Introduction
Gastric cancer is one of the most common types of
cancer worldwide. The global incidence of gastric cancer has
declined over recent decades (1);
however, due to aging in the world population, the absolute number
of new cases per year is increasing. Furthermore, there is a recent
upward trend in young patients with gastric cancer (2); therefore, gastric cancer remains an
important cause of cancer-associated mortality.
Cytotoxin-associated gene A (CagA) has been reported
to be associated with gastric diseases, particularly peptic ulcer
disease (3). In total, 85–100%
patients with duodenal ulcers are infected with CagA+
Helicobacter pylori (H. pylori) strains (4). The origin of H. pylori strains
has been reported to serve an important role in the rates of
gastric cancer, which may be associated with differences in the
capacity of different strains to express CagA. Notably, the
incidence of precancerous lesions and gastric cancer is higher in
patients with CagA+ strains (5). Furthermore, 88% of animals infected
with a rodent-adapted strain of CagA-producing human H.
pylori developed gastric dysplasia within 4 weeks, and 75% of
animals developed gastric adenocarcinoma by week 8 (6). In addition, specific amino acid
sequences in the CagA protein have been revealed to be associated
with an increased risk of malignancy (7); however, the mechanism by which CagA
affects the expression of genes in gastric cancer remains
elusive.
The phosphatase and tensin homolog (PTEN) gene,
which is located on chromosome 10q23, is a tumor-suppressor gene
(8–10). It has been reported to regulate the
phosphoinositide-3-kinase-protein kinase B (AKT) and mechanistic
target of rapamycin signaling pathways, which serve important roles
in apoptosis, cell cycle progression and cell proliferation. Loss
of function of PTEN has been reported to result in oncogenesis and
somatic mutations in various malignancies (11). Tet methylcytosine dioxygenase
(Tet)1 has been reported to exert its tumor-suppressor function in
gastric cancer through interactions with the p53-enhancer of zeste
2 polycomb repressive complex 2 subunit (EZH2) signaling pathway.
Tet1 suppresses cancer formation via inhibition of the oncogenic
protein EZH2 and activation of p53, possibly via DNA demethylation
(12). However, the associations
between CagA, PTEN and Tet1 in gastric cancer remain unclear.
Therefore, the present study aimed to investigate the interactions
between CagA, PTEN and Tet1 in gastric cancer.
Materials and methods
Reagents
Primers, TRIzol® reagent, SuperScript III
Reverse Transcriptase, SYBR Green I and DEPC H2O were
purchased from Invitrogen (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). The RNase inhibitor was purchased from Fermentas (Thermo
Fisher Scientific, Inc., Pittsburgh, PA, USA). Platinum Taq DNA
polymerase, oligo dT/primer and 100 mM dNTPs were purchased from
Invitrogen (Thermo Fisher Scientific, Inc.) and used for PCR.
Dulbecco modified Eagle's medium (DMEM) was purchased from Gibco
(Thermo Fisher Scientific, Inc.).
Construction of recombinant plasmids
and lentiviral packaging
HEK 293T cells (American Type Culture Collection,
Manassas, VA, USA) and human gastric cancer cell line HGC-27 cells
were cultivated in Dulbecco modified Eagle's medium (DMEM,
Invitrogen; Thermo Fisher Scientific, Inc.) with 10% of fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) in a
humidified incubator with 5% CO2 at 37°C. The human
gastric cancer cell line HGC-27 was purchased from RIKEN
Bioresource Center (Tsukuba, Japan); this cell line is derived from
a metastatic lymph node, and is a highly metastatic cell line due
to its undifferentiated characteristics. The Tet1 catalytic domain
(NM_030625.2) was used to design three short hairpin RNAs (shRNAs)
(Table I). Upon annealing, shRNA
was treated with T4 DNA polymerase in the presence of dATP, and
then annealed with the pds019-pl6.3-SHRNA-BSD vector (Novagen, EMD
Millipore, Billerica, MA, USA). Then, the annealed complex was
transformed into E. coli host strain. The recombinant
plasmids were sequenced, and named CL946-1, CL946-2 and CL946-3. In
addition, the Tet1 catalytic domain was cloned into a
PDS087_pL6-TO-V5-GIM vector (Novagen; EMD Millipore) using the
restriction enzyme site NheI/AscI. The recombinant
plasmid was named CL981_pL6-TO-V5-tet1-CD. Similarly, a CagA
(AB015416.1) overexpression vector was generated. Subsequently,
packaging mix (9 µg; Thermo Fisher Scientific, Inc.) and
recombinant lentiviral plasmids (3 µg) were added into Opti-Minimum
Essential Medium (MEM, Thermo Fisher Scientific, Inc.) and mixed.
In addition, Lipofectamine® 2000 (36 µl; Invitrogen;
Thermo Fisher Scientific, Inc.) was mixed with Opti-MEM (1.5 ml),
and incubated at room temperature for 5 min. The plasmid solution
and diluted Lipofectamine® 2000 were then mixed, and
incubated at room temperature for 5 min. The mixture was added into
a culture dish with HEK 293T cells, and the cells were cultured for
48 h in the conditions described above. The cell supernatant was
then collected, centrifuged at 1,500 × g for 10 min at room
temperature, and filtered. The virus solution was then condensed by
centrifuging at 50,000 × g for 2 h at 4°C, and resuspended in DMEM.
Tet1 interference (VL946_PDS19-shTET1), Tet1 interference
(VL946_PDS19-shTET2), Tet1 interference (VL946_PDS19-shTET3), Tet1
overexpression (LV576_pL6-TO-V5-tet1-CD), CagA overexpression and
negative control viruses were derived. HGC-27 cells were maintained
in DMEM with 10% of fetal bovine serum in a humidified incubator
with 5% CO2 at 37°C and were plated into cell plates (60
mm) at 4×105 cells/well overnight. HGC-27 cells were
infected with CL946-1, CL946-2 and CL946-3 viruses for 3 days and
quantitative polymerase chain reaction (qPCR) was used to detect
interference efficiency. To determine the viruses transduction
efficiency in HGC-27 cells, GFP expression was examined by
microscopy at different multiplicities of infection (MOIs) on day 3
following infection.
 | Table I.Sequences of shRNAs used for vector
construction and primers used for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of shRNAs used for vector
construction and primers used for reverse
transcription-quantitative polymerase chain reaction.
| Primer/shRNA | Sequence
(5′-3′) |
|---|
| Sh1-Tet1-F |
CACCGCAGCTAATGAAGGTCCCCAGAACGAATTCTGGACCTTCATTAGCTGC |
| Sh1-Tet1-R |
AAAAGCAGCTAATGAAGGTCCAGAATTCGTTCTGGACTAGCTGCCTTCAT |
| Sh2-Tet1-F |
CACCG*CCCAGAAGATTTAGAATTGATCGAAATCAATTCTAAATCTTCTGGG |
| Sh2-Tet1-R |
AAAACCCAGAAGATTTAGAATTGATTTCGATCAATTCTAAATCTTCTGGGC* |
| Sh3-Tet1-F |
CACCG*CCTCCAGTCTTAATAAGGTTACGAATAACCTTATTAAGACTGGAGG |
| Sh3-Tet1-R |
AAAACCTCCAGTCTTAATAAGGTTATTCGTAACCTTATTAAGACTGGAGGC |
| Tet1-F |
AGTGGTGACTATGCCAGTGC |
| Tet1-R |
CAGACCCCACATCGCTTTCT |
| Negative
control-F |
TAATTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTTT |
| Negative
control-R |
TCGAGAAAAAAAATTCTCCGAAGCGTGTCACGTTCTCTTGAAACTGACACGTTCGGAGTTA |
| PTEN-F |
CTCAGCCGTTACCTGTGTGT |
| PTEN-R |
AGGTTTCCTCTGGTCCTGGT |
| CagA-F |
CTGCCAAACAATCTTTTGCAG |
| CagA-R |
CTGGTTCTTGATTGATTGCTTC |
| Ras-F |
ACTGGGGAGGGCTTTCTTTG |
| Ras-R |
GCATCATCAACACCCTGTCT |
| c-met-F |
GTCCTGCAGTCAATGCCTCT |
| c-met-R |
GTCAGCCTTGTCCCTCCTTC |
| c-myc-F |
CATCAGCACAACTACGCAGC |
| c-myc-R |
GCTGGTGCATTTTCGGTTGT |
| ESR2-F |
GTCCCTGGTGTGAAGCAAGA |
| ESR2-R |
CCTTCACACGACCAGACTCC |
| Tet2-F |
TAGCCACACCCCAGCTTTAG |
| Tet2-R |
TCTGGATAAACGCCATGTGTCT |
| Tet3-F |
GTGCTCCGGAAGAGTTTCCA |
| Tet3-R |
ACGGTGCACCCATTGTAGAG |
| ACRC-F |
CAGCACTGGTGAGATGTGGT |
| ACRC-R |
CATCAATCAGCCAGGAGGCA |
| AICDA-F |
AGCCTCTTGATGAACCGGAG |
| AICDA-R |
TGTCACGCCTCTTCACTACG |
| ALKBH1-F |
ACTGGGTGAAACAGTGCCTT |
| ALKBH1-R |
AAACTTCGGGGTCTCCGTTT |
| ALKBH2-F |
GCTGGGCTGACCTACACATT |
| ALKBH2-R |
CGATGTGGTCACAGCCATCT |
| APEX1-F |
TTACGGCATAGGCGATGAGG |
| APEX1-R |
TGTGCCACATTGAGGTCTCC |
| APOBEC2-F |
CGTGTGCTGACCGCATTATC |
| APOBEC2-R |
CAGTTTACAGCCAGCCTCCT |
| APOBEC3A-F |
GGTCAAGATGGACCAGCACA |
| APOBEC3A-R |
AGGAACCAGGTCCAAGAAGC |
| APOBEC3C-F |
ACCTATGGGAAGCCAACGAT |
| APOBEC3C-R |
AGAGAGGAAGCACCTTTCTGC |
| APOBEC3F-F |
GTCTGGCTGTGCTACGAAGT |
| APOBEC3F-R |
CACATTTCTGCGTGGTGCTC |
| FTO-F |
ACCCATGGCTCAACTGGAAG |
| FTO-R |
AGCAGGTAATGTTCGGGCAA |
| TDG-F |
GAGAACGCGGGCAGCTATTC |
| TDG-R |
TGGGGCTGGAACTTCTTCTG |
Cell viability assay
Cell viability was measured using the Cell Counting
kit 8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). Briefly, HGC-27 cells (5×103 cells/well) were
seeded into 96-well plates and cultured overnight. Subsequently,
cells were infected with TET1 interference or CagA overexpression
viruses for 24, 48 or 72 h. The medium was then removed and 10 µl
CCK-8 solution was added to each well. After 4 h, cell viability
was detected using a plate reader at a wavelength of 450 nm.
Reverse transcription (RT)-qPCR
The mRNA expression levels of PTEN, Tet1,
apolipoprotein B mRNA editing enzyme catalytic subunit (APOBEC)3A,
APOBEC3C, APOBEC3F, Ras, c-myc, c-met, estrogen receptor 2 (ESR2),
Tet2, Tet3, acidic repeat containing (ACRC), activation-induced
cytidine deaminase (AICDA), alkB homolog 1, histone H2A dioxygenase
(ALKBH1), alkB homolog 2, α-ketoglutarate dependent dioxygenase
(ALKBH2), apurinic/apyrimidinic endodeoxyribonuclease 1 (APEX1),
APOBEC2, FTO, α-ketoglutarate dependent dioxygenase (FTO) and
thymine DNA glycosylase (TDG) in HGC-27 human gastric carcinoma
cells were detected by RT-qPCR. All these genes have been reported
to have close associations with gene methylation/demethylation
(13). Total RNA was extracted
using TRIzol® reagent, according to the manufacturer's
protocol. A universal cDNA synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for RT. Each reaction contained 0.5 µl
random primers (0.2 µg/µl) and 1 µl SuperScript III reverse
transcriptase (200 U/µl). The specific primers used for RT-qPCR are
listed in Table I. PCR was
performed using a SYBR qPCR kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The PCR conditions were as follows:
Predenaturation at 95°C for 2 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec and
polymerization at 70°C for 45 sec, followed by 1 cycle of melting
curve at 95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec. PCR
was performed using a CFX96 Touch™ Real-time PCR Detection system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Gene expression
was determined and normalized to β-actin. The primers for β-actin
were as follows: Forward, 5′-AGGGAAATCGTGCGTGAC-3′ and reverse,
5′-CGCTCATTGCCGATAGTG-3′. The 2−ΔΔCq method was used to
analyze PCR results (14). The
fluorophore used for qPCR was included in the SYBR qPCR kit.
Detection of methylation status of
PTEN by methylation-specific PCR
Promoter methylation of the PTEN gene was determined
by methylation-specific PCR. Genomic DNA from HGC-27 cells as well
as control specimens were subjected to bisulfite modifications
using an EZ DNA Methylation-Gold kit (Zymo Research Corp., Irvine,
CA, USA) in accordance with the manufacturer's protocol. DNA (500
ng) was treated with sodium hydrogen sulfite at 50°C for 16 h.
Unmethylated cytidine was deaminated and converted into uracil,
whereas methylated cytidine remained unchanged. Based on these
alterations, primers to amplify methylated and unmethylated PTEN
sequences by PCR were designed using MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi;
Table II). PCR was performed
using the following conditions: An initial melting step of 10 min
at 95°C; followed by 50 cycles of 20 sec at 95°C, 20 sec at 59°C
and 45 sec at 72°C with a final elongation step of 4 min at 72°C in
a CFX96 Touch™ Real-time PCR Detection system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). 3.5% Agarose gel
electrophoresis (Thermo Fisher Scientific, Inc.) was used to detect
methylation status.
 | Table II.Primers used for methylation-specific
polymerase chain reaction. |
Table II.
Primers used for methylation-specific
polymerase chain reaction.
| Primer | Sequences
(5′-3′) |
|---|
| PTEN-M-F |
GTATTTCGAGTAAAGGAAGAAGACG |
| PTEN-M-R |
GATAAAAAACTACAACCCAACGAA |
| PTEN-U-F |
TATTTTGAGTAAAGGAAGAAGATGA |
| PTEN-U-R |
CAATAAAAAACTACAACCCAACAAA |
Western blot analysis
The protein expression levels of PTEN and Tet1 were
detected by western blot analysis. Cells or tissues were lysed in
lysis buffer (cat. no. P0013; Beyotime Institute of Biotechnology,
Haimen, China) with protease and phosphatase inhibitors (cat. no.
P1045; Beyotime Institute of Biotechnology) at 4°C. The lysis
mixture was centrifuged at 10,000 × g for 10 min at 4°C, and the
supernatant containing cellular proteins was used in subsequent
experiments. The protein concentration was determined using a
bicinchoninic acid kit. Subsequently, proteins (40 µg/lane) were
separated by 10% SDS-PAGE (120 V) and the separated proteins were
transferred to polyvinylidene fluoride membranes (cat. no. FFP24;
Beyotime Institute of Biotechnology) at 100 V for 2 h. After
blocking with 5% non-fat milk for 1 h at room temperature, the
membranes were incubated with primary antibodies against PTEN
(1:400, cat. no. ab32199), Tet1 (1:400, cat. no. ab191698) and
GAPDH (1:400, cat. no. ab8245) at 4°C overnight. The membranes were
then washed with Tris-buffered saline containing 0.5% Tween 20, and
were incubated with horseradish peroxidase-conjugated goat
anti-rabbit secondary antibody (1:5,000, cat. no. ab150077) at room
temperature for 1 h. All antibodies were purchased from Abcam
(Cambridge, MA, USA). The membranes were then washed and incubated
in enhanced chemiluminescence solution (cat. no. P0018A; Beyotime
Institute of Biotechnology), and the images were captured on film
(FF057; Beyotime Institute of Biotechnology) in a dark room. The
experiments were repeated three times. Blot images were
semi-quantified in grayscale using ImageJ (version 2.0; National
Institutes of Health, Bethesda, MD, USA).
Gastric cancer tissue sample
collection
A total of 12 patients with gastric cancer were
enrolled between April 2017 and May 2018 for the study at the
Affiliated Hospital of Jining Medical University (Jining, China),
and gastric cancer was confirmed pathologically. Patient tumor
tissues were obtained during surgery. All patients provided written
informed consent and the study was approved by the Institutional
Review Board/Ethics Committee of the Affiliated Hospital of Jining
Medical University. The clinical information of the patients is
provided in Table III.
 | Table III.Clinical information of patients with
gastric cancer. |
Table III.
Clinical information of patients with
gastric cancer.
| Sample no. | Sex | Age (years) | TNM stage | CagA |
|---|
| 1 | Male | 45 | T3N1M0 | + |
| 2 | Male | 71 | T2N0M0 | − |
| 3 | Male | 52 | T4N3M0 | + |
| 4 | Female | 55 | T4N3M0 | + |
| 5 | Male | 64 | T3N1M0 | − |
| 6 | Male | 59 | T2N0M0 | − |
| 7 | Male | 71 | T3N2M0 | + |
| 8 | Male | 53 | T4N3M0 | + |
| 9 | Male | 74 | T4N3M0 | + |
| 10 | Female | 40 | T2N0M0 | − |
| 11 | Male | 72 | T3N2M0 | + |
| 12 | Male | 74 | T2N0M0 | − |
Statistical analysis
Statistical data were analyzed by GraphPad Prism
version 5.0 software (GraphPad Software, Inc., La Jolla, CA, USA).
Data are presented as the means ± standard deviation. Differences
between more than two groups were compared by one-way analysis of
variance followed by the Bonferroni post-hoc test. Differences
between two groups were compared by Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Tet1 interference and CagA
overexpression in HGC-27 cells
Initially, HGC-27 cells were infected with viruses
carrying Tet1 interference or CagA overexpression vectors. Relative
expression levels of Tet1 and CagA were detected by RT-qPCR. The
expression levels of Tet1 in HGC-27 cells were significantly
decreased by Tet1 interference compared with in the negative
control group (P<0.001; Fig.
1A). Since Tet1 interference-sh1 exerted the best inhibitory
effect on the cells, it was used in subsequent experiments. In
addition, compared with in the negative control group, infection of
the cells with CagA or Tet1 overexpression vectors markedly
increased the expression of CagA and Tet1, respectively
(P<0.001; Fig. 1B and C).
Furthermore, Tet1 interference or CagA overexpression significantly
enhanced HGC-27 cell viability compared with in the control group
(P<0.01; Fig. 1D). These
results indicated that Tet1 interference, Tet1 overexpression and
CagA overexpression were successfully achieved.
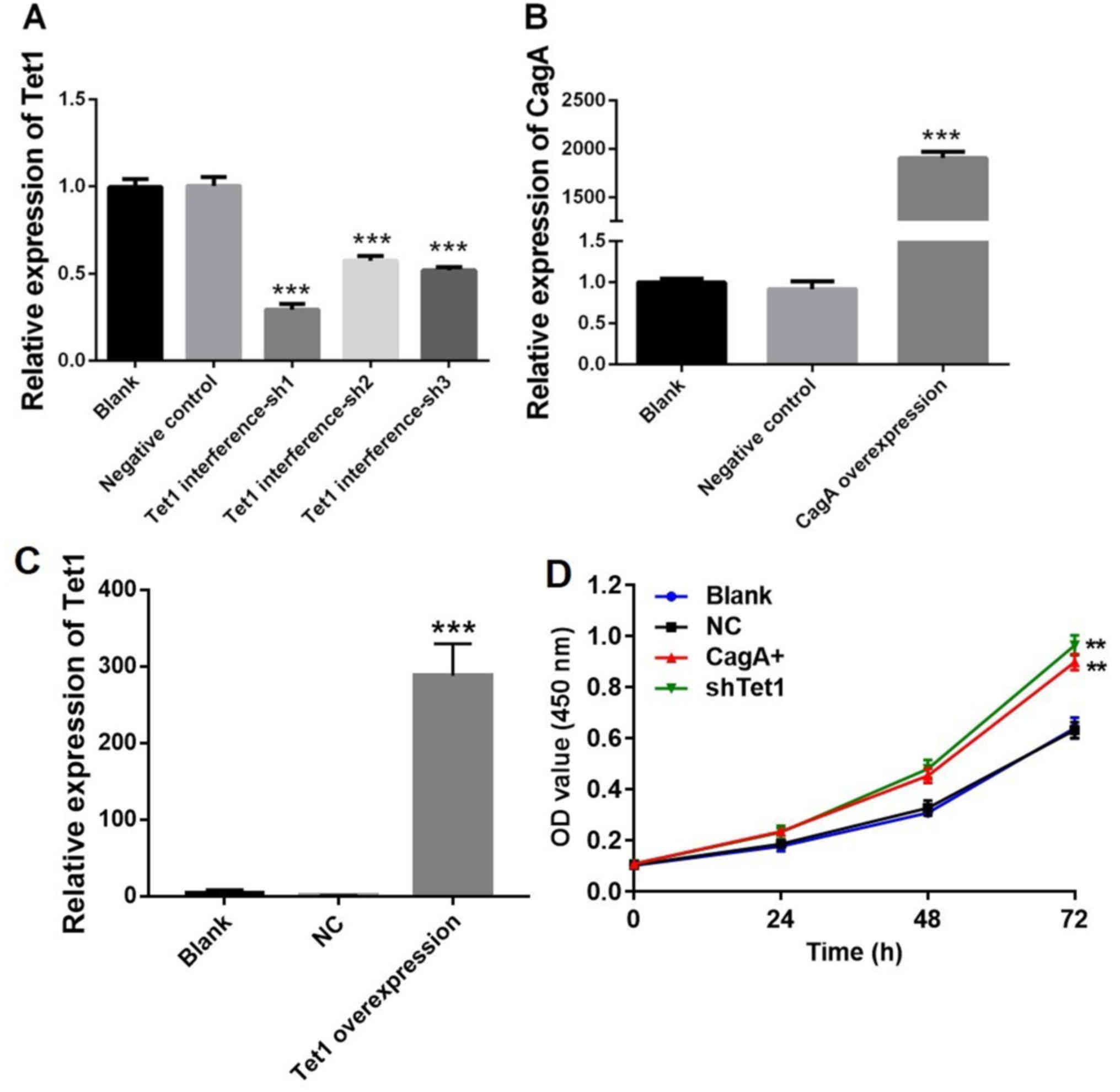 | Figure 1.Confirmation of Tet1 interference,
Tet1 overexpression and CagA overexpression in HGC-27 cells. (A)
HGC-27 cells were infected with viruses carrying Tet1 interference
vectors. Relative expression was detected by RT-qPCR. Expression
levels of Tet1 were significantly decreased in HGC-27 cells upon
Tet1 interference. (B and C) HGC-27 cells were infected with
viruses carrying a CagA or Tet1 overexpression vector. Relative
expression levels of CagA and Tet1 were detected by RT-qPCR.
Compared with in the NC group, CagA and Tet1 expression was
markedly increased in the CagA and Tet1 overexpression groups,
respectively. (D) HGC-27 cells were infected with viruses carrying
Tet1 interference or CagA overexpression vectors for 24, 48 or 72
h. Cell viability was detected using the Cell Counting kit 8 kit.
These results indicated that Tet1 interference, Tet1 overexpression
and CagA overexpression were successfully established.
***P<0.001, **P<0.01 vs. the NC group (n=3). CagA,
cytotoxin-associated gene A; NC, negative control; OD, optical
density; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; sh, short hairpin RNA; Tet1, tet methylcytosine
dioxygenase 1. |
Expression levels of PTEN, Tet1,
APOBEC3A, APOBEC3C and APOBEC3F are significantly decreased in the
CagA overexpression group
RT-qPCR was used to detect gene expression in HGC-27
cells under the condition of CagA overexpression. As shown in
Fig. 2A, the mRNA expression
levels of PTEN, Tet1, APOBEC3A, APOBEC3C and APOBEC3F were
significantly decreased in the CagA overexpression group compared
with in the negative control group (P<0.01). However, there were
no significant differences regarding the expression of Ras, c-myc,
c-met, ESR2, Tet2, Tet3, ACRC, AICDA, ALKBH1, ALKBH2, APEX1,
APOBEC2, FTO or TDG in the cells (Fig.
2B).
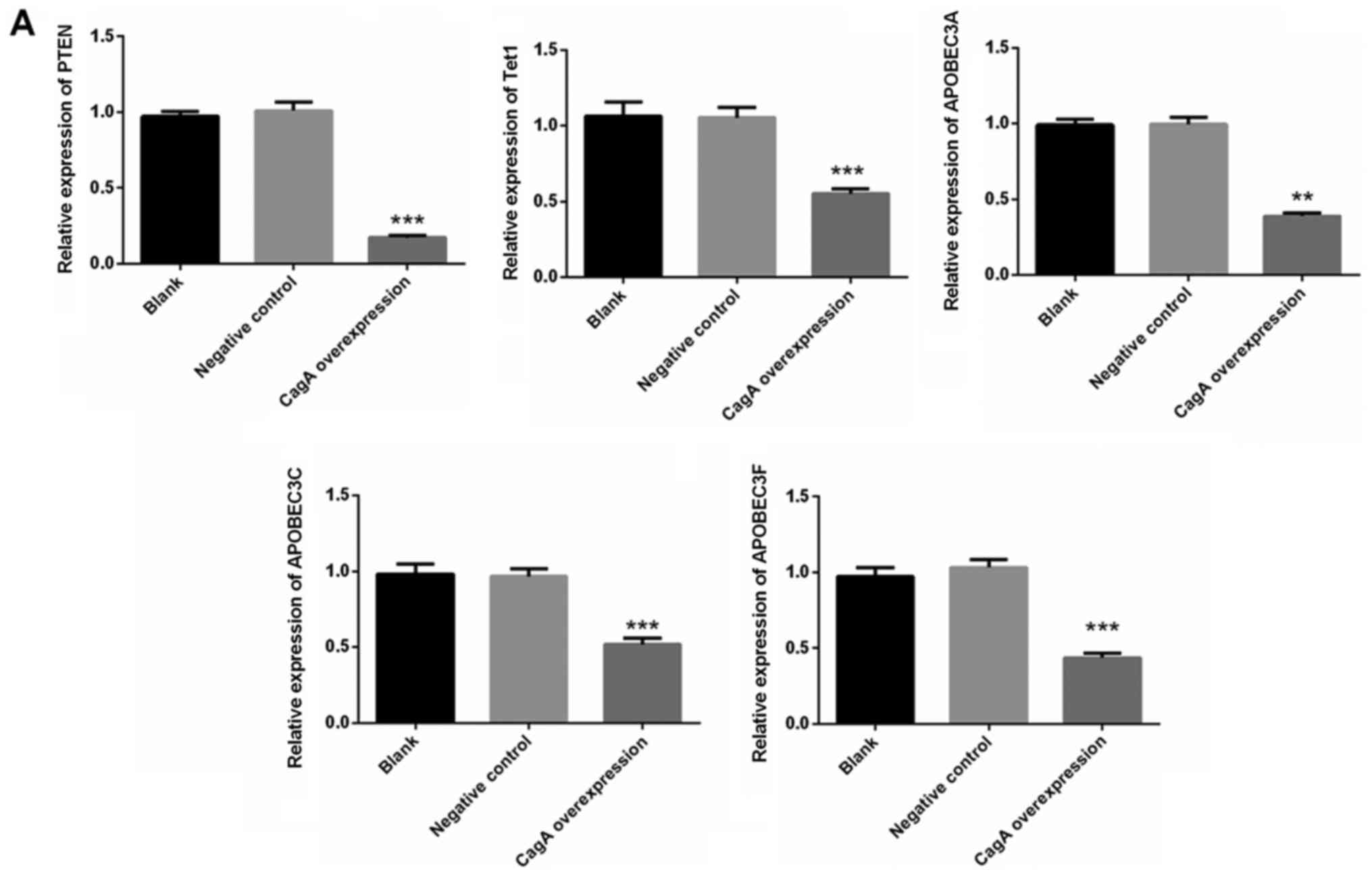 | Figure 2.mRNA expression levels of PTEN, Tet1,
APOBEC3A, APOBEC3C and APOBEC3F are significantly decreased in the
CagA overexpression group. (A) HGC-27 cells were infected with
viruses carrying a CagA overexpression vector. The expression
levels of PTEN, Tet1, APOBEC3A, APOBEC3C and APOBEC3F were detected
by reverse transcription-quantitative polymerase chain reaction.
**P<0.01, ***P<0.001 vs. with the negative control group
(n=3). APOBEC, apolipoprotein B mRNA editing enzyme catalytic
subunit; CagA, cytotoxin-associated gene A; PTEN, phosphatase and
tensin homolog; Tet1, tet methylcytosine dioxygenase 1. mRNA
expression levels of PTEN, Tet1, APOBEC3A, APOBEC3C and APOBEC3F
are significantly decreased in the CagA overexpression group. (B)
Ras, c-myc, c-met, ESR2, Tet2, Tet3, ACRC, AICDA, ALKBH1, ALKBH2,
APEX1, APOBEC2, FTO and TDG levels were detected following CagA
overexpression in the cells. ACRC, acidic repeat containing; AICDA,
activation-induced cytidine deaminase; ALKBH1, alkB homolog 1,
histone H2A dioxygenase; ALKBH2, alkB homolog 2, α-ketoglutarate
dependent dioxygenase; APOBEC, apolipoprotein B mRNA editing enzyme
catalytic subunit; CagA, cytotoxin-associated gene A; ESR2,
estrogen receptor 2; FTO, FTO, α-ketoglutarate dependent
dioxygenase; PTEN, phosphatase and tensin homolog; Tet1, tet
methylcytosine dioxygenase 1; TDG, thymine DNA glycosylase. |
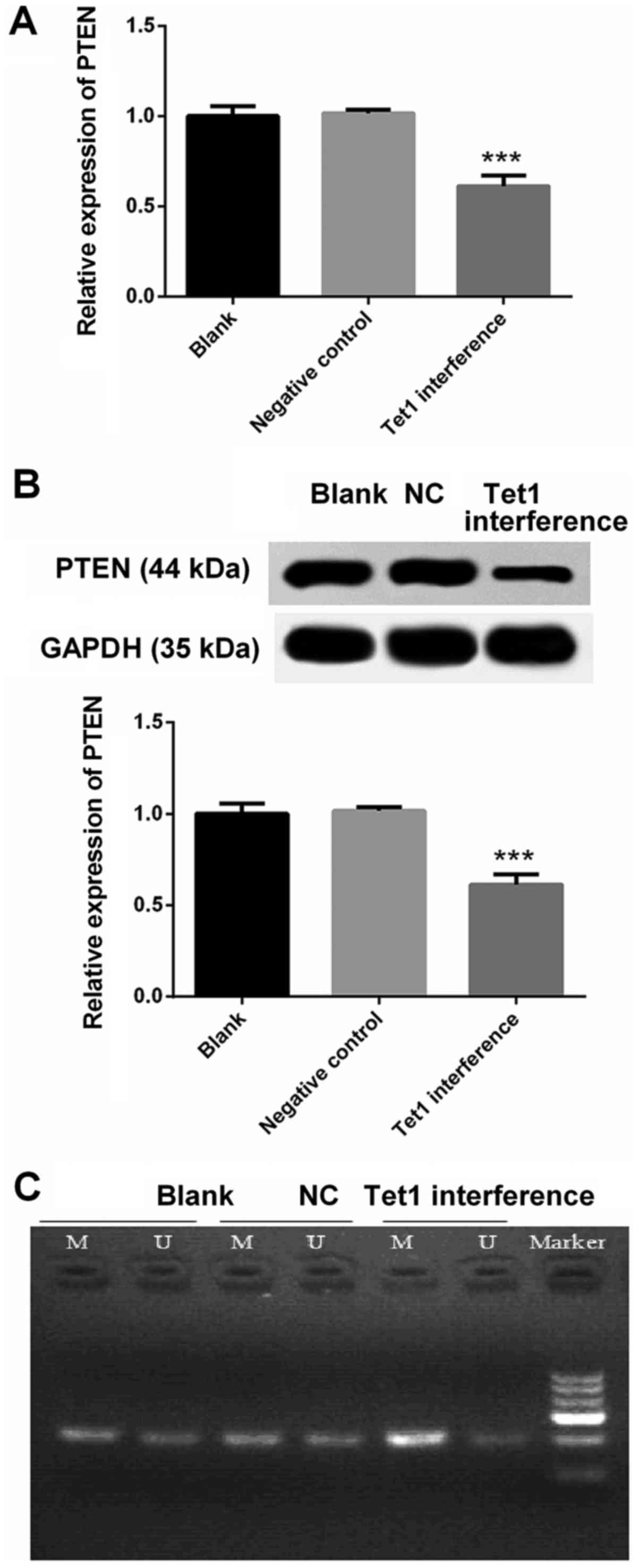
Decreased expression of PTEN in HGC-27
cells with Tet1 interference is associated with its increased
methylation
RT-qPCR and western blotting were used to detect the
mRNA and protein expression levels of PTEN, respectively. Compared
with in the negative control group, the mRNA and protein expression
levels of PTEN were markedly decreased in HGC-27 cells with Tet1
interference (P<0.001; Fig. 3A and
B). The decreased expression of PTEN in the Tet1 interference
group was associated with its increased methylation (Fig. 3C).
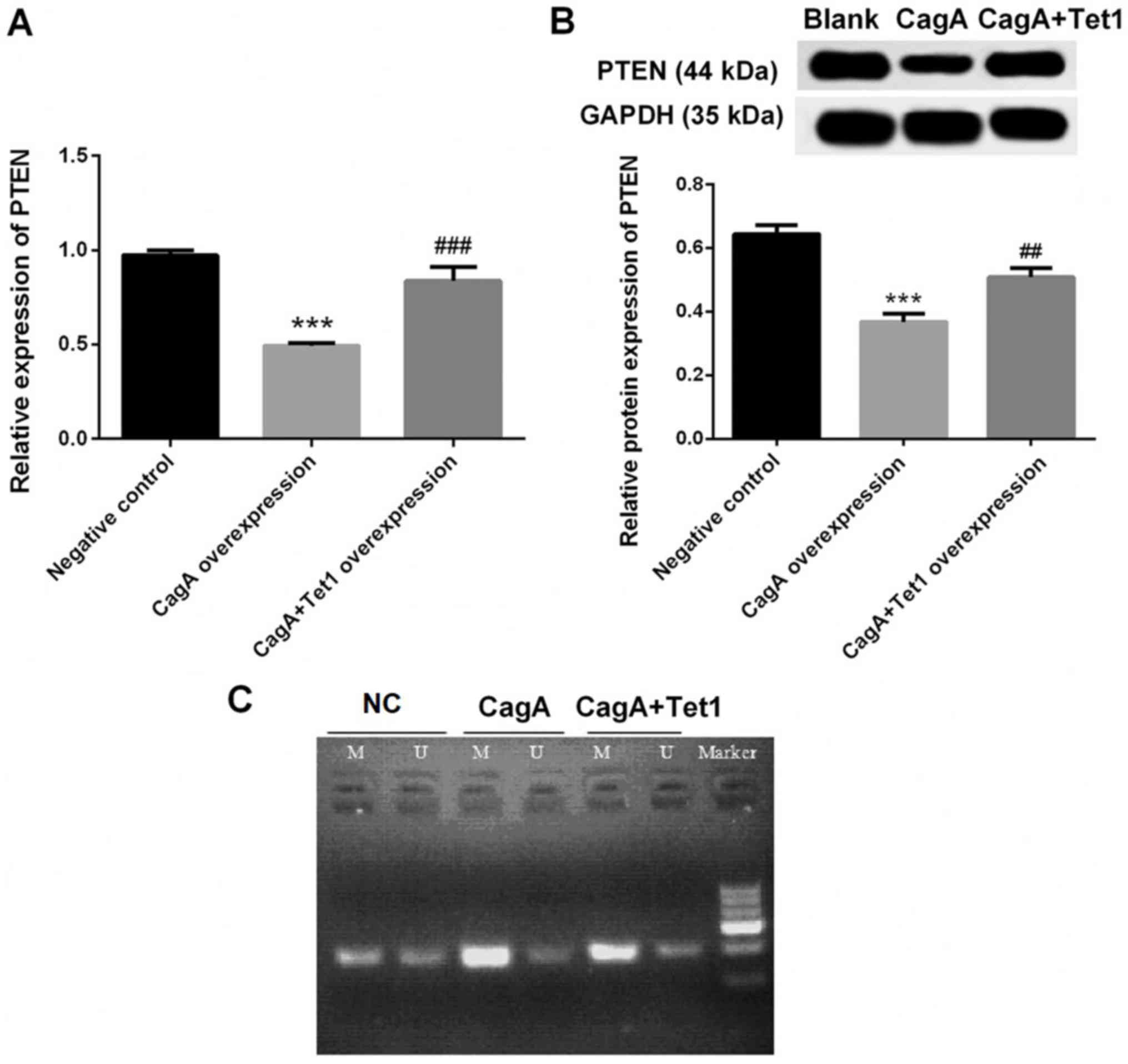
Decreased expression of PTEN in HGC-27
cells with CagA overexpression is reversed by Tet1
overexpression
Compared with in the negative control group, the
mRNA and protein expression levels of PTEN were markedly decreased
in HGC-27 cells with CagA overexpression (P<0.001; Fig. 4A and B), which was significantly
reversed by Tet1 overexpression (P<0.01; Fig. 4A and B). In addition, the decreased
expression of PTEN in the CagA overexpression group was associated
with its increased methylation in the cells, whereas Tet1
overexpression attenuated this methylation (Fig. 4C).
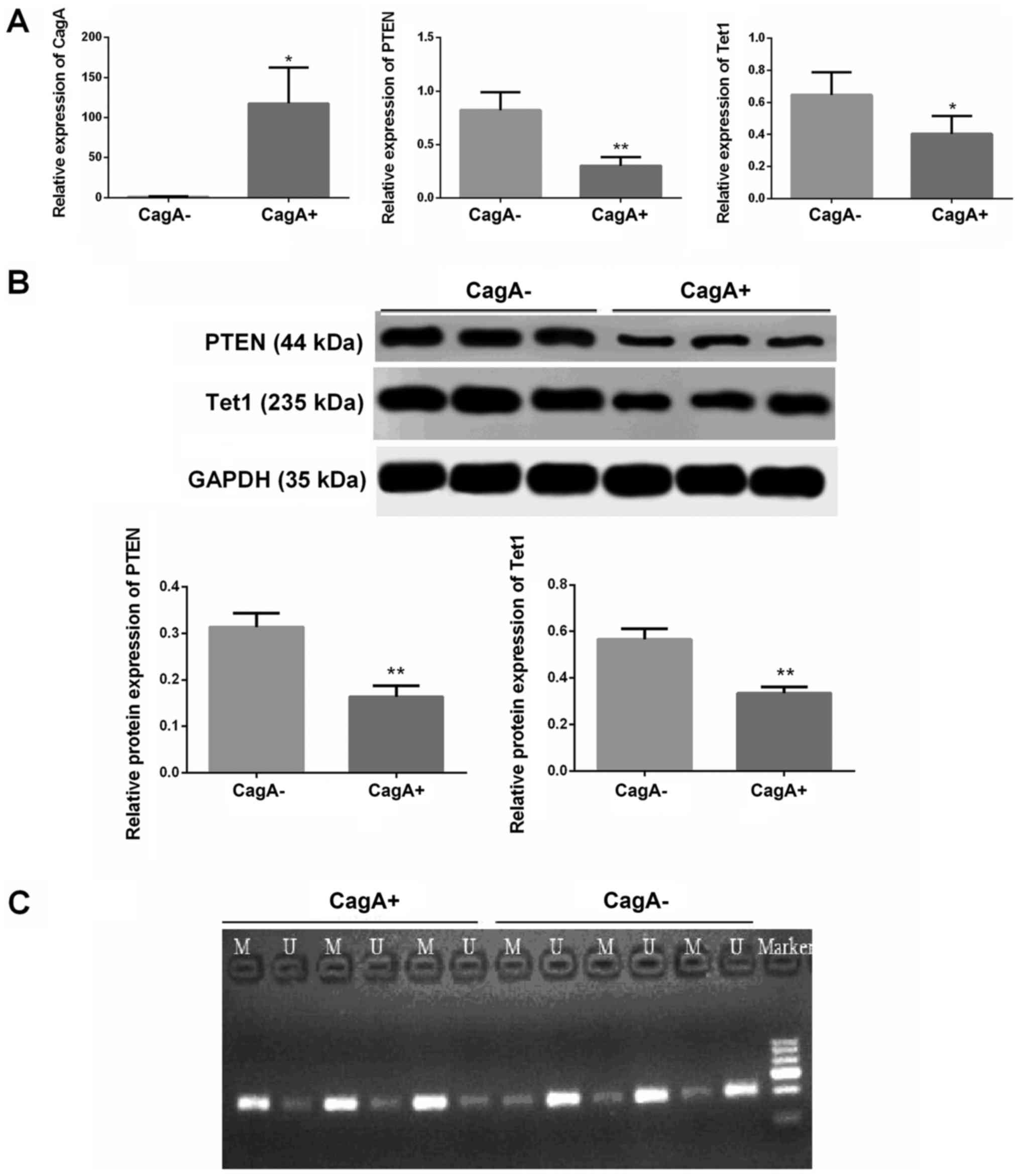
Expression of PTEN and Tet1 is
markedly decreased in CagA+ gastric cancer tissues
According to the expression of CagA, gastric cancer
tissues were grouped into a CagA− or a CagA+
group (Fig. 5A).
Immunohistochemistry was used to detect the expression of CagA. If
CagA was expressed in the gastric cancer tissues, it was designated
the CagA+ group. If CagA was not expressed in the
gastric cancer tissues, it was designated the CagA−
group (data not shown). The results indicated that the expression
levels of PTEN and Tet1 were significantly decreased in the
CagA+ gastric cancer tissues compared with in
CagA− gastric cancer tissues (Fig. 5A and B; P<0.05). In addition,
the decreased expression of PTEN in CagA+ gastric cancer
tissues was associated with its increased methylation (Fig. 5C).
Discussion
The present study demonstrated that CagA may
significantly decrease the expression of PTEN, Tet1, APOBEC3A,
APOBEC3C and APOBEC3F in human gastric cancer. The results
suggested that CagA decreased PTEN expression by increasing
methylation levels, and decreased expression of PTEN was
significantly attenuated by Tet1.
The risk of gastric cancer differs among various
strains of H. pylori. Strains with virulence factors,
including CagA and vacuolating cytotoxin A, are more likely to be
associated with precancerous lesions and gastric cancer (15–17).
A specific motif (AATAAGATA) has been revealed to be linked to the
capacity of H. pylori to express CagA (18). Highly virulent strains of H.
pylori produce CagA-modulated c-met receptor signal
transduction pathways in vitro, which may influence the
initiation and progression of gastric cancer (19).
Gastric carcinogenesis has been proposed to involve
an initial stage of dedifferentiation, including gastric atrophy,
followed by abnormal redifferentiation, including intestinal
metaplasia. This process is mediated by the effects of H.
pylori infection, particularly CagA+ strains, on
β-catenin (20,21). Apical junctions are disrupted by
CagA expression, and mechanisms that maintain the differentiation
of normal epithelium, including cell adhesion, cell polarity and
inhibition of migration, are also disturbed by CagA expression
in vitro (22). In
addition, proteolytic cleavage of E-cadherin is induced, and
E-cadherin-dependent cell-to-cell contact is disrupted by CagA
(23). Formation of the
E-cadherin/β-catenin complex is also impaired by CagA, thus leading
to the accumulation of β-catenin in the cytoplasm and nucleus, and
constitutive transcription of intestinal differentiation markers.
Nuclear localization of β-catenin has also been reported to be
present in a carcinogenic human gastric cell line, but not in a
non-carcinogenic clinical isolate (6,24).
In the present study, a highly metastatic gastric
cancer cell line, HGC-27, which was initially isolated from an
Asian patient with gastric cancer, was used. This cell line is
derived from a metastatic lymph node and may provide a novel
perspective for the role of CagA in gastric cancer metastasis. The
results of the present study demonstrated that CagA significantly
decreased the expression levels of PTEN and Tet1 in this human
gastric cancer cell line. In addition, it was revealed that CagA
decreased PTEN expression by increasing its methylation level,
which was significantly attenuated by Tet1. PTEN serves an
important role in apoptosis, cell cycle progression and cell
proliferation, and functions as a tumor-suppressor gene. It has
been reported that Tet1 exerts its tumor-suppressor function in
gastric cancer possibly via DNA demethylation (12). Tet1 has also been reported to
inhibit the growth and metastasis of gastric cancer by
demethylation and re-expression of PTEN. Tet1 suppresses the cell
growth, migration and invasion of gastric cancer via demethylation
of CpG islands in the PTEN promoter region, by increasing
5-hydroxymethylcytosine. The re-expressed PTEN has been revealed to
downregulate the activity of AKT and focal adhesion kinase
(25). This may explain how Tet1
attenuated CagA-induced methylation and PTEN downregulation in
gastric cancer in the present study. The present findings have
provided novel mechanisms for CagA-induced gastric
carcinogenesis.
The present study demonstrated that CagA
significantly decreased the expression levels of APOBEC3A, APOBEC3C
and APOBEC3F in human gastric cancer. APOBEC3A has been reported to
possess anticancer and antiviral effects via inhibiting the
expression of human papillomavirus E6 and E7 in cervical cancer
(26). The deletion of APOBEC3A is
also associated with a young age at diagnosis among patients with
lung and prostate cancer (27).
APOBEC3C has been reported to be a potent inhibitor of replication
of simian immunodeficiency virus (28), and APOBEC3F restricts the
proliferation of human immunodeficiency virus (29). However, evidence regarding the
roles of APOBEC3A, APOBEC3C and APOBEC3F in human gastric cancer is
scarce. To the best of our knowledge, the present study is the
first to reveal that CagA decreased the expression levels of
APOBEC3A, APOBEC3C and APOBEC3F in human gastric cancer. Although
more research is required to elucidate the roles of APOBEC3A,
APOBEC3C and APOBEC3F in gastric cancer, the present findings
provide another explanation for CagA-induced gastric
carcinogenesis. However, the results derived from studies of one
cell line are not directly transferable to patients.
In conclusion, the present study demonstrated that
CagA significantly decreased the expression of PTEN, Tet1,
APOBEC3A, APOBEC3C and APOBEC3F in human gastric cancer cells. In
addition, CagA decreased PTEN expression by increasing its
methylation levels, which was significantly reversed by Tet1
upregulation. Further investigations are required to provide
information on additional molecular mechanisms; however, the
present study may precede future therapeutic approaches targeting
human gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81602160),
the Shandong Provincial Natural Science Foundation (grant no.
ZR2016HL30), the Scientific Research Foundation for PhD, Affiliated
Hospital of Jining Medical University (grant no. 2016-BS-006), and
the Project of Health and Family Planning Commission of Shandong
province (grant no. 2017WS513).
Availability of data and materials
All data sets used in this study are available from
the corresponding author on reasonable request.
Authors' contributions
DG and SL designed the study. BZ, XZ, MJ and LH
performed the experiments and MZ, WQ, SW and BL analyzed the data.
BZ, XZ and MJ were the major contributors in developing the first
draft of this manuscript. DG and SL reviewed and approved the final
draft of the manuscript prior to submission.
Ethics approval and consent to
participate
All patients provided written informed consent and
the study was approved by the Institutional Review Board/Ethics
Committee of the Affiliated Hospital of Jining Medical
University.
Patient consent for publication
All patients provided written informed consent for
the publication of all associated data in this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhu AL and Sonnenberg A: Is gastric cancer
again rising? J Clin Gastroenterol. 46:804–806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Correa P: Gastric cancer: Two epidemics?
Dig Dis Sci. 56:1585–1586; author reply, 1586. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jeyamani L, Jayarajan J, Leelakrishnan V
and Swaminathan M: CagA and VacA genes of Helicobacter
pylori and their clinical relevance. Indian J Pathol Microbiol.
61:66–69. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weel JF, van der Hulst RW, Gerrits Y,
Roorda P, Feller M, Dankert J, Tytgat GN and van der Ende A: The
interrelationship between cytotoxin-associated gene A, vacuolating
cytotoxin, and Helicobacter pylori-related diseases. J
Infect Dis. 173:1171–1175. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang JQ, Zheng GF, Sumanac K, Irvine EJ
and Hunt RH: Meta-analysis of the relationship between cagA
seropositivity and gastric cancer. Gastroenterology. 125:1636–1644.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franco AT, Israel DA, Washington MK,
Krishna U, Fox JG, Rogers AB, Neish AS, Collier-Hyams L,
Perez-Perez GI, Hatakeyama M, et al: Activation of beta-catenin by
carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA.
102:10646–10651. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Basso D, Zambon CF, Letley DP, Stranges A,
Marchet A, Rhead JL, Schiavon S, Guariso G, Ceroti M, Nitti D, et
al: Clinical relevance of Helicobacter pylori cagA and vacA
gene polymorphisms. Gastroenterology. 135:91–99. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Keniry M and Parsons R: The role of PTEN
signaling perturbations in cancer and in targeted therapy.
Oncogene. 27:5477–5485. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krymskaya VP and Goncharova EA:
PI3K/mTORC1 activation in hamartoma syndromes: Therapeutic
prospects. Cell Cycle. 8:403–413. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sansal I and Sellers WR: The biology and
clinical relevance of the PTEN tumor suppressor pathway. J Clin
Oncol. 22:2954–2963. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stambolic V, Suzuki A, de la Pompa JL,
Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM,
Siderovski DP and Mak TW: Negative regulation of PKB/Akt-dependent
cell survival by the tumor suppressor PTEN. Cell. 95:29–39. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu HL, Ma Y, Lu LG, Hou P, Li BJ, Jin WL
and Cui DX: TET1 exerts its tumor suppressor function by
interacting with p53-EZH2 pathway in gastric cancer. J Biomed
Nanotechnol. 10:1217–1230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pastor WA, Aravind L and Rao A: TETonic
shift: Biological roles of TET proteins in DNA demethylation and
transcription. Nat Rev Mol Cell Biol. 14:341–356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Blaser MJ, Perez-Perez GI, Kleanthous H,
Cover TL, Peek RM, Chyou PH, Stemmermann GN and Nomura A: Infection
with Helicobacter pylori strains possessing cagA is
associated with an increased risk of developing adenocarcinoma of
the stomach. Cancer Res. 55:2111–2115. 1995.PubMed/NCBI
|
|
16
|
Miehlke S, Kirsch C, Agha-Amiri K, Günther
T, Lehn N, Malfertheiner P, Stolte M, Ehninger G and Bayerdörffer
E: The Helicobacter pylori vacA s1, m1 genotype and cagA is
associated with gastric carcinoma in Germany. Int J Cancer.
87:322–327. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Plummer M, van Doorn LJ, Franceschi S,
Kleter B, Canzian F, Vivas J, Lopez G, Colin D, Muñoz N and Kato I:
Helicobacter pylori cytotoxin-associated genotype and
gastric precancerous lesions. J Natl Cancer Inst. 99:1328–1334.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Loh JT, Shaffer CL, Piazuelo MB, Bravo LE,
McClain MS, Correa P and Cover TL: Analysis of cagA in
Helicobacter pylori strains from Colombian populations with
contrasting gastric cancer risk reveals a biomarker for disease
severity. Cancer Epidemiol Biomarkers Prev. 20:2237–2249. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Churin Y, Al-Ghoul L, Kepp O, Meyer TF,
Birchmeier W and Naumann M: Helicobacter pylori CagA protein
targets the c-Met receptor and enhances the motogenic response. J
Cell Biol. 161:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Faller G and Kirchner T: Immunological and
morphogenic basis of gastric mucosa atrophy and metaplasia.
Virchows Arch. 446:1–9. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hlubek F, Spaderna S, Schmalhofer O, Jung
A, Kirchner T and Brabletz T: Wnt/FZD signaling and colorectal
cancer morphogenesis. Front Biosci. 12:458–470. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bagnoli F, Buti L, Tompkins L, Covacci A
and Amieva MR: Helicobacter pylori CagA induces a transition
from polarized to invasive phenotypes in MDCK cells. Proc Natl Acad
Sci USA. 102:16339–16344. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weydig C, Starzinski-Powitz A, Carra G,
Löwer J and Wessler S: CagA-independent disruption of adherence
junction complexes involves E-cadherin shedding and implies
multiple steps in Helicobacter pylori pathogenicity. Exp
Cell Res. 313:3459–3471. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Murata-Kamiya N, Kurashima Y, Teishikata
Y, Yamahashi Y, Saito Y, Higashi H, Aburatani H, Akiyama T, Peek RM
Jr, Azuma T and Hatakeyama M: Helicobacter pylori CagA
interacts with E-cadherin and deregulates the beta-catenin signal
that promotes intestinal transdifferentiation in gastric epithelial
cells. Oncogene. 26:4617–4626. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pei YF, Tao R, Li JF, Su LP, Yu BQ, Wu XY,
Yan M, Gu QL, Zhu ZG and Liu BY: TET1 inhibits gastric cancer
growth and metastasis by PTEN demethylation and re-expression.
Oncotarget. 7:31322–31335. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen S, Li X, Qin J, Chen Y, Liu L and
Zhang D, Wang M, Wang M and Zhang D: APOBEC3A possesses anticancer
and antiviral effects by differential inhibition of HPV E6 and E7
expression on cervical cancer. Int J Clin Exp Med. 8:10548–10557.
2015.PubMed/NCBI
|
|
27
|
Gansmo LB, Romundstad P, Hveem K, Vatten
L, Nik-Zainal S, Lønning PE and Knappskog S: APOBEC3A/B deletion
polymorphism and cancer risk. Carcinogenesis. 39:118–124. 2017.
View Article : Google Scholar :
|
|
28
|
Yu Q, Chen D, König R, Mariani R, Unutmaz
D and Landau NR: APOBEC3B and APOBEC3C are potent inhibitors of
simian immunodeficiency virus replication. J Biol Chem.
279:53379–53386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ara A, Love RP, Follack TB, Ahmed KA,
Adolph MB and Chelico L: Mechanism of enhanced HIV restriction by
virion coencapsidated cytidine deaminases APOBEC3F and APOBEC3G. J
Virol. 91:e02230–16. 2016.
|