
lung cancer cells. Cancer Lett. 420:80–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Shi SB, Zhu Y, Qian TT and Wang
HL: Long non-coding RNA ASAP1-IT1 promotes cell proliferation,
invasion and metastasis through the PTEN/AKT signaling axis in
non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 22:142–149.
2018.PubMed/NCBI
|
|
20
|
Xi J, Feng J and Zeng S: Long noncoding
RNA lncBRM facilitates the proliferation, migration and invasion of
ovarian cancer cells via upregulation of Sox4. Am J Cancer Res.
7:2180–2189. 2017.PubMed/NCBI
|
|
21
|
Kong Q and Qiu M: Long noncoding RNA
SNHG15 promotes human breast cancer proliferation, migration and
invasion by sponging miR-211-3p. Biochem Biophys Res Commun.
495:1594–1600. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang C, Yang Y, Yang Y, Guo L, Huang J,
Liu X, Wu C and Zou J: LncRNA-HOTAIR affects tumorigenesis and
metastasis of non-small cell lung cancer by up-regulating miR-613.
Oncol Res. 26:725–734. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiang P, Chen A, Wu X, Zhou M, Ul Haq I,
Mariyam Z and Feng Q: NEAT1 acts as an inducer of cancer stem
cell-like phenotypes in NSCLC by inhibiting EGCG-upregulated CTR1.
J Cell Physiol. 233:4852–4863. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang A, Lakshmanan J, Motameni A and
Harbrecht BG: MicroRNA-203 suppresses proliferation in liver cancer
associated with PIK3CA, p38 MAPK, c-Jun, and GSK3 signaling. Mol
Cell Biochem. 441:89–98. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ren Y, Shang J, Li J, Liu W, Zhang Z, Yuan
J and Yang M: The long noncoding RNA PCAT-1 links the microRNA
miR-215 to oncogene CRKL-mediated signaling in hepatocellular
carcinoma. J Biol Chem. 292:17939–17949. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang Z, Yang Y and Zhang X: MiR-770
inhibits tumorigenesis and EMT by targeting JMJD6 and regulating
WNT/β-catenin pathway in non-small cell lung cancer. Life Sci.
188:163–171. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Wang Y and Wang J: MicroRNA-584
inhibits cell proliferation and invasion in non-small cell lung
cancer by directly targeting MTDH. Exp Ther Med. 15:2203–2211.
2018.PubMed/NCBI
|
|
28
|
Lu Q, Shan S, Li Y, Zhu D, Jin W and Ren
T: Long noncoding RNA SNHG1 promotes non-small cell lung cancer
progression by up-regulating MTDH via sponging miR-145-5p. FASEB J.
32:3957–3967. 2018. View Article : Google Scholar : PubMed/NCBI
|