Introduction
Oral squamous cell carcinoma (OSCC) is one of the
most common types of head and neck neoplasms, and accounts for ~3%
of all recently diagnosed tumor patients (1). Despite considerable advances in OSCC
treatment, the prognosis of patients with OSCC has demonstrated no
notable improvements in recent decades (2). The overall 5-year survival rate of
patients with OSCC remains <50% (3). The poor prognosis of this disease is
mainly due to late stage diagnosis, radiation resistance and
recurrence; distant metastases have been reported following
combined treatment regimens (4,5). The
occurrence and development of OSCC is a complex process involving
numerous genetic and epigenetic alterations and dynamic alterations
in the expression of coding and non-coding RNAs (6–8);
however, the specific mechanism underlying the pathogenesis of OSCC
remains unknown. Thus, an enhanced understanding of the mechanisms
underlying the carcinogenesis and progression of OSCC is critical
to the development of novel and effective therapeutic methods to
improve the treatment outcomes of this disease.
MicroRNAs (miRNAs) are a group of naturally
occurring, non-coding short RNA molecules of 18 to 25 nucleotides
in length (9). miRNAs mediate gene
expression at the post-transcriptional or translational levels
through their complete or partial complementarity with the
3′-untranslated regions (UTRs) of target genes (10). At present, 2,588 mature human
miRNAs have been detected in the human genome and are estimated to
modulate the expression of >30% of all the protein-coding genes
(11). Previously, miRNAs have
been reported to be abnormally expressed in almost all types of
human cancers (12–14). In addition, studies have indicated
that numerous miRNAs are dysregulated in OSCC, including miRNA
(miR)-433 (15), miR-195-5p
(16), miR-27b (17) and miR-373-3p (18). These aberrantly expressed miRNAs
may function as tumor suppressors or as oncogenes, depending on the
type of tumor and the biological role of their target genes
(19,20). Therefore, the identification of
additional dysregulated miRNAs may provide novel insight into the
function of miRNAs in the onset and the development of OSCC, and
may contribute to the identification of novel therapeutic targets
for the treatment of OSCC.
miR-495 was previously demonstrated to be
dysregulated in human cancers, including medulloblastoma (21), esophageal squamous cell carcinoma
(22), glioma (23,24)
and osteosarcoma (25); however,
the expression, functional roles and underlying molecular mechanism
by which miR-495 regulates the progression of OSCC remain unclear.
The present study aimed to detect the expression of miR-495 in OSCC
tissues and cell lines, and determine its effects on OSCC cells. In
addition, the underlying molecular mechanisms by which miR-495 may
affect the progression of OSCC were investigated.
Materials and methods
Tissue samples and cell lines
The present study was approved by the Ethics
Committee of the Yidu Central Hospital of Weifang (Weifang, China);
written informed consent was obtained from all patients who
participated in the present study. Surgically removed OSCC tissues
and matched adjacent normal tissues were obtained from 23 patients
(15 males, 8 females; age range, 47–69 years) who were diagnosed
with OSCC, and who underwent surgical resection at the Yidu Central
Hospital of Weifang between February 2015 and December 2016. None
of the patients with OSCC had received with chemotherapy,
radiotherapy or other specialized treatment prior to surgery. All
tissues were immediately snap frozen and stored in liquid nitrogen
for further use.
Human oral keratinocytes (HOK) were purchased from
ScienCell Research Laboratories, Inc. (San Diego, CA, USA) and
maintained in an oral keratinocyte medium (ScienCell Research
Laboratories, Inc.). The OSCC cell lines Tca8113, CAL-27 and SCC-9
were acquired from the American Type Culture Collection (Manassas,
VA, USA) and cultured in Dulbecco's modified Eagle's medium/F-12
(DMEM/F-12) supplemented with 10% fetal bovine serum (FBS) and 1%
streptomycin/penicillin mix (all from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). All cell lines were cultured
in a humidified chamber at 37°C under 5% CO2.
Cell transfections
miR-495 mimics and mimic negative controls (miR-NC)
were obtained from Guangzhou Rui Bo Biological Technology, Co.,
Ltd. (Guangzhou, China). The miR-495 mimics sequence was
5′-AAACAAACAUGGUGGACUUCUU-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. To knock down endogenous Notch1
expression, a small interfering RNA (siRNA) targeting Notch1
(si-Notch1) and a negative control (si-NC) were chemically
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The
Notch1 siRNA sequence was 5′-ACGAAGAACAGAAGCACAAAGGCGG-3′ and the
NC siRNA sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. The Notch1
overexpression plasmid (pcDNA3.1-Notch1) and control empty
(pcDNA3.1) plasmid were synthesized at the Chinese Academy of
Sciences (Changchun, China). Cells were inoculated into 6-well
plates and cultured to 60–70% confluence. miRNA mimics (100 pmol),
siRNAs (100 pmol) or plasmid (4 µg) were transfected into cells
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols.
Co-transfection of miRNA mimics (20 pmol) and plasmid (0.8 µg) was
also performed using Lipofectamine® 2000. Following
incubation for 6 h, the transfection mixture was removed and fresh
DMEM/F-12 containing 10% FBS was added into each well. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis and the Matrigel invasion assay was performed after 48 h.
CCK-8 and western blot analysis were conducted at 24 and 72 h
post-transfection, respectively.
RT-qPCR
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract the total RNA from
tissue samples (100 mg) or cell lines (1.5×106 cells),
according to the manufacturer's protocols. The quality and
concentration of total RNA was determined using a NanoDrop 2000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Pittsburgh, PA, USA). For the detection of miR-495, total RNA
(100 ng) was converted into cDNA using the TaqMan microRNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). miR-495 expression levels were detected by qPCR using the
TaqMan microRNA Assay kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The cycling conditions were as follows: 50°C for
2 min and 95°C for 10 min; 40 cycles of denaturation at 95°C for 15
sec; and annealing/extension at 60°C for 60 sec. U6 small nuclear
RNA was used as an internal reference for measuring relative
miR-495 expression. To analyze Notch1 mRNA expression, cDNA was
synthesized from total RNA (100 ng) using a PrimeScript 1st Strand
cDNA Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China),
and subjected to amplification with a SYBR® Premix Ex
Taq II (Takara Biotechnology Co., Ltd.). All reactions were
performed on the Applied Biosystems 7500 Real-Time PCR system
(Thermo Fisher Scientific, Inc.). The cycling conditions were as
follows: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec
and 65°C for 45 sec. β-actin was used as the internal control for
measuring relative Notch1 mRNA expression. The primers were
designed as follows: miR-495, 5′-TCCGATTCTTCACGTGGTAC-3′ (forward)
and 5′-GTGCAGGGTCCGAGGT-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); Notch1,
5′-GTGACTGCTCCCTCAACTTCAAT-3′ (forward) and
5′-CTGTCACAGTGGCCGTCACT-3′ (reverse); and β-actin,
5′-AGTGTGACGTGGACATCCGCAAAG-3′ (forward) and
5′-ATCCACATCTGCTGGAAGGTGGAC-3′ (reverse). Relative gene expression
was calculated using the 2−ΔΔCq method (26).
Cell Counting Kit-8 (CCK-8) assay
A CCK-8 assay was performed to evaluate the
proliferative ability of transfected cells. In brief, transfected
cells were collected and seeded (3×103 cells/well) in
96-well plates in triplicate. Following 0, 24, 48 and 72 h of cell
culture, 10 µl of CCK-8 solution (Beyotime Institute of
Biotechnology, Haimen, China) was added directly into each well,
and the 96-well plates were incubated at 37°C under 5%
CO2 for a further 2 h. The optical density was measured
at a wavelength of 450 nm using a multifunctional microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Matrigel invasion assay
The invasive capacity of transfected cells was
assessed using 24-well Transwell chambers (Corning Incorporated,
Corning, NY, USA) coated with Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA). The upper compartment of each Transwell chamber
was seeded with transfected cells (1×105 cells/well)
suspended in 200 µl FBS-free DMEM/F-12 medium. The lower chambers
were filled with 500 µl DMEM/F-12 containing 10% FBS. Cells were
incubated in a humidified incubator at 37°C under 5% CO2
for 24 h. The non-invasive cells were removed from the upper
chambers with cotton swabs. The invasive cells were then fixed in
95% ethanol at room temperature for 20 min and stained in 0.5%
crystal violet at room temperature for 20 min. The invasive cells
in the lower chambers were imaged and quantified under an Olympus
IX51 inverted microscope (×200 magnification; Olympus Corporation,
Tokyo, Japan) in five randomly selected fields per chamber.
miR-495 target prediction and
confirmation
Computational analysis was conducted to predict the
potential targets of miR-495 using TargetScan (release 7.2;
http://www.targetscan.org) and PicTar
(http://pictar.mdc-berlin.de). Notch1 was
predicted as a potential target of miR-495. The wild-type (WT) and
mutant (Mut) putative miR-495-binding sites in the 3′-UTR region of
Notch1 were amplified by Shanghai GenePharma Co., Ltd., and
inserted into the psiCHECK-2 reporter vector (restriction sites:
XhoI and NheI) (Promega Corporation, Madison, WI,
USA), which are henceforth referred to as WT-Notch1 and Mut-Notch1,
respectively. Cells were cultured in 24-well plates with a density
of 1.0×105 cells and co-transfected with miR-495 mimics
(50 pmol) or miR-NC (50 pmol) and WT-Notch1 (0.8 µg) or Mut-Notch1
(0.8 µg) using Lipofectamine 2000. At 48 h post-transfection,
luciferase activity was quantified with a Dual Luciferase Assay kit
(Promega Corporation). Firefly luciferase activity was detected
with a multifunctional microplate reader (Bio-Rad Laboratories,
Inc.), and normalized to Renilla luciferase activity.
Western blot analysis
Cells (1.5×106 cells) or tissue samples
(250 mg) were lysed with radioimmunoprecipitation assay buffer
(Beyotime Institute of Biotechnology). Total protein concentration
was determined using a BCA Protein Assay kit (Beyotime Institute of
Biotechnology). An equal amount of protein (20 µg) was separated by
10% SDS-PAGE and transferred to polyvinylidene fluoride membranes
(EMD Millipore, Billerica, MA, USA). Following blocking at room
temperature for 2 h with 5% fat-free milk in TBS containing 0.1%
Tween-20 (TBST), the membranes were incubated overnight at 4°C with
primary antibodies against Notch1 (cat. no. ab52627; 1:1,000
dilution; Abcam, Cambridge, UK) or GAPDH (cat. no. ab181603;
1:1,000 dilution; Abcam). Following three washes with TBST, the
membranes were incubated with horseradish peroxidase-conjugated
goat anti-rabbit secondary antibody (cat. no. ab205718; 1:5,000
dilution; Abcam) at room temperature for 2 h. An Enhanced
Chemiluminescence Detection System (Pierce; Thermo Fisher
Scientific, Inc.) was used to visualize the signal, according to
the manufacturer's protocols. Densitometric analysis of the
relative expression levels was performed using Quantity One
software version 4.3.0 (Bio-Rad Laboratories, Inc.), and Notch1
expression was normalized to that of GAPDH.
Statistical analysis
Data are presented as the median ± standard
deviation, and were analyzed with SPSS software (version 17; SPSS,
Inc., Chicago, IL, USA). Differences between groups were analyzed
with a two-tailed Student's t-test or one-way analysis of variance,
followed by the Student-Newman-Keuls post hoc test. The correlation
between miR-495 and Notch1 mRNA expression levels in OSCC tissues
was evaluated with Spearman's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-495 expression is downregulated in
human OSCC tissues and cell lines
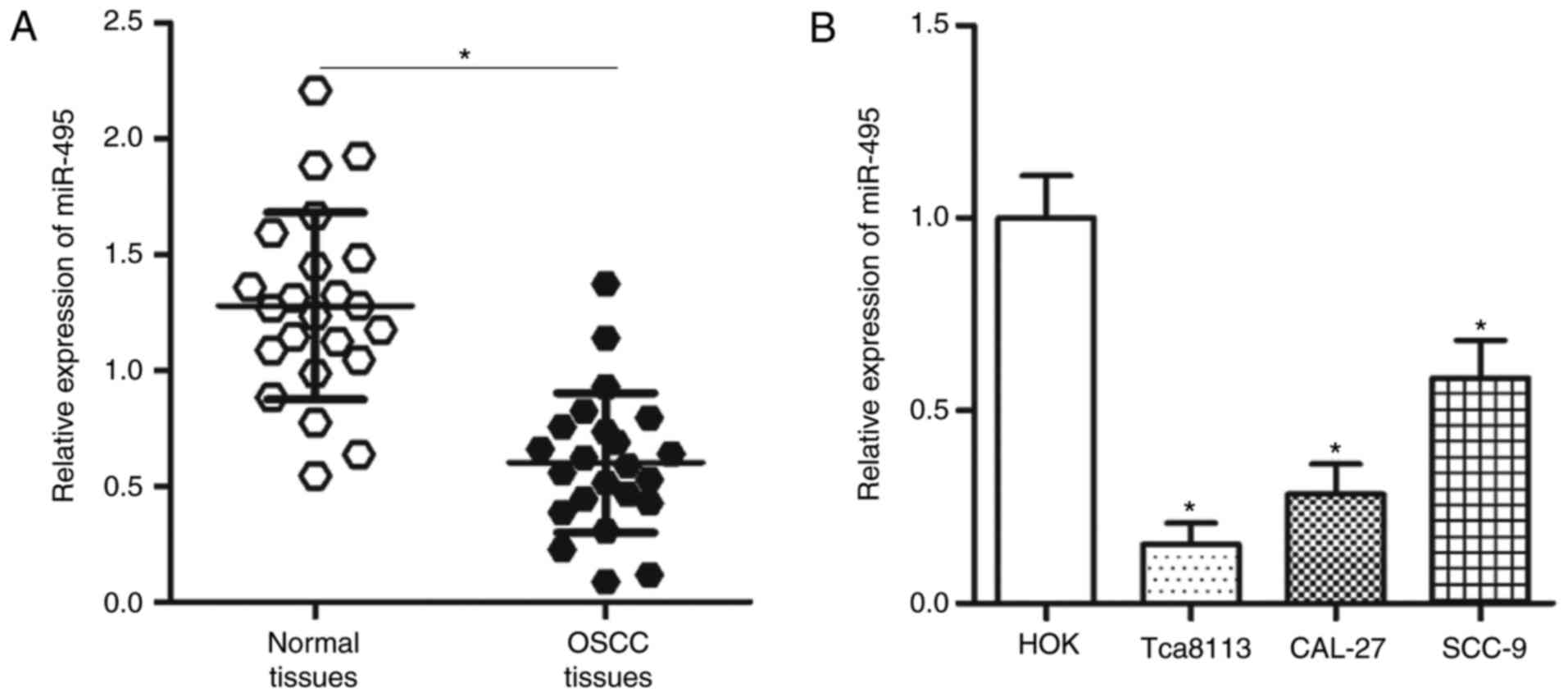
To identify the role of miR-495 in OSCC, the
expression levels of miR-495 in 23 pairs of OSCC and matched
adjacent normal tissues were analyzed. Results from RT-qPCR
revealed that miR-495 expression levels were significantly lower in
OSCC tissues compared with the expression levels in the adjacent
normal tissues (P<0.05; Fig.
1A). RT-qPCR was also performed to evaluate the expression
levels of miR-495 in three OSCC cell lines, including Tca8113,
CAL-27 and SCC-9. Compared with HOK cells, the expression levels of
miR-495 were significantly lower in the three OSCC cell lines
(P<0.05; Fig. 1B). Tca8113 and
CAL-27 cell lines exhibited the lowest relative miR-495 expression
among the three OSCC cell lines; therefore, these two cell lines
were employed for further analysis. These results suggested that
miR-495 may be involved in the development of OSCC.
Overexpression of miR-495 prohibits
the proliferation and invasion of OSCC cells
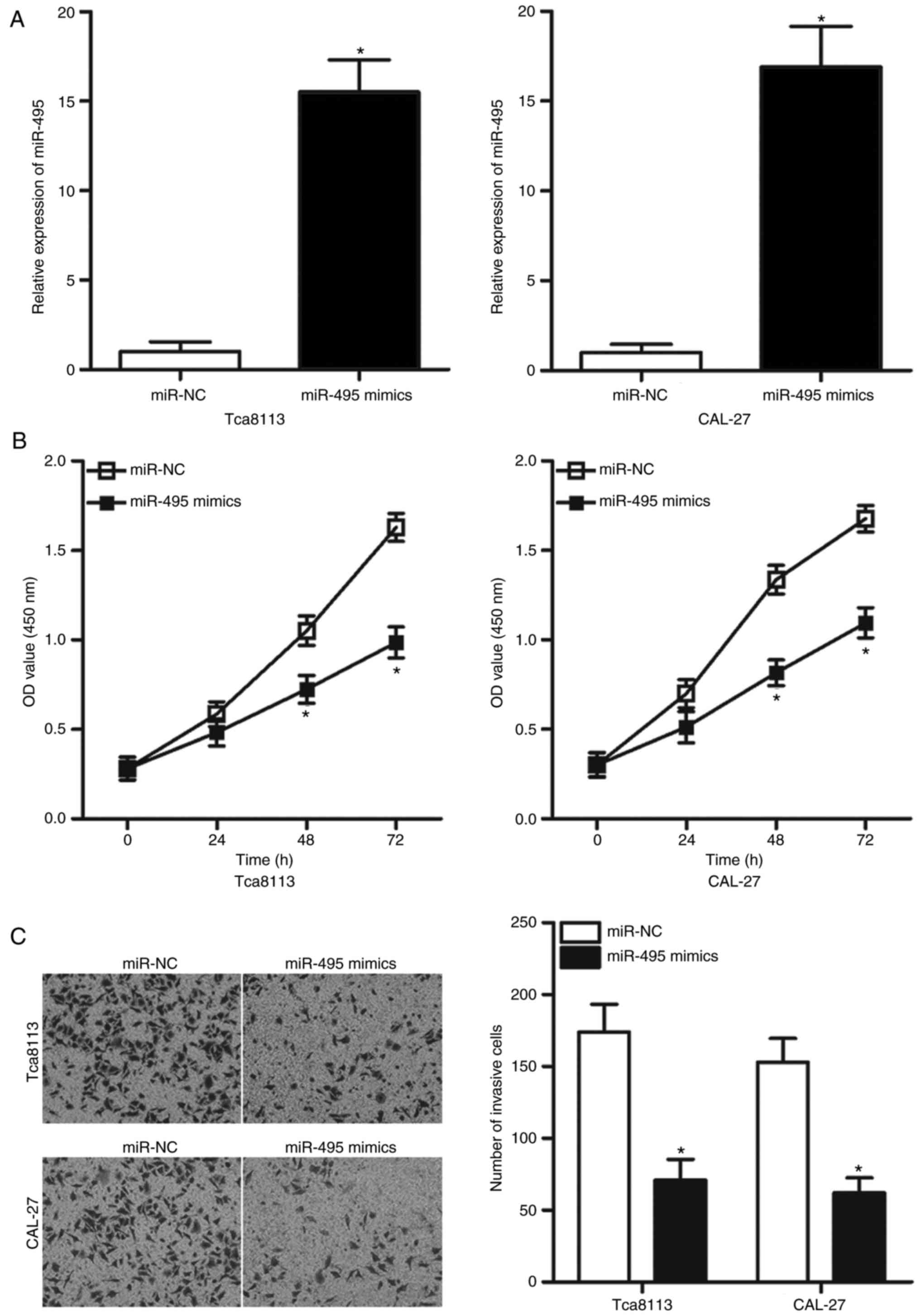
To investigate whether the dysregulation of miR-495
affected the progression of OSCC, Tca8113 and CAL-27 cells were
transfected with miR-495 mimics or miR-NC. RT-qPCR analysis
revealed that transfection of miR-495 mimics resulted in a
significant increase in miR-495 expression in Tca8113 and CAL-27
cells compared with in the miR-NC group (P<0.05; Fig. 2A). As demonstrated by the CCK-8
assay, upregulation of miR-495 resulted in significant suppression
of Tca8113 and CAL-27 cell proliferation compared with the
proliferation rates in the miR-NC group (P<0.05; Fig. 2B). A Matrigel invasion assay was
performed to determine the effects of miR-495 overexpression on
cell invasive ability. The number of invasive cells was
significantly decreased in Tca8113 and CAL-27 cells overexpressing
miR-495 compared with the miR-NC-transfected group (P<0.05;
Fig. 2C). These findings suggested
that miR-495 may serve a tumor suppressive role in OSCC.
Notch1 is a direct target of miR-495
in OSCC
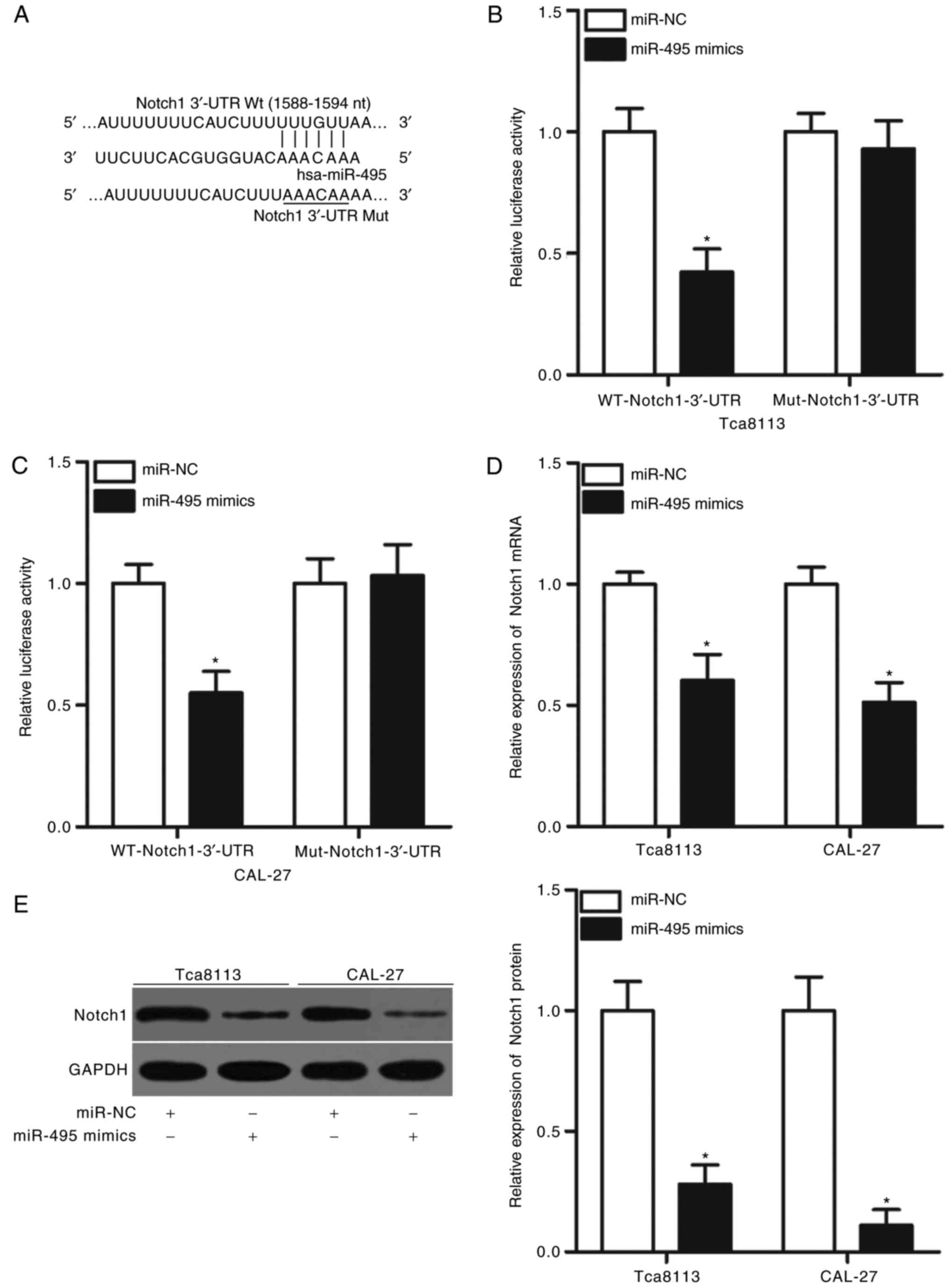
To further clarify the mechanism underlying the role
of miR-495 in OSCC, computational analysis was conducted to predict
the potential targets of miR-495. The 3′-UTR of Notch1 was
identified as containing a highly conserved binding site for
miR-495 (nucleotides 1588–1594; Fig.
3A). Notch1 was previously reported to be involved in the onset
and progression of OSCC (27–31)
and was therefore selected as a candidate for further confirmation
in the present study. A luciferase reporter assay was performed to
determine whether the 3′-UTR of Notch1 may be directly targeted by
miR-495. The luciferase activity of the reporter containing the WT
3′-UTR of Notch1 was significantly reduced in Tca8113 and CAL-27
cells transfected with miR-495 mimics compared with in the miR-NC
group (P<0.05; Fig. 3B and C);
however, miR-495 overexpression did not notably affect the
luciferase activity of the reporter harboring a mutated miR-495
binding site in the 3′-UTR of Notch1 (Fig. 3B and C). To investigate the role of
miR-495 in endogenous Notch1 regulation, RT-qPCR and western blot
analyses were performed to detect Notch1 mRNA and protein
expression levels, respectively, in Tca8113 and CAL-27 cells
following transfection with miR-495 mimics or miR-NC. The results
indicated that overexpression of miR-495 significantly decreased
the expression levels of Notch1 mRNA and protein in Tca8113 and
CAL-27 cells compared with the respective expression levels in the
miR-NC-transfected group (P<0.05; Fig. 3D and E). These results indicted
Notch1 as a direct target gene of miR-495 in OSCC.
Inhibition of Notch1 mimics the
inhibitory effects of miR-495 overexpression in OSCC cells
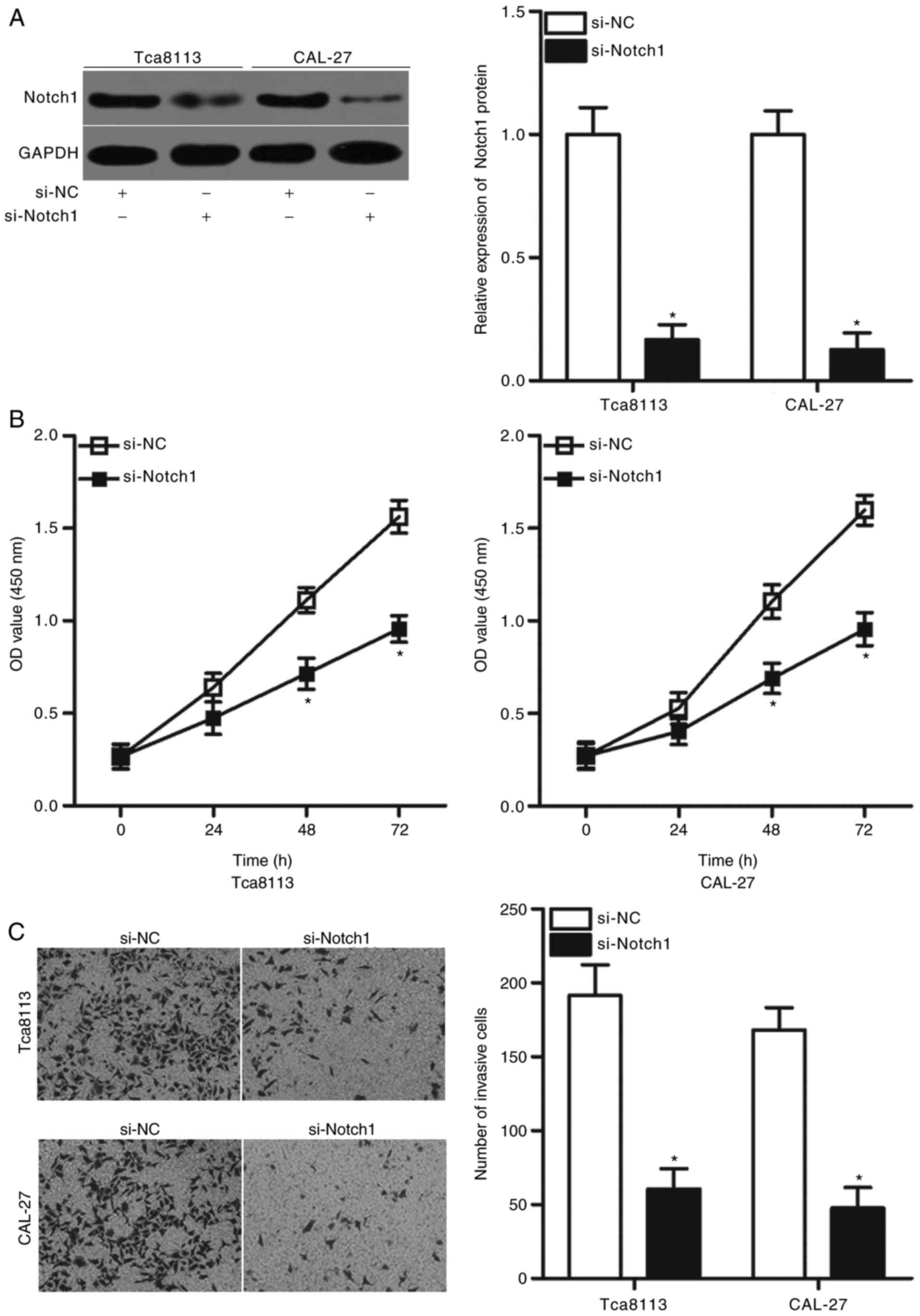
Providing that Notch1 may be a direct target of
miR-495 in OSCC as predicted in the present study, the inhibitory
effects of miR-495 in OSCC cell proliferation and invasion may be
replicated via the downregulation of Notch1. To confirm this
hypothesis, siRNAs targeting Notch1 were transfected into Tca8113
and CAL-27 cells to knock down Notch1 expression. Following
transfection, western blot analysis revealed that the expression
levels of Notch1 protein were significantly downregulated in
Tca8113 and CAL-27 cells transfected with si-Notch1 compared with
cells transfected with si-NC (P<0.05; Fig. 4A). CCK-8 and Matrigel invasion
assays indicated that Notch1 knockdown significantly prohibited the
proliferation (P<0.05; Fig. 4B)
and the invasion rates of Tca8113 and CAL-27 cells compared with
si-NC-transfected cells (P<0.05; Fig. 4C). These results corresponded with
those obtained following miR-495 overexpression, which further
indicated that Notch1 may a functional target of miR-495 in
OSCC.
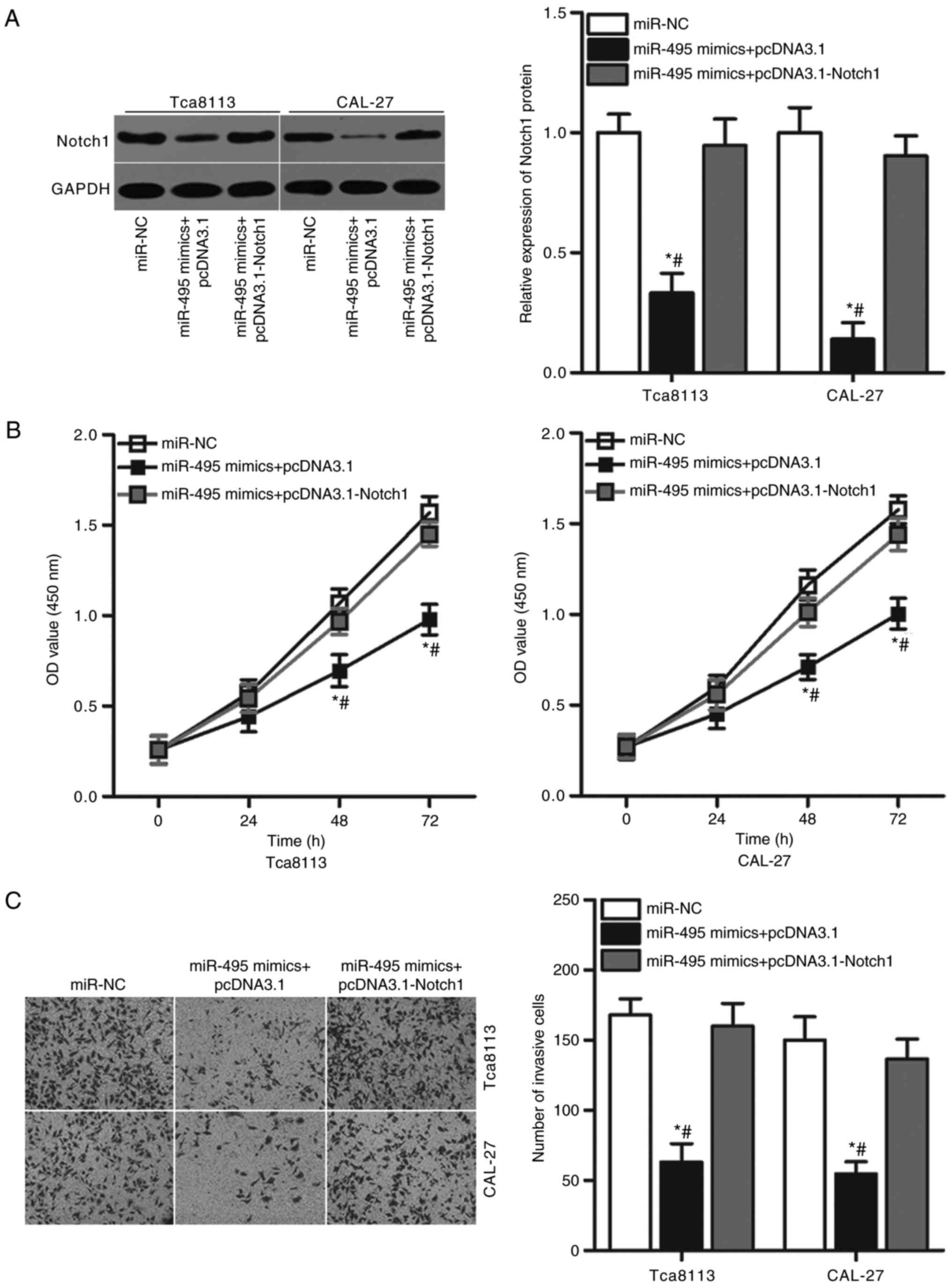
Overexpression of Notch1 reverses the
suppressive role of miR-495 in OSCC cells
To further investigate whether Notch1 mediates the
biological role of miR-495 in OSCC cells a series of rescue
experiments were performed. miR-495-overexpressing Tca8113 and
CAL-27 cells were transfected with pcDNA3.1-Notch1 or the pcDNA3.1
empty plasmid control. Western blot analysis confirmed that the
significantly decreased expression levels of Notch1 protein induced
by miR-495 overexpression were recovered by co-transfection with
pcDNA3.1-Notch1 in Tca8113 and CAL-27 cells (P<0.05; Fig. 5A). Functional experiments indicated
that the restoration of Notch1 expression rescued the suppressive
effects of miR-495 mimics on proliferation (P<0.05; Fig. 5B) and invasion (P<0.05; Fig. 5C) of Tca8113 and CAL-27 cells.
These results suggested that miR-495 may serve a tumor-suppressing
role in OSCC cell proliferation and invasion, at least partly
through the inhibition of Notch1 expression.
Discussion
miRNAs have been associated with the initiation and
progression of OSCC by regulating a variety of cancer-associated
behaviors, including cell proliferation, apoptosis, cell cycle,
invasion, metastasis and angiogenesis (32–34).
Therefore, fully understanding the regulatory mechanism of miRNAs
in the pathogenesis of OSCC may provide novel, promising approaches
for the development of prognostic biomarkers and therapeutic
targets. In the present study, it was demonstrated that miR-495 was
downregulated in OSCC tissues and cell lines. Exogenous expression
of miR-495 inhibited the proliferative and invasive abilities of
OSCC cells. In addition, Notch1 was determined to be a direct
target gene of miR-495 in OSCC cells. Results from the present
study also demonstrated that Notch1 knockdown imitated the
inhibitory effects of miR-495 on OSCC cells. Furthermore,
restoration of expression of Notch1 counteracted the suppressive
effects of miR-495 overexpression on OSCC cell proliferation and
invasion. The results of the present study suggested that miR-495
may act as a tumor suppressor in OSCC, and may therefore be
considered as a therapeutic target for patients with OSCC.
Downregulation of miR-495 is frequently observed in
various types of human cancer (21–24).
For example, miR-495 is downregulated in medulloblastoma tissues
and cell lines; patients with medulloblastoma and decreased
expression levels of miR-495 exhibit poorer prognosis compared with
those expressing higher miR-495 levels (21). Furthermore, miR-495 has been
reported as an independent predictor of overall survival in
patients with medulloblastoma (21). In esophageal squamous cell
carcinoma, the expression levels of miR-495 were reported to be
reduced in tumor tissues and significantly associated with lymph
node metastasis, invasion and tumor, node and metastasis (TNM)
stage (22). miR-495 was observed
to be expressed at low levels in glioma (23,24),
osteosarcoma (25), colorectal
(35,36), gastric (37–39),
prostate (40) and endometrial
cancers (41), and renal cell
carcinoma (42). miR-495 has been
reported to be highly expressed in bladder (43) and breast (44) cancers; miR-495 upregulation in
bladder cancer has been strongly correlated with larger tumor
sizes, advanced TNM stages and lymph node metastasis (43). These observations suggested that
the expression pattern of miR-495 in human cancers is tissue
specific, and may therefore be employed as a diagnostic biomarker
and prognostic predictor for patients with these types of
cancer.
miR-495 has been associated with the malignant
progression of several types of human malignancy. For instance,
upregulation of miR-495 has been reported to lead to the reduction
of cell proliferation and invasion in glioma (23,24).
Another study revealed that restoration of miR-495 expression
attenuated cell proliferation and invasion, and promoted the
apoptosis of osteosarcoma cells (25). Ectopic expression of miR-495 was
demonstrated to prohibit proliferation, colony formation,
metastasis and epithelial-mesenchymal transition (EMT), and induce
the apoptosis of colorectal cancer cells (35,36).
Numerous studies have also demonstrated that miR-495 acts as a
tumor suppressor in gastric cancer by regulating cell growth,
motility and chemotherapy resistance (37–39,45).
Overexpression of miR-495 suppressed prostate cancer cell
proliferation, migration and invasion in vitro, and tumor
growth in vivo (40).
Restoration of the expression of miR-495 reduces endometrial cancer
cell growth and migration, and increases apoptosis (41) and decreased renal cell carcinoma
cell proliferation and migration, and induced G0/G1 phase arrest
(42). Additionally, miR-495 has
been reported to serve as an oncogene in bladder (43) and breast cancers (44,46),
by promoting cell proliferation, colony formation, migration and
invasion in vitro, and tumorigenesis in vivo. These
findings suggested that miR-495 may possess the potential as an
effective target for cancer therapy.
Numerous miR-495 targets have been previously
identified, including: Cyclin-dependent kinase 6 and MYB in glioma
(23,24); high mobility group (HMG) nucleosome
binding domain 5 in osteosarcoma (25); family with sequence similarity 83
member D and Annexin A3 in colorectal cancer (35,36);
phosphatase of regenerating liver-3, HMG AT-hook 2 and ATP binding
cassette subfamily B member 1 in gastric cancer (38,39,45);
protein kinase B in prostate cancer (40); forkhead box C1 in endometrial
cancer (41); stabilin 1 in renal
cell carcinoma (42); phosphatase
and tensin homolog in bladder cancer (43); and junctional adhesion molecule-A
in breast cancer (44). Notch1, a
member of the Notch receptors, was predicted as a novel direct
target gene of miR-495 in OSCC in the present study. Notch1 was
reported to be overexpressed in OSCC, which was correlated with the
T-stage, clinical stage, differentiation, lymph node metastasis,
depth of invasion and locoregional recurrence (27–29).
Notch1 activation has been reported to promote OSCC cell
proliferation, apoptosis, migration, invasion and EMT (29–31).
These data suggested that targeting Notch1 may be a useful approach
for treating OSCC.
In conclusion, to the best of our knowledge, the
present study demonstrated for the first time that miR-495 was
downregulated in OSCC, which may exert an inhibitory effect on the
proliferation and invasion of OSCC cells, at least in part by
directly targeting Notch1. These findings suggested that the
association between miR-495 and Notch1 may be a potential
therapeutic target for the treatment of OSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HJ and LL designed this research. LL and QW
performed the RT-qPCR, CCK-8 and Matrigel invasion assays. Western
blot analysis and the luciferase reporter assay were conducted by
YY. All authors have read and approved the final draft.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Yidu Central Hospital of Weifang (Weifang, China).
Written informed consent was obtained from all patients who
participated in the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brocklehurst PR, Baker SR and Speight PM:
Oral cancer screening: What have we learnt and what is there still
to achieve? Future Oncol. 6:299–304. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hui Y, Li Y, Jing Y, Feng JQ and Ding Y:
miRNA-101 acts as a tumor suppressor in oral squamous cell
carcinoma by targeting CX chemokine receptor 7. Am J Transl Res.
8:4902–4911. 2016.PubMed/NCBI
|
|
3
|
Pérez-Sayáns M, Suárez-Peñaranda JM,
Padin-Iruegas ME, Gayoso-Diz P, Reis-De Almeida M, Barros-Angueira
F, Gándara-Vila P, Blanco-Carrión A and García-García A: The loss
of p16 expression worsens the prognosis of OSCC. Appl
Immunohistochem Mol Morphol. 23:724–732. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Walden MJ and Aygun N: Head and neck
cancer. Semin Roentgenol. 48:75–86. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sinha N, Mukhopadhyay S, Das DN, Panda PK
and Bhutia SK: Relevance of cancer initiating/stem cells in
carcinogenesis and therapy resistance in oral cancer. Oral Oncol.
49:854–862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo T and Califano JA: Molecular biology
and immunology of head and neck cancer. Surg Oncol Clin N Am.
24:397–407. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu MJ, Jan CI, Tsay YG, Yu YH, Huang CY,
Lin SC, Liu CJ, Chen YS, Lo JF and Yu CC: Elimination of head and
neck cancer initiating cells through targeting glucose regulated
protein78 signaling. Mol Cancer. 9:2832010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hitt R and Echarri MJ: Molecular biology
in head and neck cancer. Clin Transl Oncol. 8:776–779. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
A microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
An L, Liu Y, Wu A and Guan Y: microRNA-124
inhibits migration and invasion by down-regulating ROCK1 in glioma.
PLoS One. 8:e694782013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhai L, Li Y, Lan X and Ai L:
MicroRNA-10a-5p suppresses cancer proliferation and division in
human cervical cancer by targeting BDNF. Exp Ther Med.
14:6147–6151. 2017.PubMed/NCBI
|
|
14
|
Huang J, He Y, McLeod HL, Xie Y, Xiao D,
Hu H, Chen P, Shen L, Zeng S, Yin X, et al: miR-302b inhibits
tumorigenesis by targeting EphA2 via Wnt/β-catenin/EMT signaling
cascade in gastric cancer. BMC Cancer. 17:8862017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang YJ, Zhang ZF, Fan SH, Zhuang J, Shan
Q, Han XR, Wen X, Li MQ, Hu B, Sun CH, et al: MicroRNA-433 inhibits
oral squamous cell carcinoma cells by targeting FAK. Oncotarget.
8:100227–100241. 2017.PubMed/NCBI
|
|
16
|
Wang T, Ren Y, Liu R, Ma J, Shi Y, Zhang L
and Bu R: miR-195-5p suppresses the proliferation, migration, and
invasion of oral squamous cell carcinoma by targeting TRIM14.
Biomed Res Int. 2017:73781482017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu B, Chen W, Cao G, Dong Z, Xu J, Luo T
and Zhang S: MicroRNA-27b inhibits cell proliferation in oral
squamous cell carcinoma by targeting FZD7 and Wnt signaling
pathway. Arch Oral Biol. 83:92–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Weng J, Zhang H, Wang C, Liang J, Chen G,
Li W, Tang H and Hou J: miR-373-3p targets DKK1 to promote
EMT-induced metastasis via the Wnt/β-catenin pathway in
tongue squamous cell carcinoma. Biomed Res Int. 2017:60109262017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ryan BM, Robles AI and Harris CC: Genetic
variation in microRNA networks: The implications for cancer
research. Nat Rev Cancer. 10:389–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang C, Yun Z, Zhao T, Liu X and Ma X:
MiR-495 is a predictive biomarker that downregulates GFI1
expression in medulloblastoma. Cell Physiol Biochem. 36:1430–1439.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mao Y, Li L, Liu J, Wang L and Zhou Y:
MiR-495 inhibits esophageal squamous cell carcinoma progression by
targeting Akt1. Oncotarget. 7:51223–51236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen SM, Chen HC, Chen SJ, Huang CY, Chen
PY, Wu TW, Feng LY, Tsai HC, Lui TN, Hsueh C and Wei KC:
MicroRNA-495 inhibits proliferation of glioblastoma multiforme
cells by downregulating cyclin-dependent kinase 6. World J Surg
Oncol. 11:872013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang B, Yuan F, Liu J, Li Y, Zhou F, Liu
X, Hao Z, Li Q, Zheng Y and Wang W: Hsa-miR-495 acts as a tumor
suppressor gene in glioma via the negative regulation of MYB. Mol
Med Rep. 14:977–982. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang W, Zheng J, Yu T and Wang J:
Overexpression of microRNA-495 suppresses the proliferation and
invasion and induces the apoptosis of osteosarcoma cells by
targeting high-mobility group nucleosome-binding domain 5. Oncol
Rep. 38:1099–1107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang S, Fan H, Xu J and Zhao E: Prognostic
implication of NOTCH1 in early stage oral squamous cell cancer with
occult metastases. Clin Oral Investig. 22:1131–1138. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yoshida R, Nagata M, Nakayama H,
Niimori-Kita K, Hassan W, Tanaka T, Shinohara M and Ito T: The
pathological significance of Notch1 in oral squamous cell
carcinoma. Lab Invest. 93:1068–1081. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gan RH, Wei H, Xie J, Zheng DP, Luo EL,
Huang XY, Xie J, Zhao Y, Ding LC, Su BH, et al: Notch1 regulates
tongue cancer cells proliferation, apoptosis and invasion. Cell
Cycle. 17:216–224. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Natsuizaka M, Whelan KA, Kagawa S, Tanaka
K, Giroux V, Chandramouleeswaran PM, Long A, Sahu V, Darling DS,
Que J, et al: Interplay between Notch1 and Notch3 promotes EMT and
tumor initiation in squamous cell carcinoma. Nat Commun.
8:17582017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhong R, Bao R, Faber PW, Bindokas VP,
Bechill J, Lingen MW and Spiotto MT: Notch1 activation or loss
promotes HPV-induced oral tumorigenesis. Cancer Res. 75:3958–3969.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rastogi B, Kumar A, Raut SK, Panda NK,
Rattan V, Joshi N and Khullar M: Downregulation of miR-377 promotes
oral squamous cell carcinoma growth and migration by targeting
HDAC9. Cancer Invest. 35:152–162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yan Y, Wang X, Venø MT, Bakholdt V,
Sørensen JA, Krogdahl A, Sun Z, Gao S and Kjems J: Circulating
miRNAs as biomarkers for oral squamous cell carcinoma recurrence in
operated patients. Oncotarget. 8:8206–8214. 2017.PubMed/NCBI
|
|
34
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yan L, Yao J and Qiu J: miRNA-495
suppresses proliferation and migration of colorectal cancer cells
by targeting FAM83D. Biomed Pharmacother. 96:974–981. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bai Z, Wang J, Wang T, Li Y, Zhao X, Wu G,
Yang Y, Deng W and Zhang Z: The miR-495/Annexin A3/p53 axis
inhibits the invasion and EMT of colorectal cancer cells. Cell
Physiol Biochem. 44:1882–1895. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eun JW, Kim HS, Shen Q, Yang HD, Kim SY,
Yoon JH, Park WS, Lee JY and Nam SW: MicroRNA-495-3p functions as a
tumor suppressor by regulating multiple epigenetic modifiers in
gastric carcinogenesis. J Pathol. 244:107–119. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, Zhu
G, Liu Z, Tu Y, Peng G, et al: miR-495 and miR-551a inhibit the
migration and invasion of human gastric cancer cells by directly
interacting with PRL-3. Cancer Lett. 323:41–47. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang H, Jiang Z, Chen H, Wu X, Xiang J and
Peng J: MicroRNA-495 inhibits gastric cancer cell migration and
invasion possibly via targeting high mobility group AT-Hook 2
(HMGA2). Med Sci Monit. 23:640–648. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li JZ, Wang ZL, Xu WH, Li Q, Gao L and
Wang ZM: MicroRNA-495 regulates migration and invasion in prostate
cancer cells via targeting Akt and mTOR signaling. Cancer Invest.
34:181–188. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu YY, Tian J, Hao Q and Yin LR:
MicroRNA-495 downregulates FOXC1 expression to suppress cell growth
and migration in endometrial cancer. Tumour Biol. 37:239–251. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lv C, Bai Z, Liu Z, Luo P and Zhang J:
MicroRNA-495 suppresses human renal cell carcinoma malignancy by
targeting SATB1. Am J Transl Res. 7:1992–1999. 2015.PubMed/NCBI
|
|
43
|
Tan M, Mu X, Liu Z, Tao L, Wang J, Ge J
and Qiu J: microRNA-495 promotes bladder cancer cell growth and
invasion by targeting phosphatase and tensin homolog. Biochem
Biophys Res Commun. 483:867–873. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cao M, Nie W, Li J, Zhang Y, Yan X, Guan
X, Chen X, Zen K, Zhang CY, Jiang X and Hou D: MicroRNA-495 induces
breast cancer cell migration by targeting JAM-A. Protein Cell.
5:862–872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zou Z, Zou R, Zong D, Shi Y, Chen J, Huang
J, Zhu J, Chen L, Bao X, Liu Y, et al: miR-495 sensitizes MDR
cancer cells to the combination of doxorubicin and taxol by
inhibiting MDR1 expression. J Cell Mol Med. 21:1929–1943. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hwang-Verslues WW, Chang PH, Wei PC, Yang
CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY and Lee WH: miR-495
is upregulated by E12/E47 in breast cancer stem cells, and promotes
oncogenesis and hypoxia resistance via downregulation of E-cadherin
and REDD1. Oncogene. 30:2463–2474. 2011. View Article : Google Scholar : PubMed/NCBI
|